Exploration of the Antibacterial and Anti-Inflammatory Activity of a Novel Antimicrobial Peptide Brevinin-1BW
Abstract
1. Introduction
2. Results
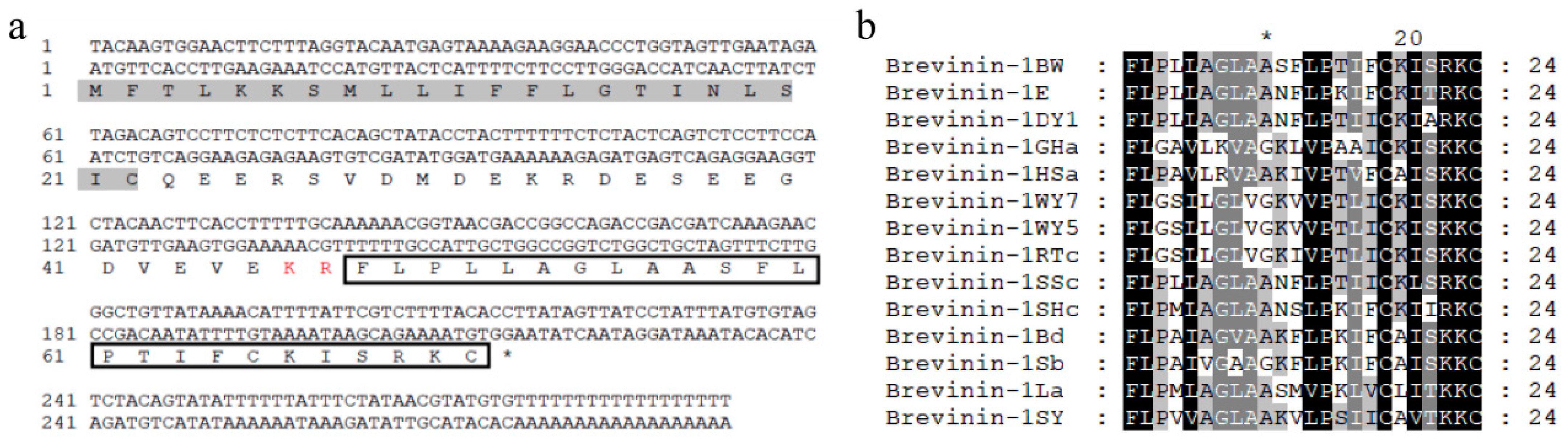
2.1. Identification and Synthesis of Brevinin-1BW
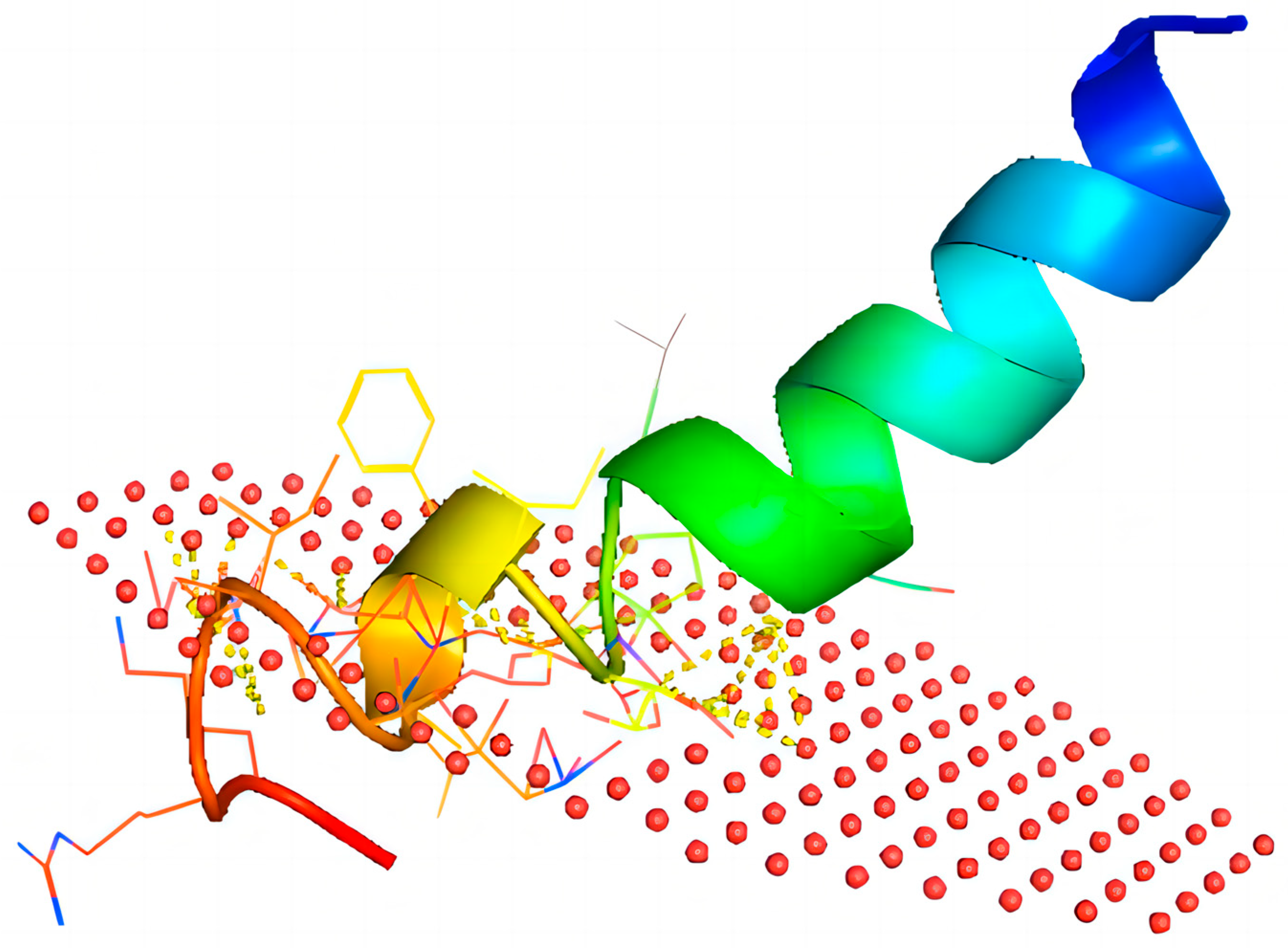
2.2. Structural Analysis of Brevinin-1BW
2.3. Antibacterial Activity Analysis
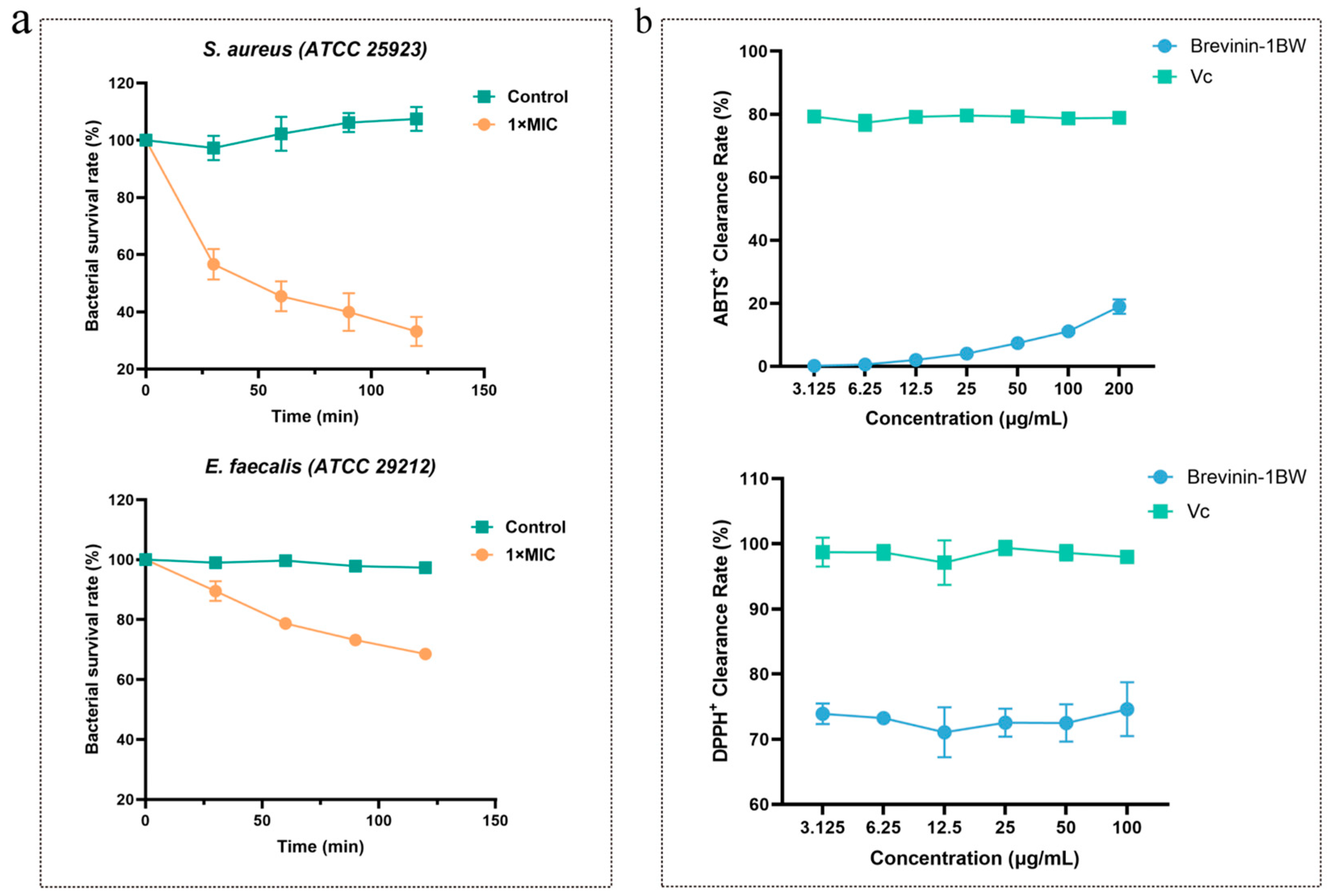
2.4. Time-Killing Assay
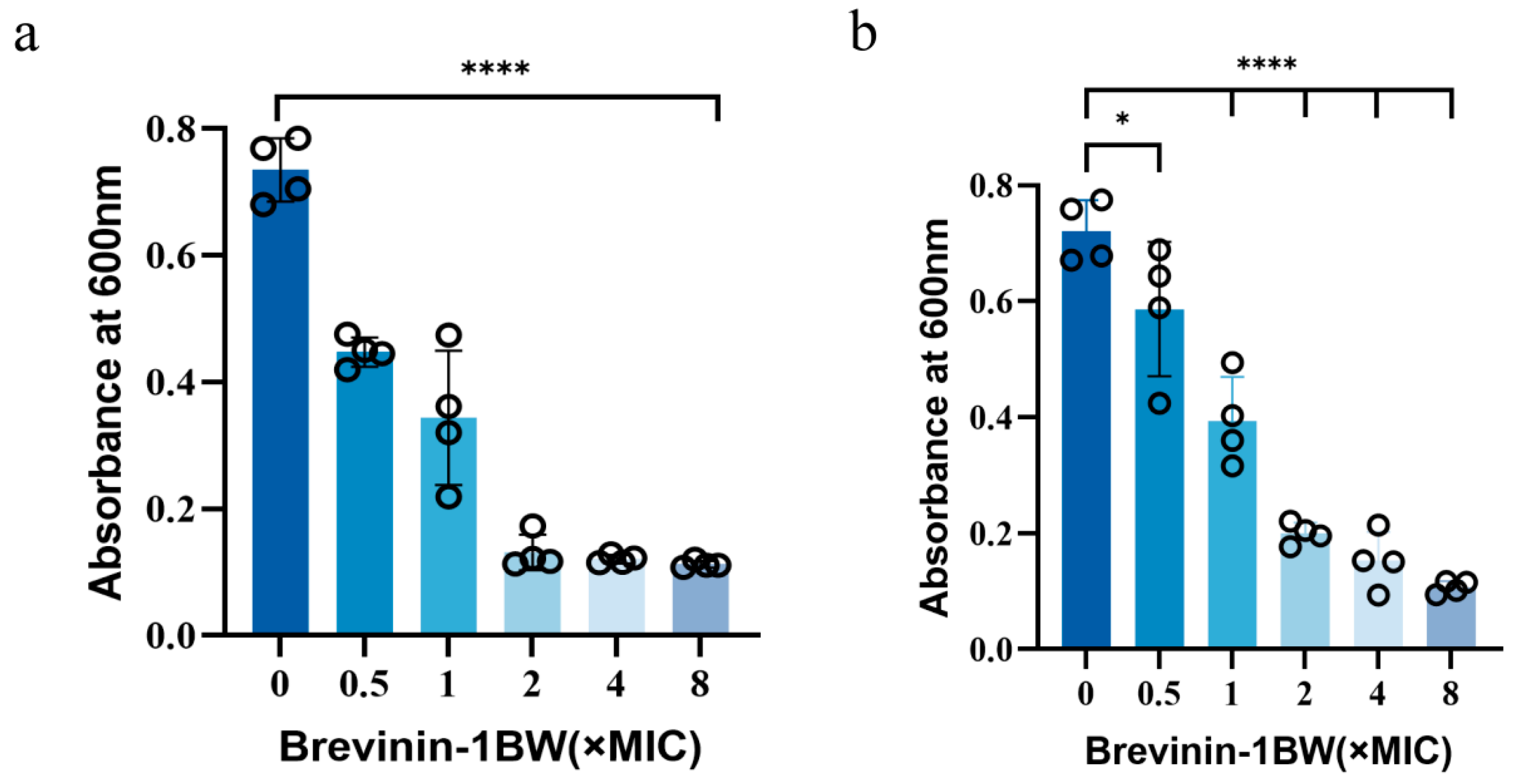
2.5. Inhibition and Elimination of Biofilms by Brevinin-1BW
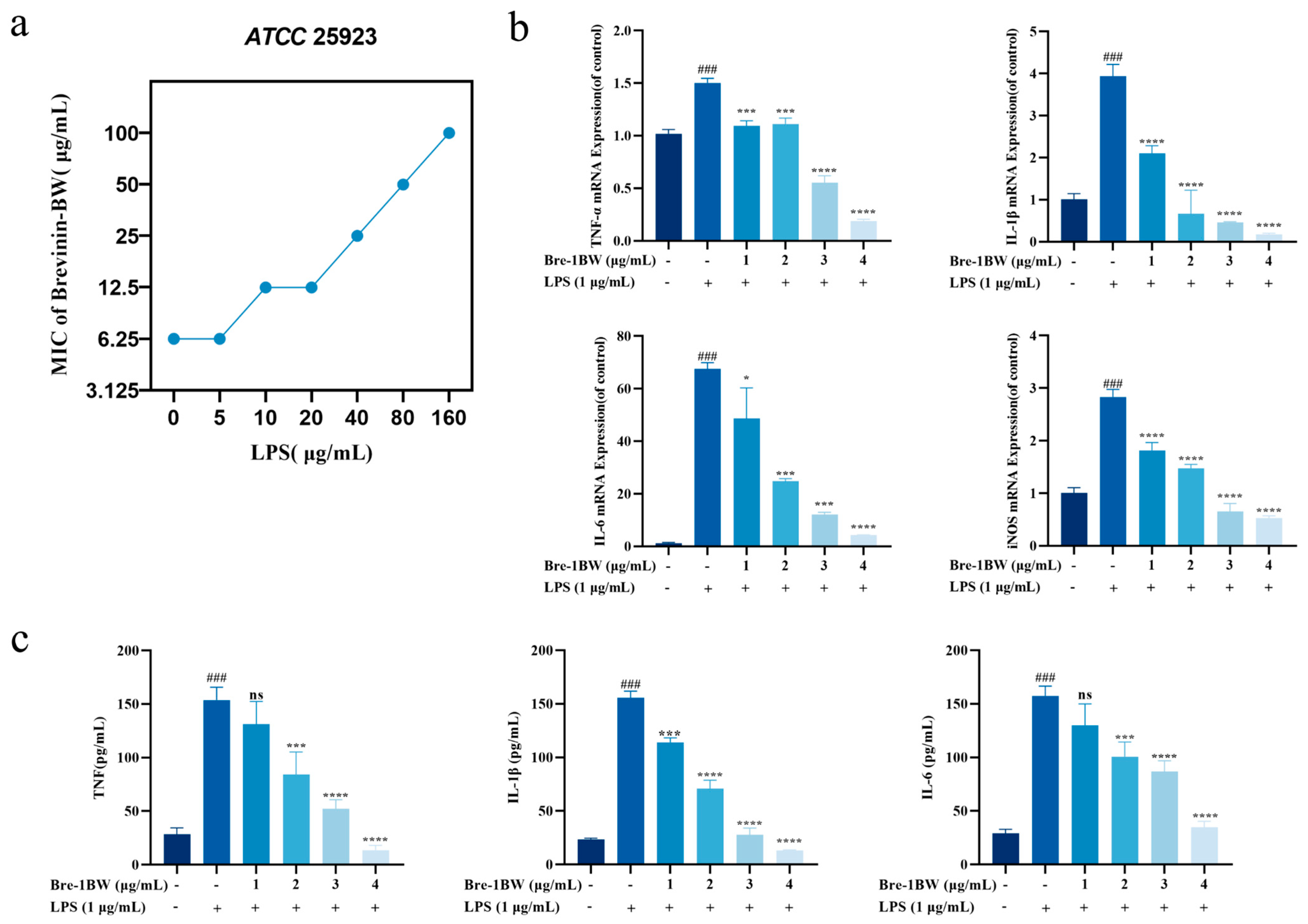
2.6. Effect of Temperature, Serum, and pH on Brevinin-1BW
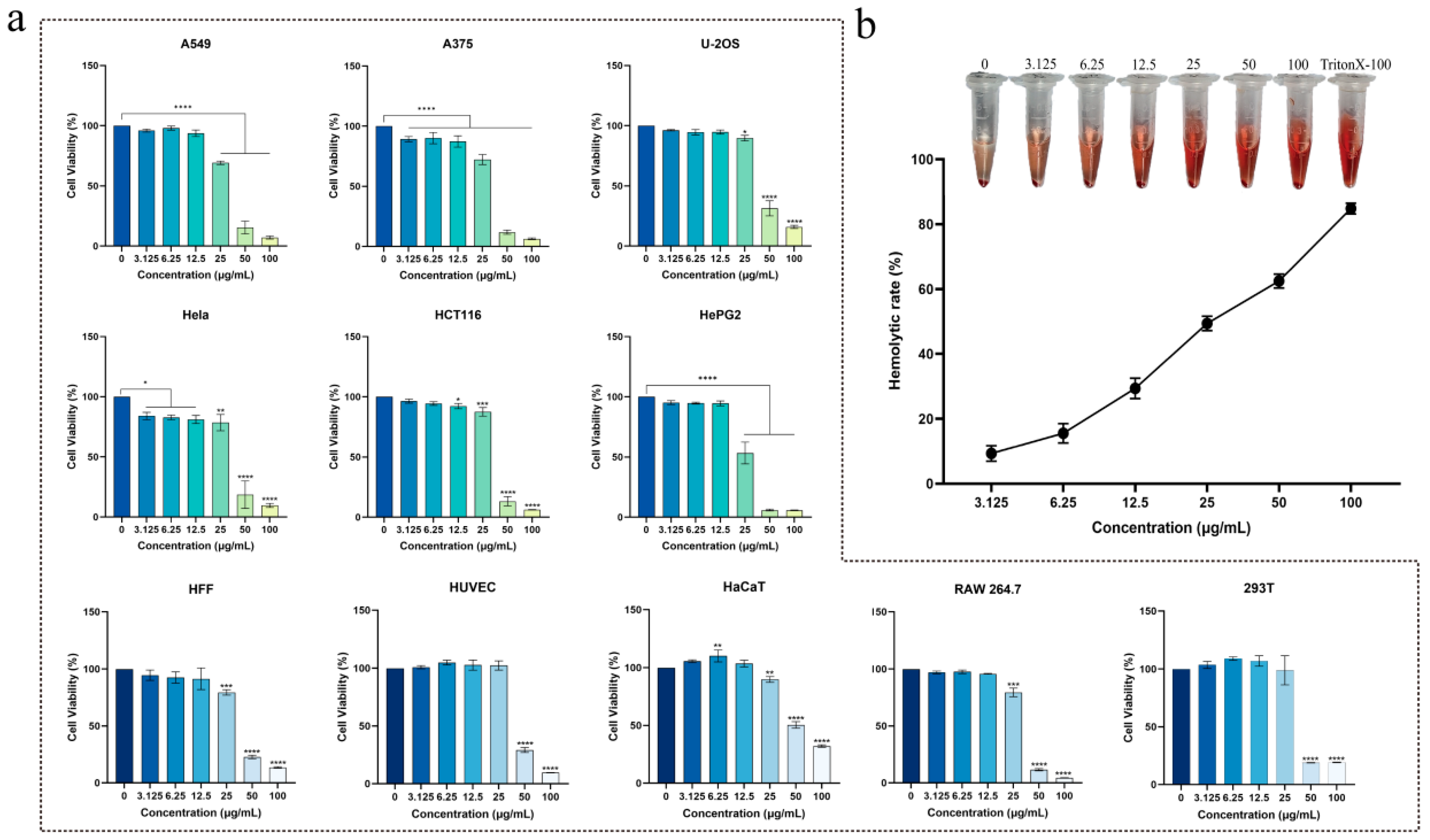
2.7. Cytotoxicity and Hemolytic Activity of Brevinin-1BW
2.8. Antioxidant Activity Analysis
2.9. LPS-Binding Assay
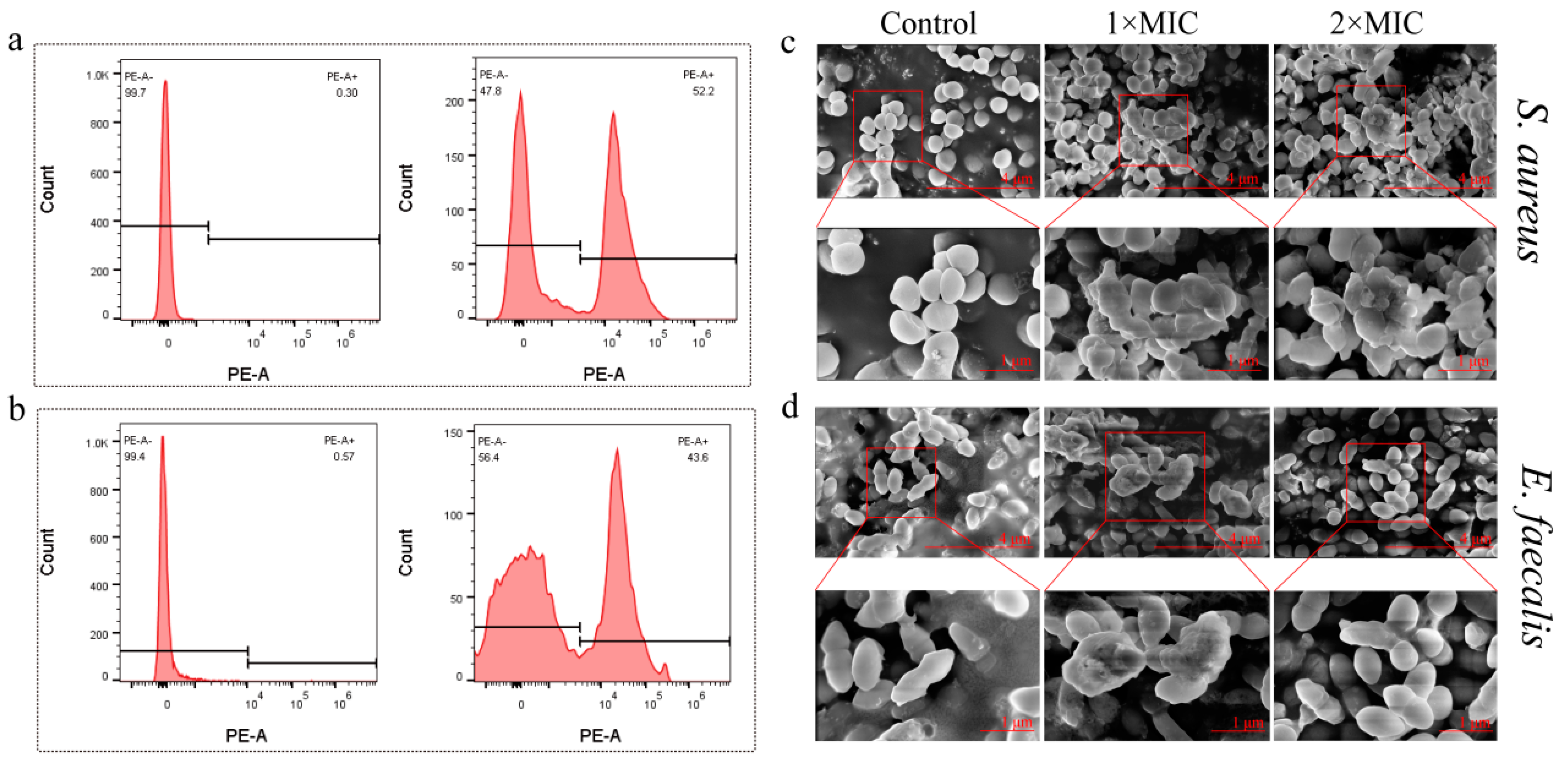
2.10. Changes in Bacterial Morphology after Brevinin-1BW Treatment
2.11. Anti-Inflammatory Effects of Brevinin-1BW
3. Discussion
4. Materials and Methods
4.1. Identification and Synthesis of Brevinin-1BW
4.2. Structural Analysis and Sequence Alignment
4.3. Biological Materials and Reagents
4.4. Antibacterial Activity Analysis
4.5. Time-Killing Assay of Brevinin-1BW against S. aureus and E. faecalis
4.6. Antibiofilm Assay
4.7. Stability Analysis of Brevinin-1BW
4.8. Hemolysis Assays
4.9. Cytotoxicity Assays
4.10. Antioxidant Activity Analysis
4.11. LPS-Binding Assay
4.12. Membrane Permeability and Morphology Change Observation
4.13. Proinflammatory Cytokine Level
4.14. Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Motta, J.-P.; Wallace, J.L.; Buret, A.G.; Deraison, C.; Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 314–334. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Kong, W.; Peng, J.; Gu, H.; Zheng, H. Analysis of main pathogenic bacteria and drug sensitivity in patients with chronic suppurative otitis media and middle ear cholesteatoma in China. Biotechnol. Lett. 2020, 42, 1559–1566. [Google Scholar] [CrossRef] [PubMed]
