Identification of Vernonia patula Merr. and Its Similar Varieties Based on a Combination of HPLC Fingerprinting and Chemical Pattern Recognition
Abstract
1. Introduction
2. Results
2.1. Validation of Fingerprint Analysis Method
2.1.1. The Results of Precision
2.1.2. The Results of Stability Test
2.1.3. The Results of Repeatability Test
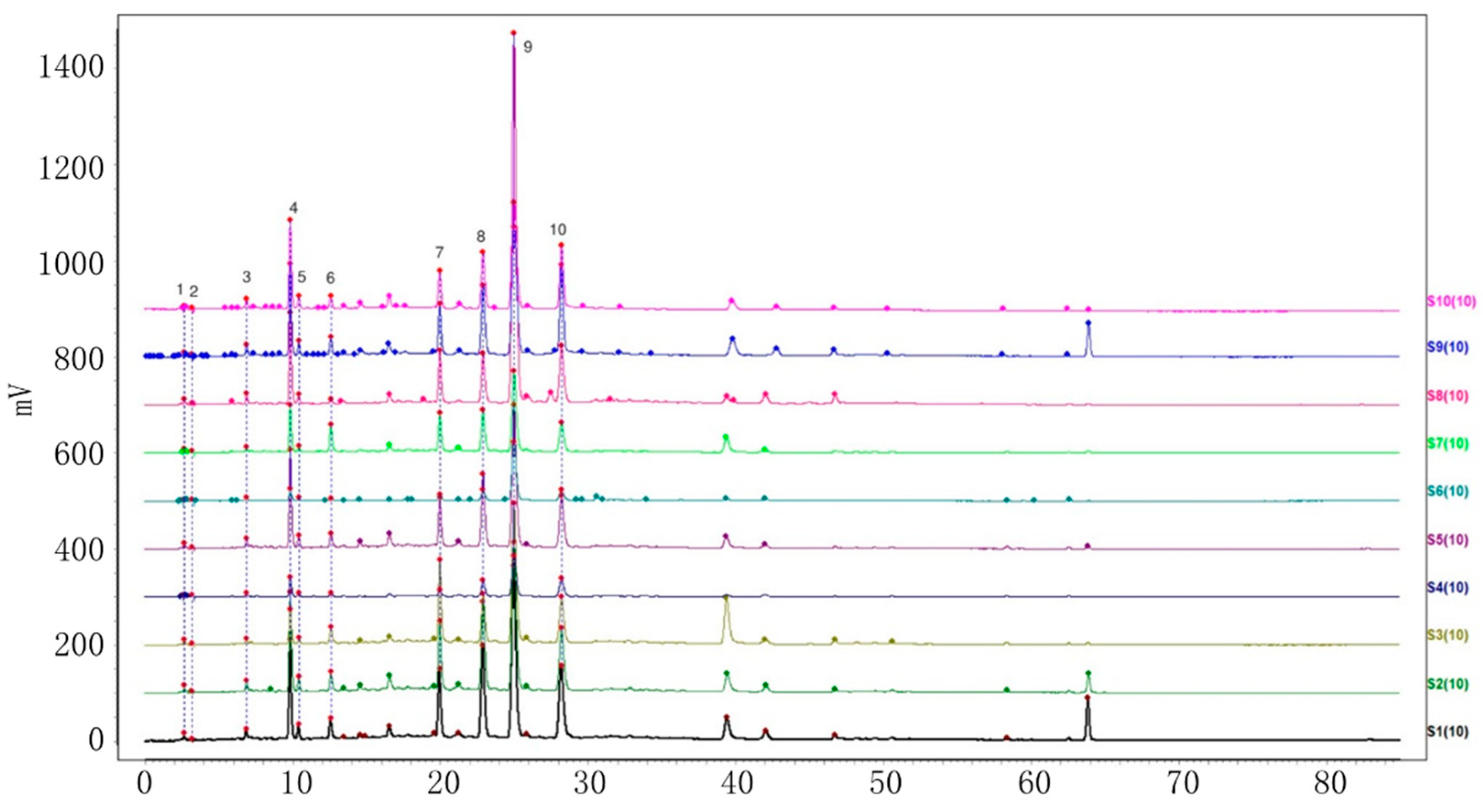
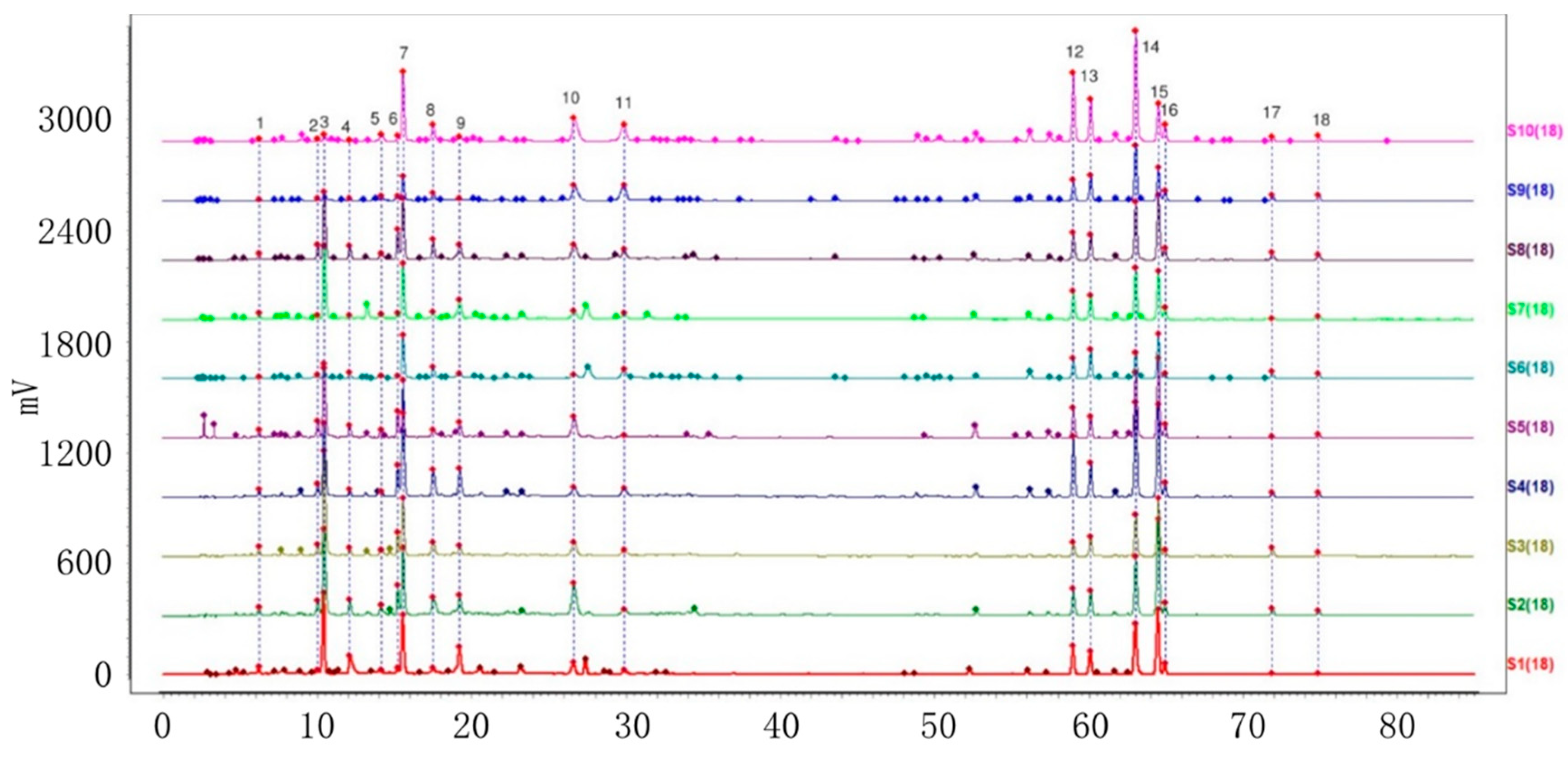
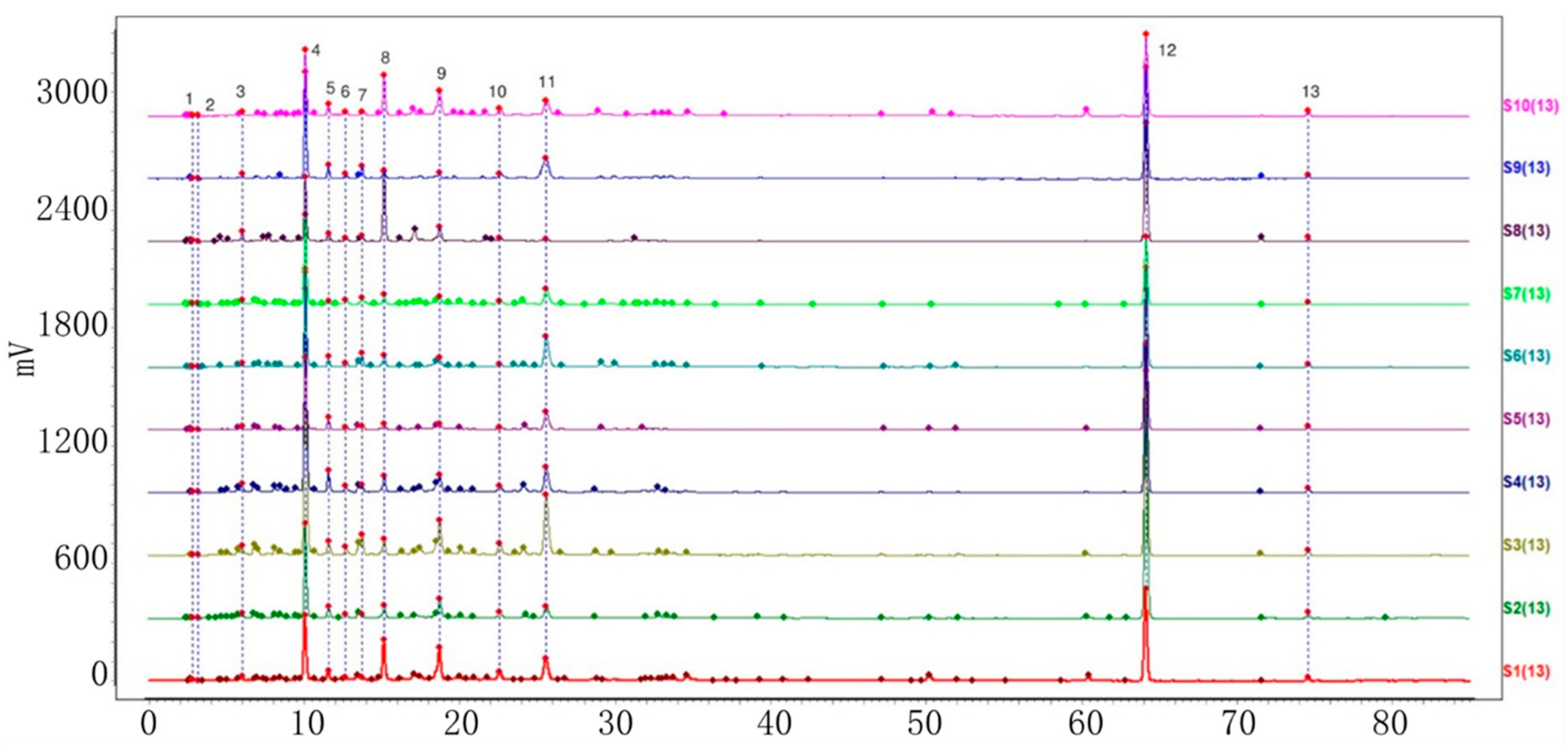
2.2. Establishment of HPLC Fingerprints
2.2.1. Identification of Common Peaks
2.2.2. Similarity Analysis of HPLC Fingerprints of Individual Variety
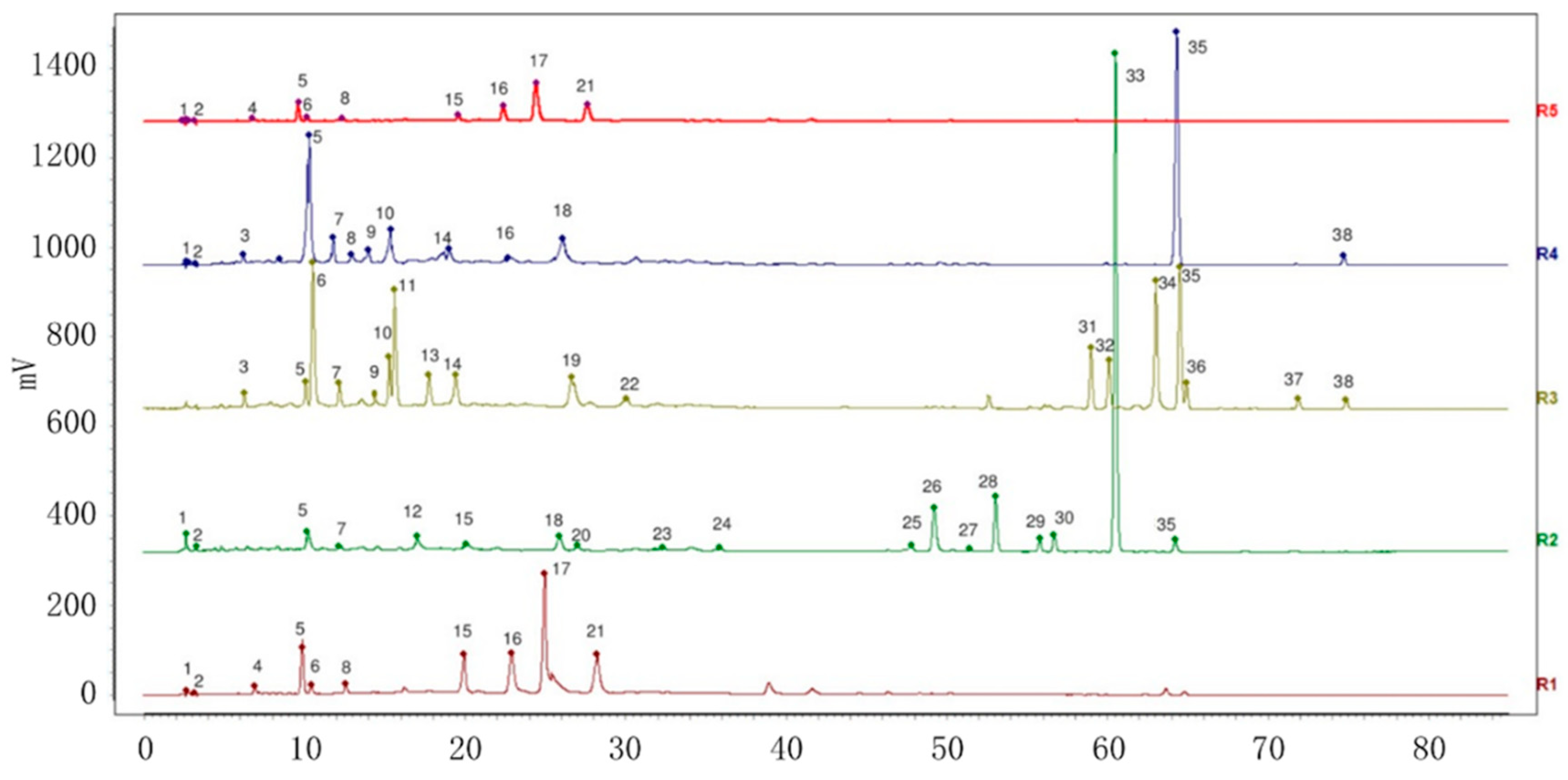
2.2.3. HPLC Fingerprint Similarity Analysis of VP and Three Similar Varieties
2.3. Chemical Pattern Recognition
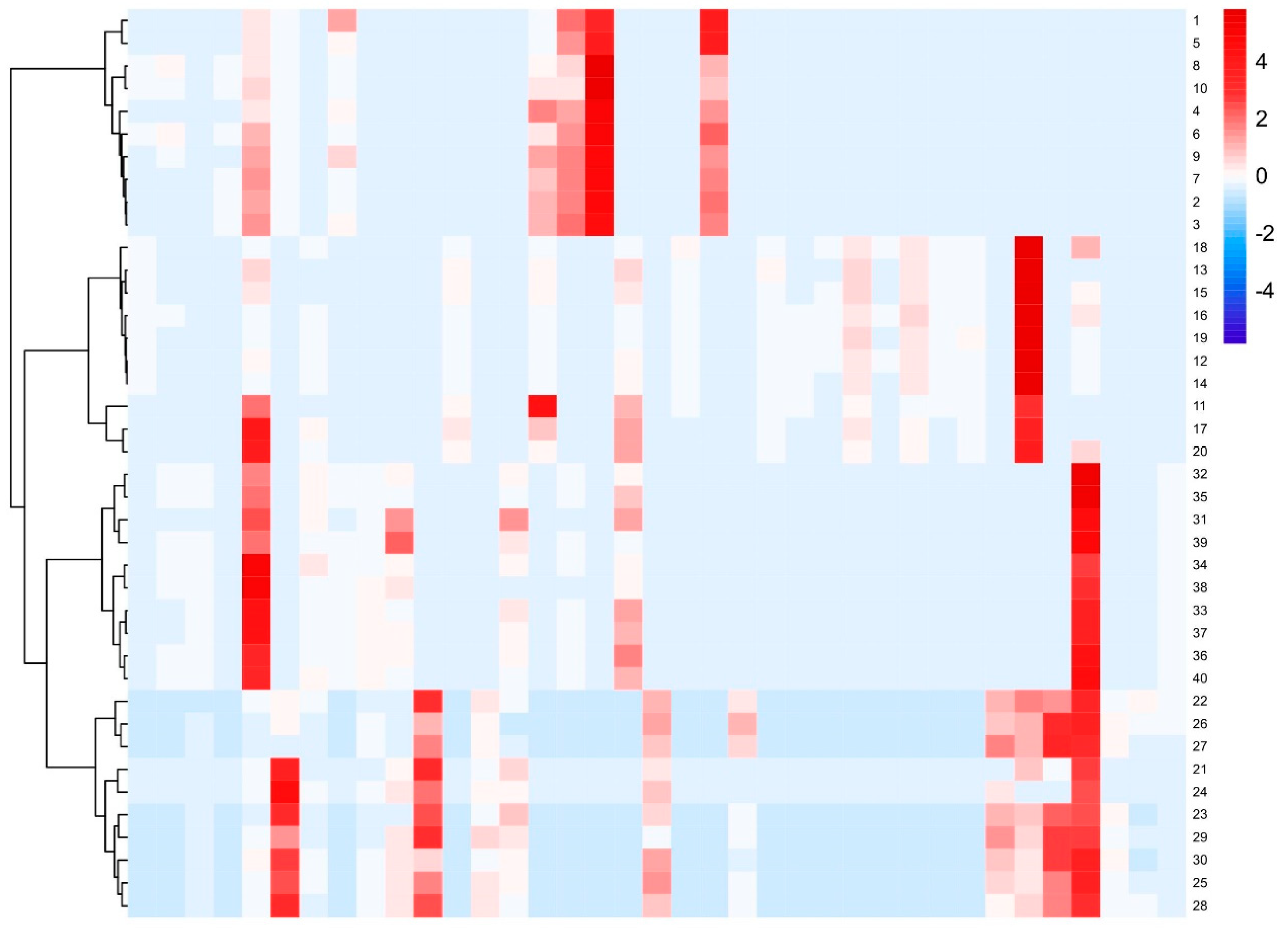
2.3.1. Cluster Analysis (CA)
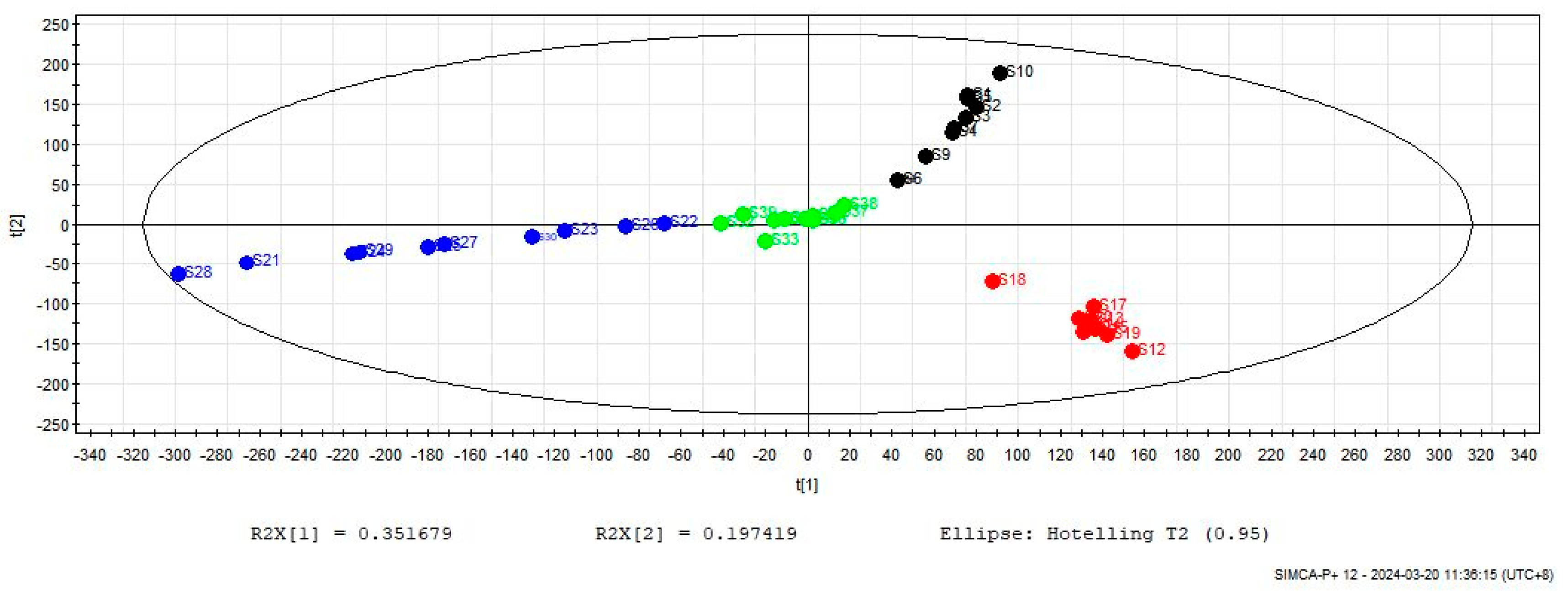
2.3.2. Partial Least Squares Discriminant Analysis (PLS-DA)
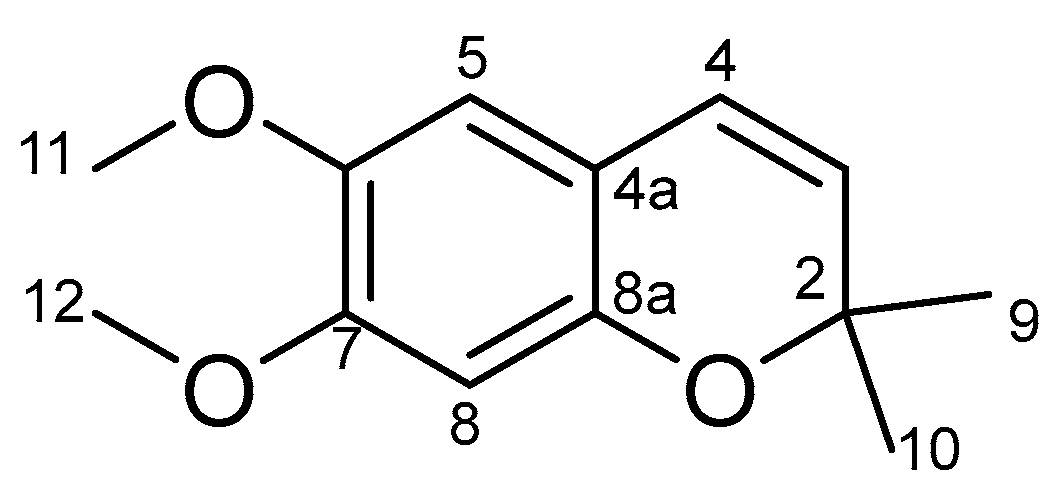
2.3.3. Separation and Identification of Peak 35
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Chemicals and Reagents
4.2. Apparatus
4.3. Fingerprints of VP and Three Similar Varieties
4.4. Validation of HPLC Methodology
4.4.1. Precision Experiment
4.4.2. Stability Test
4.4.3. Repeatability Test
