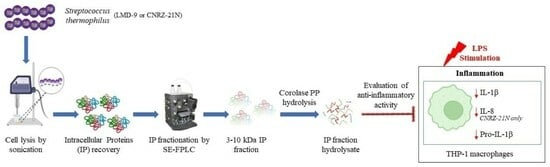
Streptococcus thermophilus: A Source of Postbiotics Displaying Anti-Inflammatory Effects in THP 1 Macrophages
Abstract
1. Introduction
2. Results and Discussion
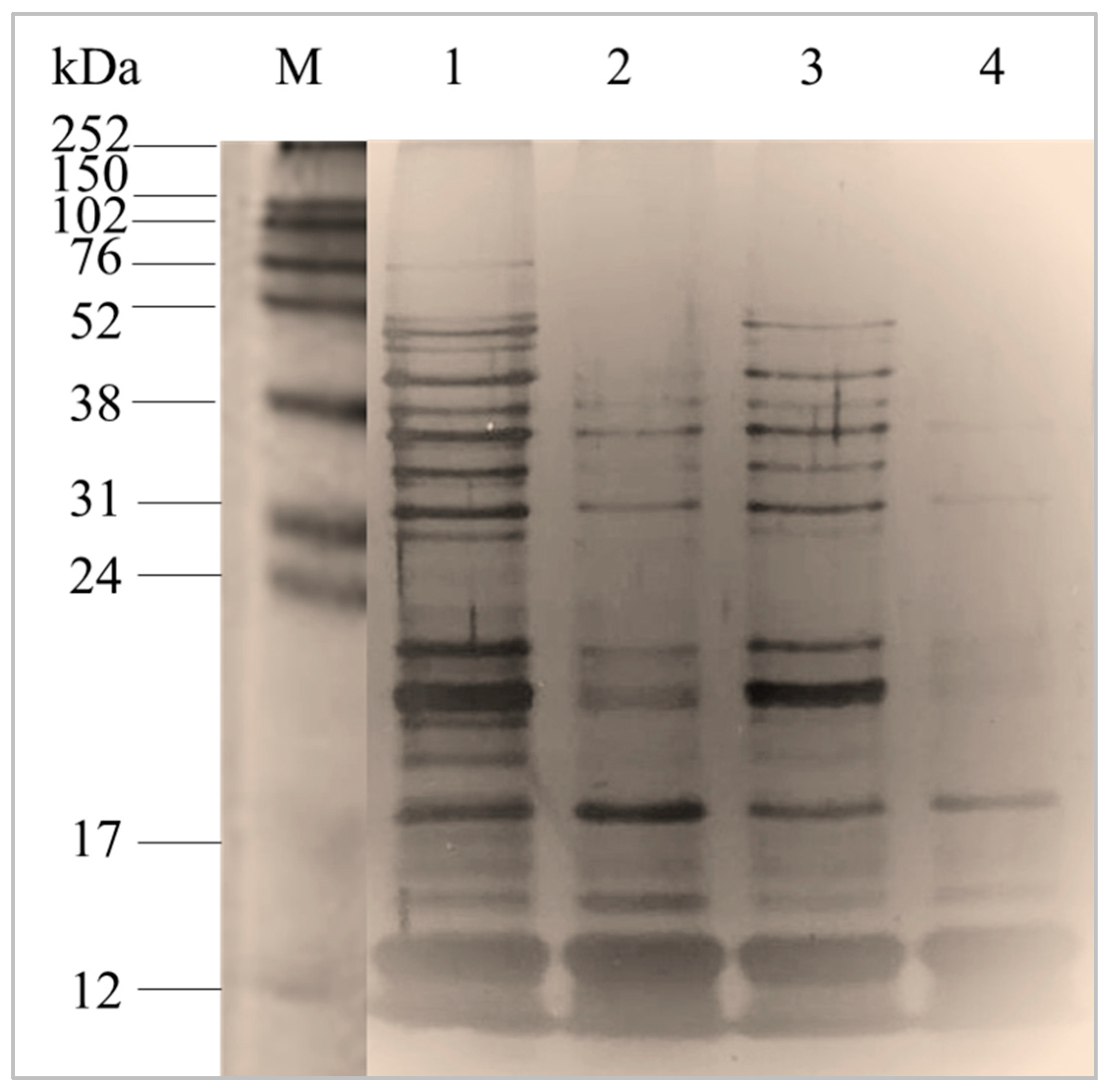
2.1. Purification of Total Intracellular Proteins from S. thermophilus Strains
2.1.1. Characterization of Total Intracellular Proteins by Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis
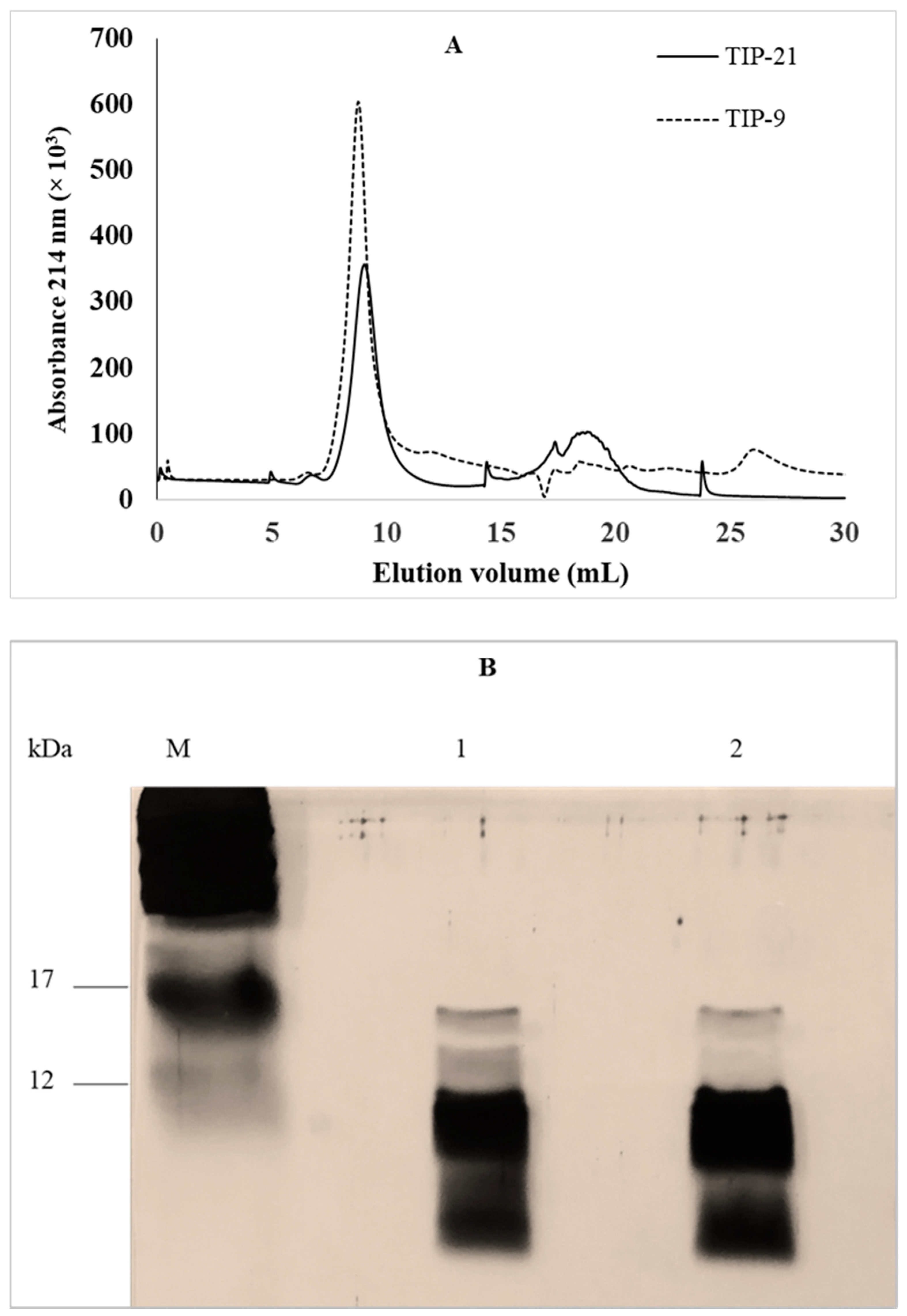
2.1.2. Fractionation of Total Intracellular Proteins by Size Exclusion Fast Protein Liquid Chromatography
2.1.3. LC-MS Analysis of IP-9 and IP-21 Fractions
2.2. Identification of Intracellular Proteins by LC-MS/MS Analysis of the Peptides Obtained after Hydrolysis of Intracellular Protein Fractions by Corolase PP
2.3. Predicted Anti-Inflammatory Activity Based on In Silico Simulation
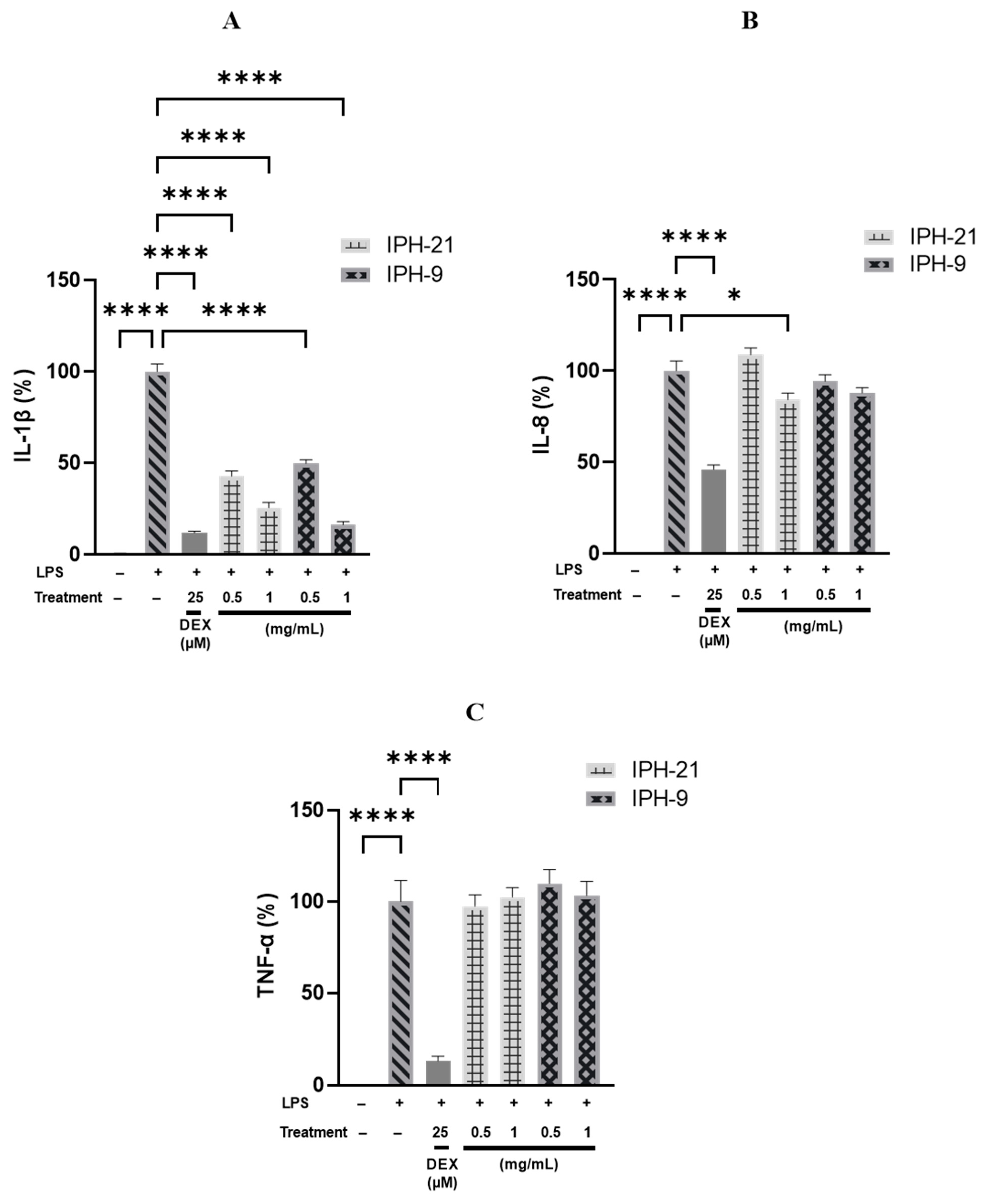
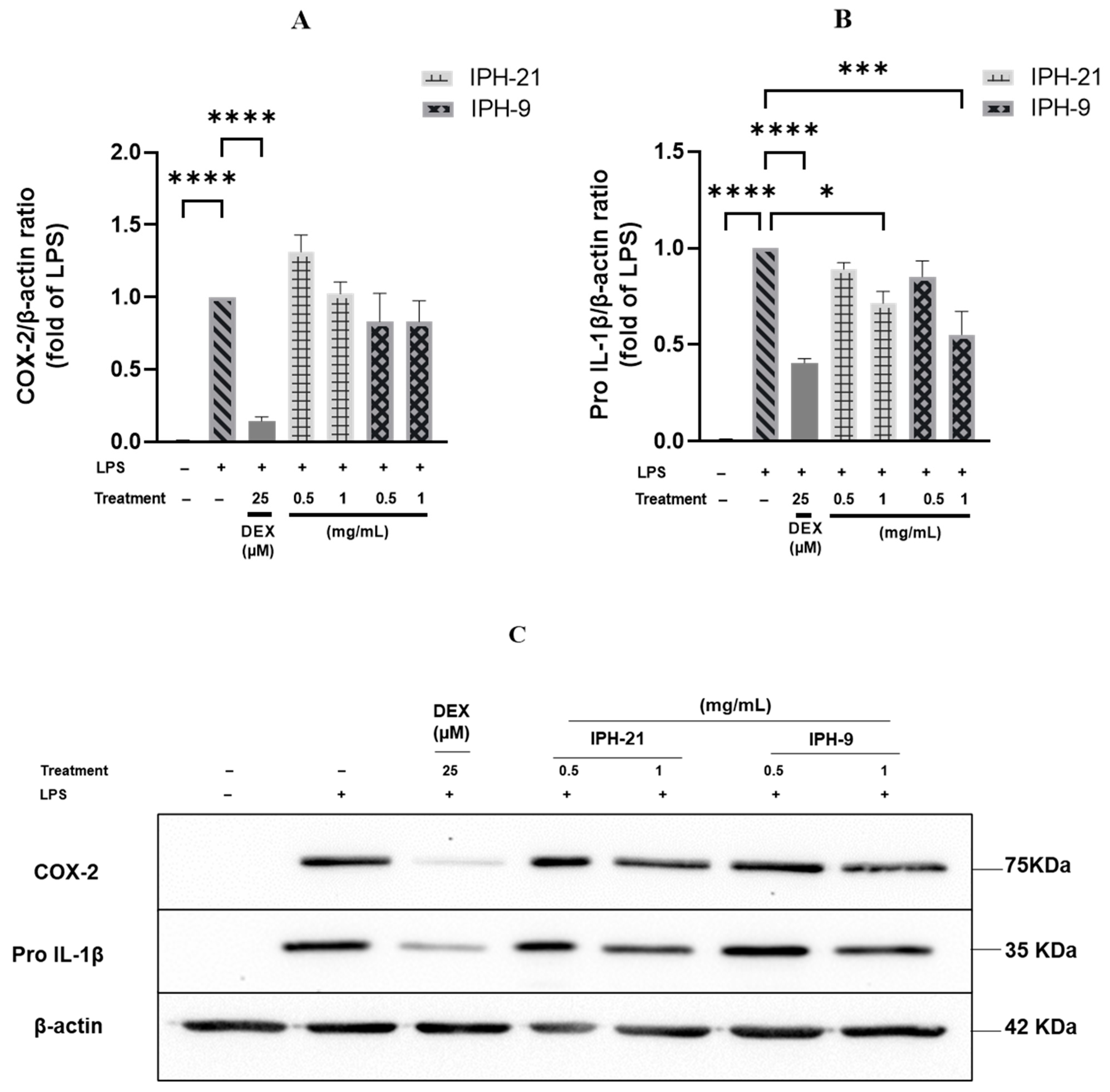
2.4. Anti-Inflammatory Activity of Intracellular Protein Hydrolysates IPH-9 and IPH-21
3. Materials and Methods
3.1. Bacterial Strains and Culture Conditions
3.2. Extraction of Total Intracellular Proteins
3.3. Protein Precipitation
3.4. Size-Exclusion Fast Protein Liquid Chromatography
3.5. SDS-PAGE Analysis of TIP
3.6. Enzymatic Hydrolysis
3.7. LC-MS and LC-MS/MS Analysis
3.8. Anti-Inflammatory Assay, Cell Cytotoxicity, ELISA and Western Blotting
3.9. In Silico Anti-Inflammatory Prediction
3.10. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uriot, O.; Denis, S.; Junjua, M.; Roussel, Y.; Dary-Mourot, A.; Blanquet-Diot, S. Streptococcus thermophilus: From Yogurt Starter to a New Promising Probiotic Candidate? J. Funct. Foods 2017, 37, 74–89. [Google Scholar] [CrossRef]
- Martinović, A.; Cocuzzi, R.; Arioli, S.; Mora, D. Streptococcus thermophilus: To Survive, or Not to Survive the Gastrointestinal Tract, That Is the Question! Nutrients 2020, 12, 2175. [Google Scholar] [CrossRef] [PubMed]