- Mazzariol, A.; Bazaj, A.; Cornaglia, G. Multi-drug-resistant Gram-negative bacteria causing urinary tract infections: A review. J. Chemother. 2017, 29, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Nasser, A.; Azimi, T.; Ostadmohammadi, S.; Ostadmohammadi, S. A comprehensive review of bacterial osteomyelitis with emphasis on Staphylococcus aureus. Microb. Pathog. 2020, 148, 104431. [Google Scholar] [CrossRef] [PubMed]
- Blumer, J. Treatment of pediatric Gram-positive multidrug-resistant infections. J. Infect. 2009, 59, S51–S58. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, E.G.; Collazos, J.; Cartón, J.A.; Camporro, D.; Asensi, V. Bacterial osteomyelitis: Microbiological, clinical, therapeutic, and evolutive characteristics of 344 episodes. Rev. Española Quimioter. 2018, 31, 217–225. [Google Scholar]
- Massa, H.M.; Cripps, A.W.; Lehmann, D. Otitis media: Viruses, bacteria, biofilms and vaccines. Med. J. Aust. 2009, 191, S44–S49. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Arason, V.A.; Sigurdsson, J.A. The problems of antibiotic overuse. Scand. J. Prim. Health Care 2010, 28, 65–66. [Google Scholar] [CrossRef][Green Version]
- Terreni, M.; Taccani, M.; Pregnolato, M. New Antibiotics for Multidrug-Resistant Bacterial Strains: Latest Research Developments and Future Perspectives. Molecules 2021, 26, 2671. [Google Scholar] [CrossRef] [PubMed]
- Hetzler, L.; Kollef, M.H.; Yuenger, V.; Micek, S.T.; Betthauser, K.D. New antimicrobial treatment options for severe Gram-negative infections. Curr. Opin. Crit. Care 2022, 28, 522–533. [Google Scholar] [CrossRef]
- Principe, L. New Antimicrobial Options in the Clinical Practice of Infections Caused by Difficult-to-Treat Pathogens: A Global Opportunity for Public Health. Antibiotics 2022, 11, 740. [Google Scholar] [CrossRef] [PubMed]
- Kardani, K.; Bolhassani, A. Antimicrobial/anticancer peptides: Bioactive molecules and therapeutic agents. Immunotherapy 2021, 13, 669–684. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, J.; Leal, E.C.; Carvalho, E. Bioactive Antimicrobial Peptides as Therapeutic Agents for Infected Diabetic Foot Ulcers. Biomolecules 2021, 11, 1894. [Google Scholar] [CrossRef]