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Editorial Committee of the Flora of China, Chinese Academy of Sciences. Flora of China; Science Press: Beijing, China, 1985; 41p. [Google Scholar]
- Guangxi Zhuang Autonomous Region Food and Drug Administration. Quality Standards for Zhuang Medicine in Guangxi Zhuang Autonomous Region (Volume III); Guangxi Science and Technology Press: Nanning, China, 2017; 155p. [Google Scholar]
- Liu, Q.; Yang, J.; Suo, M. Study on the chemical composition and pharmacological activity of sesquiterpene lactones and steroidal saponins of vernonia. China J. Chin. Mater. Med. 2007, 32, 10–17. [Google Scholar]
- Wei, Q.; Xin, C.; Hui, Z. Study on the chemical composition of Vernonia patula. J. Anhui Agric. Sci. 2018, 46, 181–183. [Google Scholar]
- Sun, L.; Ba, Y.; Yu, L.; Xu, J. Progress in the research of pharmacological activities of the genus Vernonia esulenta. Xinjiang J. Tradit. Chin. Med. 2009, 27, 82–85. [Google Scholar]
- Wu, P.S.; Jeng, J.; Yang, J.J.; Kao, V.; Yen, J.H.; Wu, M.J. Vernonia patula (Dryand.) Merr. and Leucas chinensis (Retz.) R. Brown exert anti-inflammatory activities and relieve oxidative stress via Nrf2 activation. J. Ethnopharmacol. 2020, 262, 113155. [Google Scholar] [CrossRef]