- Piqué, N.; Berlanga, M.; Miñana-Galbis, D. Health Benefits of Heat-Killed (Tyndallized) Probiotics: An Overview. Int. J. Mol. Sci. 2019, 20, 2534. [Google Scholar] [CrossRef] [PubMed]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Toalá, J.E.; Garcia-Varela, R.; Garcia, H.S.; Mata-Haro, V.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Postbiotics: An Evolving Term within the Functional Foods Field. Trends Food Sci. Technol. 2018, 75, 105–114. [Google Scholar] [CrossRef]
- Martín, R.; Langella, P. Emerging Health Concepts in the Probiotics Field: Streamlining the Definitions. Front. Microbiol. 2019, 10, 1047. [Google Scholar] [CrossRef] [PubMed]
- de Almada, C.N.; Almada, C.N.; Martinez, R.C.R.; Sant’Ana, A.S. Paraprobiotics: Evidences on Their Ability to Modify Biological Responses, Inactivation Methods and Perspectives on Their Application in Foods. Trends Food Sci. Technol. 2016, 58, 96–114. [Google Scholar] [CrossRef]
- Żółkiewicz, J.; Marzec, A.; Ruszczyński, M.; Feleszko, W. Postbiotics—A Step Beyond Pre- and Probiotics. Nutrients 2020, 12, 2189. [Google Scholar] [CrossRef] [PubMed]
- Ben Othman, M.; Sakamoto, K. Effect of Inactivated Bifidobacterium Longum Intake on Obese Diabetes Model Mice (TSOD). Food Res. Int. 2020, 129, 108792. [Google Scholar] [CrossRef]
- Miyazawa, K.; Kawase, M.; Kubota, A.; Yoda, K.; Harata, G.; Hosoda, M.; He, F. Heat-Killed Lactobacillus gasseri Can Enhance Immunity in the Elderly in a Double-Blind, Placebo-Controlled Clinical Study. Benef. Microbes 2015, 6, 441–449. [Google Scholar] [CrossRef]
- Maehata, H.; Arai, S.; Iwabuchi, N.; Abe, F. Immuno-Modulation by Heat-Killed Lacticaseibacillus Paracasei MCC1849 and Its Application to Food Products. Int. J. Immunopathol. Pharmacol. 2021, 35, 205873842110082. [Google Scholar] [CrossRef] [PubMed]