- Wang, L.; Qu, L.; Lin, S.; Yang, Q.; Zhang, X.; Jin, L.; Dong, H.; Sun, D. Biological Functions and Applications of Antimicrobial Peptides. Curr. Protein Pept. Sci. 2022, 23, 226–247. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm activity of host defence peptides: Complexity provides opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr. Opin. Pharmacol. 2006, 6, 468–472. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial peptides as therapeutic agents: Opportunities and challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef]
- Luong, H.X.; Thanh, T.T.; Tran, T.H. Antimicrobial peptides—Advances in development of therapeutic applications. Life Sci. 2020, 260, 118407. [Google Scholar] [CrossRef]
- Szadkowska, M.; Olewniczak, M.; Kloska, A.; Jankowska, E.; Kapusta, M.; Rybak, B.; Wyrzykowski, D.; Zmudzinska, W.; Gieldon, A.; Kocot, A.; et al. A Novel Cryptic Clostridial Peptide That Kills Bacteria by a Cell Membrane Permeabilization Mechanism. Microbiol. Spectr. 2022, 10, e01657-22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Fu, L.; Wan, M.; Song, J.; Gao, L.; Fang, W. Peripheral Antimicrobial Peptide Gomesin Induces Membrane Protrusion, Folding, and Laceration. Langmuir 2019, 35, 13233–13242. [Google Scholar] [CrossRef]
- Toke, O. Antimicrobial peptides: New candidates in the fight against bacterial infections. Pept. Sci. 2005, 80, 717–735. [Google Scholar] [CrossRef] [PubMed]
- Bucki, R.; Leszczyńska, K.; Namiot, A.; Sokołowski, W. Cathelicidin LL-37: A Multitask Antimicrobial Peptide. Arch. Immunol. Ther. Exp. 2010, 58, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Contreras, G.; Shirdel, I.; Braun, M.S.; Wink, M. Defensins: Transcriptional regulation and function beyond antimicrobial activity. Dev. Comp. Immunol. 2020, 104, 103556. [Google Scholar] [CrossRef]
- Bechinger, B.; Gorr, S.-U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef]
- Du, Z.; Li, B.; Shen, X.; Wang, K.; Du, J.; Yu, X.; Yuan, J. A new antimicrobial peptide isoform, Pc-crustin 4 involved in antibacterial innate immune response in fresh water crayfish, Procambarus clarkii. Fish Shellfish. Immunol. 2019, 94, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Singh, M.; Mukhopadhyay, K. Alpha-Melanocyte Stimulating Hormone: An Emerging Anti-Inflammatory Antimicrobial Peptide. BioMed Res. Int. 2014, 2014, 874610. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, S.; Yao, Z.; Zheng, X.; Li, W.; Zhang, Y.; Wang, L.; Cao, J.; Zhou, T. Antimicrobial peptide WAM-1: A promising antibacterial and anti-inflammatory drug against carbapenem-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 2022, 77, 1903–1911. [Google Scholar] [CrossRef] [PubMed]
- Neshani, A.; Zare, H.; Akbari Eidgahi, M.R.; Khaledi, A.; Ghazvini, K. Epinecidin-1, a highly potent marine antimicrobial peptide with anticancer and immunomodulatory activities. BMC Pharmacol. Toxicol. 2019, 20, 33. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Hunter, L.; Attoub, S.; Casciaro, B.; Mechkarska, M.; Abdel-Wahab, Y.H.A. Antimicrobial, cytotoxic, and insulin-releasing activities of the amphibian host-defense peptide ocellatin-3N and its L-lysine-substituted analogs. J. Pept. Sci. 2023, 29, e3463. [Google Scholar] [CrossRef] [PubMed]
- Soltaninejad, H.; Zare-Zardini, H.; Ordooei, M.; Ghelmani, Y.; Ghadiri-Anari, A.; Mojahedi, S.; Hamidieh, A.A. Antimicrobial Peptides from Amphibian Innate Immune System as Potent Antidiabetic Agents: A Literature Review and Bioinformatics Analysis. J. Diabetes Res. 2021, 2021, 2894722. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Musale, V.; Attoub, S.; Mangoni, M.L.; Leprince, J.; Coquet, L.; Jouenne, T.; Abdel-Wahab, Y.H.A.; Flatt, P.R.; Rinaldi, A.C. Cytotoxic peptides with insulin-releasing activities from skin secretions of the Italian stream frog Rana italica (Ranidae): Host-Defense Peptides from Rana italica. J. Pept. Sci. 2017, 23, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Savelyeva, A.; Ghavami, S.; Davoodpour, P.; Asoodeh, A.; Łos, M.J. An overview of brevinin superfamily: Structure, function and clinical perspectives. In Anticancer Genes; Grimm, S., Ed.; Advances in Experimental Medicine and Biology; Springer: London, UK, 2014; pp. 197–212. ISBN 978-1-4471-6458-6. [Google Scholar]