- Hira, A.; Dey, S.K.; Howlader, M.S.; Ahmed, A.; Hossain, H.; Jahan, I.A. Anti-inflammatory and antioxidant activities of ethanolic extract of aerial parts of Vernonia patula (Dryand.) Merr. Asian Pac. J. Trop. Biomed. 2013, 3, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y. Morphological characteristics of the invasive species Pseudo Stinky Grass and differences between similar species of Agastache rugosa. Guangdong For. Sci. Technol. 2012, 28, 69–71. [Google Scholar]
- Lin, M.; Kong, D.; Zhou, L. Research progress on the biological characteristics and control of Pseudomonas aeruginosa. China Plant Prot. 2020, 40, 82–85. [Google Scholar]
- Cai, Y.; Zhu, Y.; Liu, D.; Teng, J. Pharmacological identification of salted shrimp flowers. J. Chin. Med. Mater. 2006, 9, 116–118. [Google Scholar]
- Guo, H.; Liu, J.; Hu, H.; Wang, X.; Liu, W. Determination of chlorogenic acid and caffeic acid content in the Zhuang Yao medicine Vernonia patula Merr. by HPLC. Hubei Agric. Sci. 2020, 59, 188–190+195. [Google Scholar]
- Liu, W.; Guo, H.; Qin, J.; Luo, Y.; Zheng, L. Quality control of Vernonia patula Merr. based on multi-component content determination and principal component analysis. Chin. J. Hosp. Pharm. 2021, 41, 135–138. [Google Scholar]
- Zhang, L.; Huang, Z.; Lin, Z.; Zhong, X.; Cheng, R.; Huang, Z. Microscopic identification characteristics and HPLC fingerprint analysis of Patrinia villosa and its counterfeit products. Chin. Tradit. Herbal. Drugs 2024, 55, 2066–2076. [Google Scholar]