- Ménard, S.; Laharie, D.; Asensio, C.; Vidal-Martinez, T.; Candalh, C.; Rullier, A.; Zerbib, F.; Mégraud, F.; Matysiak-Budnik, T.; Heyman, M. Bifidobacterium breve and Streptococcus thermophilus Secretion Products Enhance T Helper 1 Immune Response and Intestinal Barrier in Mice. Exp. Biol. Med. (Maywood) 2005, 230, 749–756. [Google Scholar] [CrossRef]
- Dargahi, N.; Johnson, J.C.; Apostolopoulos, V. Immune Modulatory Effects of Probiotic Streptococcus thermophilus on Human Monocytes. Biologics 2021, 1, 396–415. [Google Scholar] [CrossRef]
- Han, F.; Wu, G.; Zhang, Y.; Zheng, H.; Han, S.; Li, X.; Cai, W.; Liu, J.; Zhang, W.; Zhang, X.; et al. Streptococcus thermophilus Attenuates Inflammation in Septic Mice Mediated by Gut Microbiota. Front. Microbiol. 2020, 11, 598010. [Google Scholar] [CrossRef] [PubMed]
- Junjua, M.; Kechaou, N.; Chain, F.; Awussi, A.A.; Roussel, Y.; Perrin, C.; Roux, E.; Langella, P.; Bermúdez-Humarán, L.G.; Le Roux, Y.; et al. A Large Scale in Vitro Screening of Streptococcus thermophilus Strains Revealed Strains with a High Anti-Inflammatory Potential. LWT—Food Sci. Technol. 2016, 70, 78–87. [Google Scholar] [CrossRef]
- Drouault, S.; Corthier, G.; Ehrlich, S.D.; Renault, P. Survival, Physiology, and Lysis of Lactococcus lactis in the Digestive Tract. Appl. Environ. Microbiol. 1999, 65, 4881–4886. [Google Scholar] [CrossRef] [PubMed]
- Allouche, R.; Genay, M.; Dary-Mourot, A.; Hafeez, Z.; Miclo, L. Cell Proteins Obtained by Peptic Shaving of Two Phenotypically Different Strains of Streptococcus thermophilus as a Source of Anti-Inflammatory Peptides. Nutrients 2022, 14, 4777. [Google Scholar] [CrossRef]
- Allouche, R.; Hafeez, Z.; Papier, F.; Dary-Mourot, A.; Genay, M.; Miclo, L. In Vitro Anti-Inflammatory Activity of Peptides Obtained by Tryptic Shaving of Surface Proteins of Streptococcus thermophilus LMD-9. Foods 2022, 11, 1157. [Google Scholar] [CrossRef]