- Zohrab, F.; Askarian, S.; Jalili, A.; Kazemi Oskuee, R. Biological Properties, Current Applications and Potential Therapeautic Applications of Brevinin Peptide Superfamily. Int. J. Pept. Res. Ther. 2019, 25, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.-Y.; Hong, S.-Y.; Lee, K.-H. Structure-activity analysis of brevinin 1E amide, an antimicrobial peptide from Rana esculenta. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1998, 1387, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Brown, A.O.; Singh, K.V.; Cruz, M.R.; Kaval, K.G.; Francisco, L.E.; Murray, B.E.; Garsin, D.A. Cardiac Microlesions Form During Severe Bacteremic Enterococcus faecalis Infection. J. Infect. Dis. 2021, 223, 508–516. [Google Scholar] [CrossRef]
- Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691. [Google Scholar] [CrossRef]
- Feng, Q.; Huang, Y.; Chen, M.; Li, G.; Chen, Y. Functional synergy of α-helical antimicrobial peptides and traditional antibiotics against Gram-negative and Gram-positive bacteria in vitro and in vivo. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Putker, F.; Bos, M.P.; Tommassen, J. Transport of lipopolysaccharide to the Gram-negative bacterial cell surface. FEMS Microbiol. Rev. 2015, 39, 985–1002. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Cheng, P.; Ma, C.; Xi, X.; Wang, L.; Zhou, M.; Bian, H.; Chen, T. Evaluating the Bioactivity of a Novel Broad-Spectrum Antimicrobial Peptide Brevinin-1GHa from the Frog Skin Secretion of Hylarana guentheri and Its Analogues. Toxins 2018, 10, 413. [Google Scholar] [CrossRef] [PubMed]
- Iwakoshi-Ukena, E.; Soga, M.; Okada, G.; Fujii, T.; Sumida, M.; Ukena, K. Characterization of novel antimicrobial peptides from the skin of the endangered frog Odorrana ishikawae by shotgun cDNA cloning. Biochem. Biophys. Res. Commun. 2011, 412, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Jiang, J.; Wu, Z.; Xie, F. Antimicrobial peptides from the skin of the Asian frog, Odorrana jingdongensis: De novo sequencing and analysis of tandem mass spectrometry data. J. Proteom. 2012, 75, 5807–5821. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Al-Ghaferi, N.; Abraham, B.; Sonnevend, A.; Coquet, L.; Leprince, J.; Jouenne, T.; Vaudry, H.; Iwamuro, S. Antimicrobial peptides from the skin of the Tsushima brown frog Rana tsushimensis. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2006, 143, 42–49. [Google Scholar] [CrossRef]