- Li, H.; Li, F.; Gao, B.; Guo, J.; Zhang, Y.; Jiang, G.; Yin, X. A comparative study on the HPLC fingerprint and chemical pattern recognition of Corydalis flavescens and its related medicinal herbs. Chin. Tradit. Herbal. Drugs 2024, 55, 1709–1716. [Google Scholar]
- Zheng, C.; Li, W.; Yao, Y.; Zhou, Y. Quality evaluation of Atractylodis macrocephalae rhizoma based on combinative method of HPLC fingerprint, quantitative analysis of multi-components and chemical pattern recognition analysis. Molecules 2021, 26, 7124. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Qian, Z.; Liao, G.; Zeng, J.; Huang, D.; Liu, Q.; Xie, X. HPLC coupled with chemical fingerprinting for multi-pattern recognition for identifying the authenticity of Clematidis armandii Caulis. J. Vis. Exp. 2022, 189. [Google Scholar]
- Zhang, H.J.; Li, H.R.; Feng, Z.Y.; Li, K.; Hu, Y.P.; Feng, S.X. Comparative study on HPLC fingerprints between crude and processed Ligustri lucidi Fructus. China J. Chin. Mater. Med. 2020, 45, 3871–3876. [Google Scholar]
- Wu, L.; Liang, W.; Chen, W.; Li, S.; Cui, Y.; Qi, Q.; Zhang, L. Screening and analysis of the marker components in Ganoderma lucidum by HPLC and HPLC-MSn with the aid of chemometrics. Molecules 2017, 22, 584. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhao, X.; Qiu, Z.; Sun, L.; Deng, Y.; Ren, X.; Mou, J. Comparative investigation of the stems, leaves, flowers, and roots of Centipeda minima based on fingerprinting-multivariate classification techniques. J. AOAC Int. 2022, 105, 934–940. [Google Scholar] [CrossRef] [PubMed]



| No. | 1 | 2 | 3 | 4 | 5 | 6 | RSD% |
|---|---|---|---|---|---|---|---|
| Peak 1 | 0.1044 | 0.1041 | 0.1040 | 0.1041 | 0.1041 | 0.1041 | 0.12 |
| Peak 2 | 0.2469 | 0.2455 | 0.2453 | 0.2455 | 0.2454 | 0.2454 | 0.23 |
| Peak 4 | 0.3491 | 0.3483 | 0.3483 | 0.3483 | 0.3483 | 0.3480 | 0.11 |
| Peak 5 | 0.3713 | 0.3704 | 0.3705 | 0.3706 | 0.3706 | 0.3703 | 0.090 |
| Peak 6 | 0.4500 | 0.4494 | 0.4495 | 0.4496 | 0.4496 | 0.4493 | 0.050 |
| Peak 8 | 0.6325 | 0.6322 | 0.6319 | 0.6321 | 0.6320 | 0.6320 | 0.030 |
| Peak 15 | 0.8117 | 0.8118 | 0.8118 | 0.8120 | 0.8118 | 0.8118 | 0.010 |
| Peak 16 | 0.8848 | 0.8848 | 0.8848 | 0.8848 | 0.8848 | 0.8848 | 0.00 |
| Peak 17 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.020 |
| Peak 21 | 1.4148 | 1.4139 | 1.4135 | 1.4139 | 1.4135 | 1.4133 | 0.030 |
| No. | 1 | 2 | 3 | 4 | 5 | 6 | RSD% |
|---|---|---|---|---|---|---|---|
| Peak 1 | 0.0066 | 0.0062 | 0.0062 | 0.0062 | 0.0062 | 0.0062 | 2.6 |
| Peak 2 | 0.0503 | 0.0512 | 0.0513 | 0.0513 | 0.0512 | 0.0513 | 0.71 |