- Alexandraki, V.; Kazou, M.; Blom, J.; Pot, B.; Papadimitriou, K.; Tsakalidou, E. Comparative Genomics of Streptococcus thermophilus Support Important Traits Concerning the Evolution, Biology and Technological Properties of the Species. Front. Microbiol. 2019, 10, 2916. [Google Scholar] [CrossRef]
- Han, X.; Yang, Z.; Jing, X.; Yu, P.; Zhang, Y.; Yi, H.; Zhang, L. Improvement of the Texture of Yogurt by Use of Exopolysaccharide Producing Lactic Acid Bacteria. BioMed Res. Int. 2016, 2016, 7945675. [Google Scholar] [CrossRef]
- Li, S.; Shah, N.P. Characterization, Anti-Inflammatory and Antiproliferative Activities of Natural and Sulfonated Exo-Polysaccharides from Streptococcus thermophilus ASCC 1275. J. Food Sci. 2016, 81, M1167–M1176. [Google Scholar] [CrossRef] [PubMed]
- Kanamarlapudi, S.L.R.K.; Muddada, S. Characterization of Exopolysaccharide Produced by Streptococcus thermophilus CC30. BioMed Res. Int. 2017, 2017, 4201809. [Google Scholar] [CrossRef] [PubMed]
- Coscueta, E.R.; Amorim, M.M.; Voss, G.B.; Nerli, B.B.; Picó, G.A.; Pintado, M.E. Bioactive Properties of Peptides Obtained from Argentinian Defatted Soy Flour Protein by Corolase PP Hydrolysis. Food Chem. 2016, 198, 36–44. [Google Scholar] [CrossRef]
- Hu, X.-P.; Dourado, H.; Schubert, P.; Lercher, M.J. The Protein Translation Machinery Is Expressed for Maximal Efficiency in Escherichia coli. Nat. Commun. 2020, 11, 5260. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, H.S.; Tate, W.P. A Ribosome Without RNA. Front. Ecol. Evol. 2015, 3, 129. [Google Scholar] [CrossRef]
- Tosaka, R.; Yamamoto, H.; Ohdomari, I.; Watanabe, T. Adsorption Mechanism of Ribosomal Protein L2 onto a Silica Surface: A Molecular Dynamics Simulation Study. Langmuir 2010, 26, 9950–9955. [Google Scholar] [CrossRef] [PubMed]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 Cell Line: An in Vitro Cell Model for Immune Modulation Approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.D.; Pangloli, P.; Krishnan, H.B.; Dia, V.P. BG-4, a Novel Bioactive