- Chongsiriwatana, N.P.; Patch, J.A.; Czyzewski, A.M.; Dohm, M.T.; Ivankin, A.; Gidalevitz, D.; Zuckermann, R.N.; Barron, A.E. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 2794–2799. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, Y.; Gao, Y.; Wang, Y.; Xia, Q.; Zhong, R.; Ma, C.; Zhou, M.; Xi, X.; Shaw, C.; et al. Enhanced Antimicrobial Activity of N-Terminal Derivatives of a Novel Brevinin-1 Peptide from The Skin Secretion of Odorrana schmackeri. Toxins 2020, 12, 484. [Google Scholar] [CrossRef]
- Torres, M.D.T.; Sothiselvam, S.; Lu, T.K.; De La Fuente-Nunez, C. Peptide Design Principles for Antimicrobial Applications. J. Mol. Biol. 2019, 431, 3547–3567. [Google Scholar] [CrossRef]
- Rincón-Cortés, C.A.; Bayona-Rojas, M.A.; Reyes-Montaño, E.A.; Vega-Castro, N.A. Antimicrobial Activity Developed by Scorpion Venoms and Its Peptide Component. Toxins 2022, 14, 740. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, Y.; Wang, T.; Chen, X.; Zhou, M.; Ma, C.; Xi, X.; Zhang, Y.; Chen, T.; Shaw, C.; et al. Brevinin-1GHd: A novel Hylarana guentheri skin secretion-derived Brevinin-1 type peptide with antimicrobial and anticancer therapeutic potential. Biosci. Rep. 2020, 40, BSR20200019. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Xie, Z.; Zhang, S.; Wei, H.; Song, Y.; Zhang, Y.; Wang, M. Brevinin-GR23 from frog Hylarana guentheri with antimicrobial and antibiofilm activities against Staphylococcus aureus. Biosci. Biotechnol. Biochem. 2020, 84, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Bhunia, A.; Domadia, P.N.; Torres, J.; Hallock, K.J.; Ramamoorthy, A.; Bhattacharjya, S. NMR Structure of Pardaxin, a Pore-forming Antimicrobial Peptide, in Lipopolysaccharide Micelles: Mechanism of outer membrane permeabilization 2. J. Biol. Chem. 2010, 285, 3883–3895. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.-S.; Lee, H.; Lee, J.-O. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.M.; Park, B.S.; Kim, J.-I.; Kim, S.E.; Lee, J.; Oh, S.C.; Enkhbayar, P.; Matsushima, N.; Lee, H.; Yoo, O.J.; et al. Crystal Structure of the TLR4-MD-2 Complex with Bound Endotoxin Antagonist Eritoran. Cell 2007, 130, 906–917. [Google Scholar] [CrossRef]