| Peak 4 | 0.0207 | 0.0206 | 0.0199 | 0.0219 | 0.0208 | 0.0208 | 2.8 |
| Peak 5 | 0.2156 | 0.2163 | 0.2160 | 0.2167 | 0.2162 | 0.2164 | 0.15 |
| Peak 6 | 0.0675 | 0.0676 | 0.0676 | 0.0678 | 0.0677 | 0.0679 | 0.17 |
| Peak 8 | 0.0463 | 0.0462 | 0.0465 | 0.0467 | 0.0469 | 0.0466 | 0.51 |
| Peak 15 | 0.0410 | 0.0383 | 0.0387 | 0.0392 | 0.0388 | 0.0408 | 2.6 |
| Peak 16 | 0.1962 | 0.1962 | 0.1968 | 0.1969 | 0.1968 | 0.1970 | 0.17 |
| Peak 17 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.00 |
| Peak 21 | 0.2468 | 0.2464 | 0.2465 | 0.2471 | 0.2476 | 0.2475 | 0.19 |
| No. | 1 | 2 | 3 | 4 | 5 | 6 | RSD% |
|---|---|---|---|---|---|---|---|
| Peak 1 | 0.1091 | 0.1090 | 0.1091 | 0.1090 | 0.1087 | 0.1087 | 0.17 |
| Peak 2 | 0.1202 | 0.1201 | 0.1201 | 0.1201 | 0.1198 | 0.1198 | 0.12 |
| Peak 4 | 0.2773 | 0.2770 | 0.2773 | 0.2770 | 0.2771 | 0.2769 | 0.060 |
| Peak 5 | 0.3931 | 0.3926 | 0.3928 | 0.3929 | 0.3933 | 0.3928 | 0.050 |
| Peak 6 | 0.4179 | 0.4175 | 0.4177 | 0.4177 | 0.4182 | 0.4180 | 0.060 |
| Peak 8 | 0.5118 | 0.5117 | 0.5116 | 0.5117 | 0.5098 | 0.5095 | 0.19 |
| Peak 15 | 0.8061 | 0.8066 | 0.8065 | 0.8064 | 0.8029 | 0.8037 | 0.18 |
| Peak 16 | 0.9177 | 0.9179 | 0.9179 | 0.9177 | 0.9175 | 0.9177 | 0.010 |
| Peak 17 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.00 |
| Peak 21 | 1.1269 | 1.1269 | 1.1273 | 1.1270 | 1.1282 | 1.1277 | 0.040 |