Peptide from Momordica Charantia, Inhibits Lipopolysaccharide-Induced Inflammation in THP-1 Human Macrophages. Phytomedicine 2018, 42, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Rodriguez, J.; Wee, J. Dietary Postbiotics Reduce Cytotoxicity and Inflammation Induced by Crystalline Silica in an In Vitro RAW 264.7 Macrophage Model. Foods 2022, 11, 877. [Google Scholar] [CrossRef]
- Izuddin, W.I.; Loh, T.C.; Foo, H.L.; Samsudin, A.A.; Humam, A.M. Postbiotic L. plantarum RG14 Improves Ruminal Epithelium Growth, Immune Status and Upregulates the Intestinal Barrier Function in Post-Weaning Lambs. Sci. Rep. 2019, 9, 9938. [Google Scholar] [CrossRef]
- García-Lafuente, A.; Moro, C.; Manchón, N.; Gonzalo-Ruiz, A.; Villares, A.; Guillamón, E.; Rostagno, M.; Mateo-Vivaracho, L. In Vitro Anti-Inflammatory Activity of Phenolic Rich Extracts from White and Red Common Beans. Food Chem. 2014, 161, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Garlanda, C.; Dinarello, C.A.; Mantovani, A. The Interleukin-1 Family: Back to the Future. Immunity 2013, 39, 1003–1018. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Tomé, S.; Marin, A.C.; Ortega Moreno, L.; Baldan-Martin, M.; Mora-Gutiérrez, I.; Lanas-Gimeno, A.; Moreno-Monteagudo, J.A.; Santander, C.; Sánchez, B.; Chaparro, M.; et al. Immunomodulatory Effect of Gut Microbiota-Derived Bioactive Peptides on Human Immune System from Healthy Controls and Patients with Inflammatory Bowel Disease. Nutrients 2019, 11, 2605. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, J.; Christie-Oleza, J.A.; Clair, G.; Malard, V.; Duport, C. Exoproteomics: Exploring the world around biological systems. Exp. Rev. Proteomics 2012, 9, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.; Crawford, J.; Gianazza, E. Protein Stains for Proteomic Applications:Which, When, Why? Proteomics 2006, 6, 5385–5408. [Google Scholar] [CrossRef]