- Tian, M.; Wang, K.; Liang, Y.; Chai, J.; Wu, J.; Zhang, H.; Huang, X.; Chen, X.; Xu, X. The first Brevinin-1 antimicrobial peptide with LPS-neutralizing and anti-inflammatory activities in vitro and in vivo. Front. Microbiol. 2023, 14, 1102576. [Google Scholar] [CrossRef]
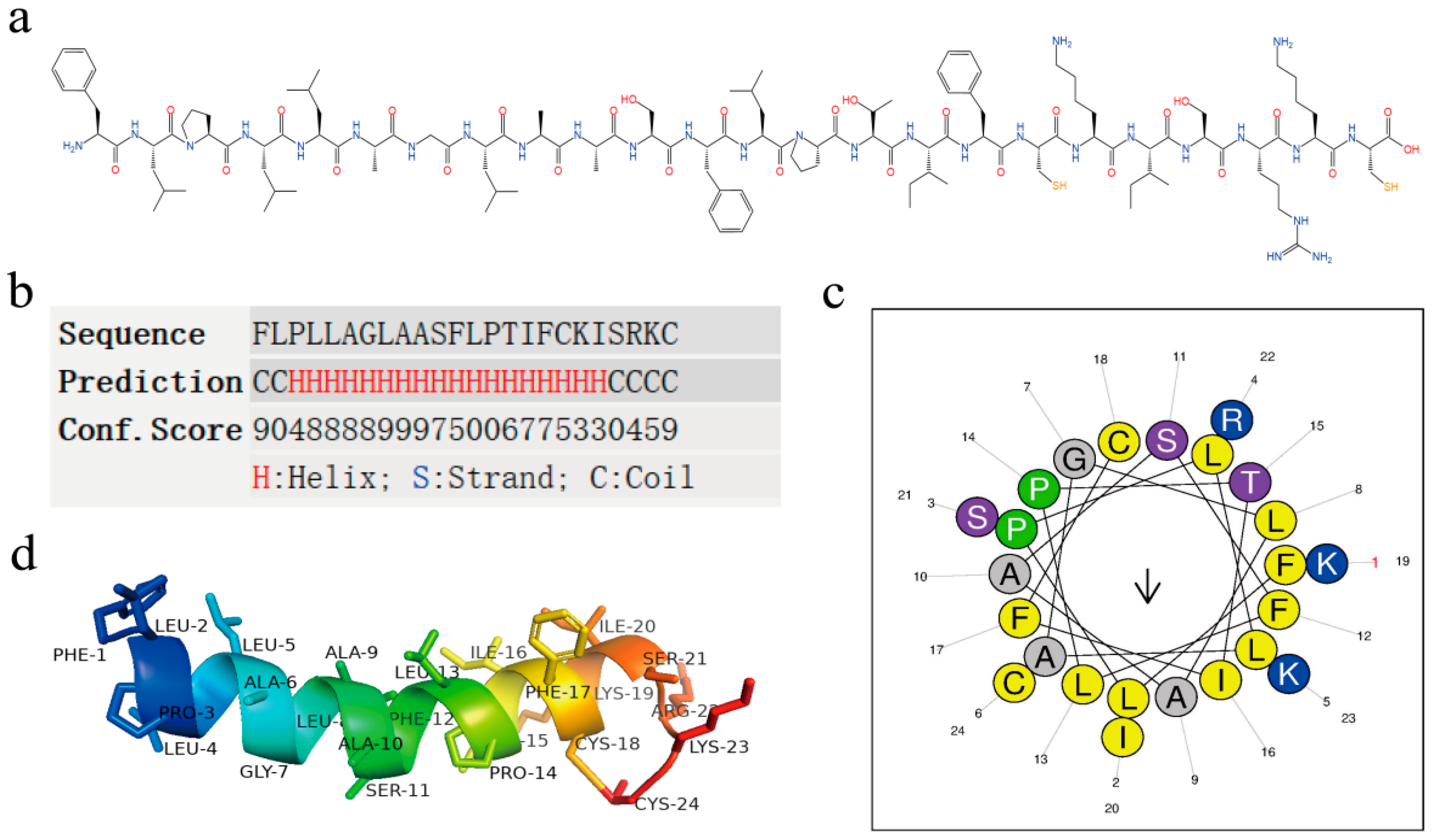
| 1FLPLLAGLAASFLPTIFCKISRKC24 | ||
| Physico-chemical properties | Polar residues + GLY | Nonpolar residues |
| Hydrophobicity <H> | Polar residues + GLY (n/%) | Nonpolar residues (n/%) |
| 0.838 | 7/29.17 | 17/70.83 |
| Hydrophobic moment <µH> | Uncharged residues + GLY | Aromatic residues |
| 0.256 | SER 2, THR 1, GLY 1 | PHE 3 |
| Net charge z | Charged residues | Special residues |
| 3 | LYS 2, ARG 1 | CYS 2, PRO 2 |
| Hydrophobic face: I A L I L A C F A | ||
| Bacterial Strains | Brevinin-1BW | Ampicillin | |
|---|---|---|---|
| MIC (μg/mL) | MBC (μg/mL) | MIC (μg/mL) | |
| Gram-positive bacteria | |||
| S. aureus (ATCC25923) | 6.25 | 6.25 | 3.125 |
| E. faecalis (ATCC29212) | 3.125 | 12.5 | 3.125 |
| S. saprophytics (ATCCBAA750) | 6.25 | 12.5 | 3.125 |
| S. aureus (MDR ATCC29213) | 6.25 | 25 | >100 |
| S. epidermidis (CI 1607BL1D39) | 12.5 | 25 | 3.125 |
| S. haemolyticus (CI 1607BLD590) | 12.5 | 25 | >100 |
| Gram-negative bacteria | |||
| E. coli (ATCC25922) | 100 | >100 | 100 |
| E. hormaechei (ATCC700323) | 100 | >100 | >100 |
| P. aeruginosa (ATCC27853) | 100 | >100 | >100 |
| K. pneumoniae (ATCC700603) | >100 | >100 | >100 |
| E. coli (MDR ATCC35218) | >100 | >100 | >100 |
| C. koseri (CI 1611SED223) | 50 | >100 | >100 |
| Conditions | MIC (µg/mL) * | Conditions | MIC (µg/mL) * | ||
|---|---|---|---|---|---|
| Temperature | −20 °C | 6.25 | Fetal bovine serum | 0 | 6.25 |
| 4 °C | 6.25 | 5% | 6.25 | ||
| 37 °C | 6.25 | 10% | 12.5 | ||
| 60 °C | 6.25 | pH | 2 | 6.25 | |
| 100 °C | 6.25 | 12 | 6.25 |
| Cancer Cells | CC50 (µg/mL) | Non-Cancer Cells | CC50 (µg/mL) |
|---|---|---|---|
| A375 | 23.79 ± 0.80 | 293T | 28.34 ± 1.08 |
| A549 | 27.64 ± 0.41 | HUVEC | 43.62 ± 0.77 |
| Hela | 37.62 ± 0.91 | HaCaT | 36.84 ± 1.44 |
| HePG2 | 25.42 ± 0.49 | HFF | 33.99 ± 0.48 |
| U-2OS | 39.20 ± 0.40 | RAW264.7 | 32.33 ± 0.47 |