| No. | 1 | 2 | 3 | 4 | 5 | 6 | RSD% |
|---|---|---|---|---|---|---|---|
| Peak 1 | 0.0270 | 0.0265 | 0.0269 | 0.0267 | 0.0238 | 0.0235 | 0.77 |
| Peak 2 | 0.0448 | 0.0440 | 0.0445 | 0.0443 | 0.0470 | 0.0462 | 0.80 |
| Peak 4 | 0.0250 | 0.0243 | 0.0248 | 0.0252 | 0.0174 | 0.0188 | 2.1 |
| Peak 5 | 0.1740 | 0.1725 | 0.1744 | 0.1736 | 0.1706 | 0.1685 | 0.90 |
| Peak 6 | 0.0215 | 0.0227 | 0.0257 | 0.0295 | 0.0313 | 0.0331 | 2.8 |
| Peak 8 | 0.0303 | 0.0333 | 0.0316 | 0.0322 | 0.0332 | 0.0357 | 0.45 |
| Peak 15 | 0.0436 | 0.0437 | 0.0437 | 0.0435 | 0.0461 | 0.0484 | 1.1 |
| Peak 16 | 0.1979 | 0.1949 | 0.1977 | 0.1954 | 0.1910 | 0.1882 | 0.34 |
| Peak 17 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.00 |
| Peak 21 | 0.2869 | 0.2779 | 0.2781 | 0.2804 | 0.2631 | 0.2564 | 2.6 |
| No. | 1 | 2 | 3 | 4 | 5 | 6 | RSD% |
|---|---|---|---|---|---|---|---|
| Peak1 | 0.1018 | 0.1018 | 0.1017 | 0.1019 | 0.1019 | 0.1017 | 0.070 |
| Peak 2 | 0.1176 | 0.1176 | 0.1176 | 0.1177 | 0.1177 | 0.1174 | 0.080 |
| Peak 4 | 0.2778 | 0.2782 | 0.2774 | 0.2774 | 0.2778 | 0.2775 | 0.10 |
| Peak 5 | 0.3952 | 0.3959 | 0.3952 | 0.3947 | 0.3950 | 0.3950 | 0.090 |
| Peak 6 | 0.4191 | 0.4196 | 0.4190 | 0.4185 | 0.4185 | 0.4189 | 0.090 |
| Peak 8 | 0.5066 | 0.5050 | 0.5062 | 0.5060 | 0.5064 | 0.5065 | 0.10 |