- Morrissey, J.H. Silver Stain for Proteins in Polyacrylamide Gels: A Modified Procedure with Enhanced Uniform Sensitivity. Anal. Biochem. 1981, 117, 307–310. [Google Scholar] [CrossRef]


| LMD-9 | CNRZ-21N | |||||||
|---|---|---|---|---|---|---|---|---|
| Protein Id | Description | pI * | MW * (kDa) | Nb Specific Sequences Identified | Coverage (%) | MW * (kDa) | Nb Specific Sequences Identified | Coverage (%) |
| STER_1896|ID:1900559|rplX| | 50S ribosomal subunit protein L24 | 9.91 | 10.83 | 78 | 99.02 | 10.86 | 126 | 99.02 |
| STER_1963|ID:1899676| | conserved protein of unknown function | 6.29 | 7.07 | 67 | 98.53 | |||
| STER_1889|ID:1900552|rpmD| | 50S ribosomal subunit protein L30 | 10.12 | 6.39 | 45 | 96.72 | 6.39 | 62 | 98.36 |
| STER_1243|ID:1900052| | Phosphocarrier protein HPr (Histidine-containing protein) | 4.88 | 8.91 | 47 | 93.18 | 8.91 | 95 | 98.86 |
| STER_1892|ID:1900555|rplF| | 50S ribosomal subunit protein L6 | 9.47 | 19.44 | 33 | 83.80 | 19.44 | 41 | 93.86 |
| STER_0456|ID:1899076|rpmA| | 50S ribosomal subunit protein L27 | 10.33 | 10.31 | 28 | 64.95 | 10.31 | 45 | 69.07 |
| STER_1317|ID:1899536| | conserved protein of unknown function | 8.86 | 6.81 | 29 | 98.46 | 6.81 | 31 | 98.46 |
| STER_1954|ID:1899671|rpmG| | 50S ribosomal subunit protein L33 | 10.35 | 5.92 | 26 | 80.00 | 5.92 | 37 | 98.00 |
| STER_1764|ID:1900458|rpsL| | 30S ribosomal subunit protein S12 | 11.66 | 15.05 | 24 | 66.67 | 15.05 | 30 | 79.71 |
| STER_0850|ID:1899831|rpsT| | 30S ribosomal subunit protein S20 | 10.68 | 8.40 | 24 | 67.09 | 8.40 | 49 | 83.54 |
| STER_1175|ID:1899990|hupB| | HU, DNA-binding transcriptional regulator, beta subunit | 9.05 | 9.60 | 22 | 85.87 | 9.60 | 24 | 85.87 |
| STER_0128|ID:1898817|rpsI| | 30S ribosomal subunit protein S9 | 10.67 | 14.23 | 21 | 53.44 | 14.23 | 11 | 63.36 |
| STER_0541|ID:1899153| | protein of unknown function | 6.85 | 9.77 | 20 | 55.81 | |||