| HCT116 | 35.44 ± 0.47 |
| Peptide Name | GRAVY | pI | MIC (µM) | ||
|---|---|---|---|---|---|
| S. aureus | MRSA/MDR S. aureus | E. faecalis | |||
| Brevinin-1BW (FLPLLAGLAASFLPTIFCKISRKC) | 1.192 | 9.50 | 2.38 | 2.38 | 1.19 |
| Brevinin1TSa (FLGSIVGALASALPSLISKIRN) | 1.073 | 11.0 | 25 | 25 | 50 |
| Brevinin-1ISa (FLPGVLRLVTKVGPAVVCAITRNC) | 1.104 | 9.70 | 3.1 | 3.1 | — |
| Brevinin-1JDa (FLPAVIRVAANVLPTVFCAISKKC) | 1.279 | 9.50 | 6 | 13 | — |
| Brevinin-1JDc (FLPAVLRVAAKVVPTVFCLISKKC) | 1.333 | 9.85 | 6 | 3 | — |
| Brevinin-1GHa (FLGAVLKVAGKLVPAAICKISKKC) | 1.054 | 9.90 | 2 | 4 | 8 |
| Brevinin-1GHd (FLGALFKVASKLVPAAICSISKKC) | 1.108 | 9.70 | 2 | 4 | — |
| Gene | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) |
|---|---|---|
| β-Actin | TATCGGACGCCTGGTTA | TGTGCCGTTGAACTTGC |
| TNF-α | GTCAACCTCCTCTCTGCCAT | CCAAAGTAGACCTGCCCAGA |
| IL-1β | CCAGGATGAGGACATGAGCA | CGGAGCCTGTAGTGCAGTTG |
| IL-6 | TCCATCCAGTTGCCTTCTTG | AAGCCTCCGACTTGTGAAGTG |
| iNOS | CATGCTACTGGAGGTGGGTG | CATTGATCTCCGTGACAGCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Wang, L.; He, D.; Liu, Q.; Han, Q.; Zhang, J.; Zhang, A.-M.; Song, Y. Exploration of the Antibacterial and Anti-Inflammatory Activity of a Novel Antimicrobial Peptide Brevinin-1BW. Molecules 2024, 29, 1534. https://doi.org/10.3390/molecules29071534
Chen Z, Wang L, He D, Liu Q, Han Q, Zhang J, Zhang A-M, Song Y. Exploration of the Antibacterial and Anti-Inflammatory Activity of a Novel Antimicrobial Peptide Brevinin-1BW. Molecules. 2024; 29(7):1534. https://doi.org/10.3390/molecules29071534
Chicago/Turabian StyleChen, Zhizhi, Lei Wang, Dongxia He, Qi Liu, Qinqin Han, Jinyang Zhang, A-Mei Zhang, and Yuzhu Song. 2024. "Exploration of the Antibacterial and Anti-Inflammatory Activity of a Novel Antimicrobial Peptide Brevinin-1BW" Molecules 29, no. 7: 1534. https://doi.org/10.3390/molecules29071534
APA StyleChen, Z., Wang, L., He, D., Liu, Q., Han, Q., Zhang, J., Zhang, A.-M., & Song, Y. (2024). Exploration of the Antibacterial and Anti-Inflammatory Activity of a Novel Antimicrobial Peptide Brevinin-1BW. Molecules, 29(7), 1534. https://doi.org/10.3390/molecules29071534