| STER_1888|ID:1900551|rplO| | 50S ribosomal subunit protein L15 | 10.50 | 15.52 | 17 | 53.74 | 15.54 | 16 | 65.99 |
| STER_1953|ID:1899670|rpmF| | 50S ribosomal subunit protein L32 | 10.49 | 6.78 | 18 | 57.38 | 6.78 | 23 | 59.02 |
| STER_1899|ID:1900562|rpmC| | 50S ribosomal subunit protein L29 | 9.35 | 7.90 | 17 | 91.30 | 7.90 | 15 | 98.55 |
| STER_1141|ID:1899514| | exported protein of unknown function | 10.84 | 6.26 | 18 | 69.49 | |||
| STER_1936|ID:1899661| | 50S ribosomal protein L28 | 11.21 | 6.91 | 16 | 60.32 | 6.91 | 25 | 60.32 |
| STER_0252|ID:1898920|groS| | Cpn10 chaperonin GroES, small subunit of GroESL | 4.56 | 9.91 | 15 | 80.21 | |||
| STER_0787|ID:1899802|ykgM| | putative ribosomal protein | 8.00 | 9.32 | 15 | 74.07 | |||
| STER_0455|ID:1899075|rplU| | 50S ribosomal subunit protein L21 | 9.86 | 11.18 | 15 | 41.91 | 11.18 | 17 | 75.24 |
| STER_1904|ID:1900567|rplB| | 50S ribosomal subunit protein L2 | 10.57 | 29.91 | 13 | 51.80 | |||
| STER_1451|ID:1900209| | 30S ribosomal protein S21 (BS-B) | 11.29 | 6.91 | 12 | 59.32 | 6.92 | 9 | 59,32 |
| STER_1902|ID:1900565|rplV| | 50S ribosomal subunit protein L22 | 11.02 | 12.41 | 9 | 53.91 | 12.41 | 11 | 54.78 |
| STER_1882|ID:1900545|rpsK| | 30S ribosomal subunit protein S11 | 11.44 | 13.36 | 11 | 53.91 | 13.36 | 10 | 67.19 |
| STER_1891|ID:1900554|rplR| | 50S ribosomal subunit protein L18 | 9.84 | 12.88 | 11 | 43.70 | |||
| STER_1506|ID:1900257|rpsP| | 30S ribosomal subunit protein S16 | 10.05 | 10.40 | 9 | 65.93 | 10.40 | 13 | 89.01 |
| STER_0568|ID:1899173|rplL| | 50S ribosomal subunit protein L7/L12 | 4.42 | 12.35 | 6 | 51.22 | |||
| STER_1798|ID:1900486|rplK| | 50S ribosomal subunit protein L11 | 9.47 | 14.79 | 6 | 51.41 | 14.79 | 21 | 67.61 |
| STER_1885|ID:1900548|infA| | translation initiation factor IF-1 | 8.06 | 8.24 | 4 | 57.53 | 8.24 | 4 | 61.64 |
| STER_1903|ID:1900566|rpsS| | 30S ribosomal subunit protein S19 | 10.04 | 10.54 | 5 | 65.59 | 10.61 | 5 | 63.44 |
| STHERMOCNRZ21N_v1_10599|ID:59659749| | conserved protein of unknown function | 6.85 | 9.77 | 26 | 60.47 | |||
| STHERMOCNRZ21N_v1_30315|ID:59660455| | conserved exported protein of unknown function | 11.45 | 6.31 | 23 | 83.05 | |||
| STHERMOCNRZ21N_v1_30185|ID:59660325| | conserved protein of unknown function | 6.29 | 7.13 | 19 | 97.06 | |||
| STHERMOCNRZ21N_v1_30920|ID:59661060|rpsR| | ribosomal protein S18 | 10.63 | 9.26 | 10 | 46.25 | |||
| STHERMOCNRZ21N_v1_30627|ID:59660767|rpsU| | ribosomal protein S21 | 11.29 | 6.92 | 9 | 59.32 | |||
| STHERMOCNRZ21N_v1_10341|ID:59659491|rpmEB| | ribosomal protein L31 | 8.89 | 10.36 | 9 | 67.42 | |||
| STHERMOCNRZ21N_v1_31101|ID:59661241|rpsNA| | ribosomal protein S14 | 10.61 | 7.05 | 7 | 53.23 | |||
| STHERMOCNRZ21N_v1_10428|ID:59659578| | conserved protein of unknown function | 9.10 | 4.92 | 4 | 68.18 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allouche, R.; Hafeez, Z.; Dary-Mourot, A.; Genay, M.; Miclo, L. Streptococcus thermophilus: A Source of Postbiotics Displaying Anti-Inflammatory Effects in THP 1 Macrophages. Molecules 2024, 29, 1552. https://doi.org/10.3390/molecules29071552
Allouche R, Hafeez Z, Dary-Mourot A, Genay M, Miclo L. Streptococcus thermophilus: A Source of Postbiotics Displaying Anti-Inflammatory Effects in THP 1 Macrophages. Molecules. 2024; 29(7):1552. https://doi.org/10.3390/molecules29071552
Chicago/Turabian StyleAllouche, Rania, Zeeshan Hafeez, Annie Dary-Mourot, Magali Genay, and Laurent Miclo. 2024. "Streptococcus thermophilus: A Source of Postbiotics Displaying Anti-Inflammatory Effects in THP 1 Macrophages" Molecules 29, no. 7: 1552. https://doi.org/10.3390/molecules29071552
APA StyleAllouche, R., Hafeez, Z., Dary-Mourot, A., Genay, M., & Miclo, L. (2024). Streptococcus thermophilus: A Source of Postbiotics Displaying Anti-Inflammatory Effects in THP 1 Macrophages. Molecules, 29(7), 1552. https://doi.org/10.3390/molecules29071552