| Peak 15 | 0.8030 | 0.8035 | 0.8042 | 0.8037 | 0.8040 | 0.8029 | 0.060 |
| Peak 16 | 0.9178 | 0.9174 | 0.9184 | 0.9179 | 0.9180 | 0.9179 | 0.030 |
| Peak 17 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.00 |
| Peak 21 | 1.132 | 1.132 | 1.133 | 1.133 | 1.133 | 1.132 | 0.060 |
| No. | 1 | 2 | 3 | 4 | 5 | 6 | RSD% |
|---|---|---|---|---|---|---|---|
| Peak1 | 0.0077 | 0.0077 | 0.0076 | 0.0076 | 0.0077 | 0.0076 | 0.70 |
| Peak 2 | 0.0519 | 0.0519 | 0.0517 | 0.0517 | 0.0524 | 0.0526 | 0.70 |
| Peak 4 | 0.0141 | 0.0145 | 0.0142 | 0.0145 | 0.0144 | 0.0150 | 2.0 |
| Peak 5 | 0.1783 | 0.1796 | 0.1805 | 0.1808 | 0.1833 | 0.1873 | 1.6 |
| Peak 6 | 0.0227 | 0.0239 | 0.0247 | 0.0242 | 0.0232 | 0.0240 | 2.7 |
| Peak 8 | 0.0331 | 0.0332 | 0.0328 | 0.0330 | 0.0339 | 0.0333 | 1.0 |
| Peak 15 | 0.0503 | 0.0505 | 0.0516 | 0.0512 | 0.0519 | 0.0515 | 1.1 |
| Peak 16 | 0.1833 | 0.1834 | 0.1852 | 0.1836 | 0.1864 | 0.1827 | 0.70 |
| Peak 17 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.00 |
| Peak 21 | 0.2674 | 0.2639 | 0.2713 | 0.2676 | 0.2710 | 0.2487 | 2.9 |
| Time (t/min) | Acetonitrile (%) | 0.2% Phosphoric Acid (%) |
|---|---|---|
| 0 | 10 | 90 |
| 14 | 20 | 80 |
| 25 | 22 | 78 |
| 27 | 25 | 75 |
| 40 | 29 | 71 |
| 56 | 45 | 55 |
| 80 | 75 | 25 |
| 85 | 75 | 25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Huang, L.; Zhu, J.; Lu, L.; Su, X.; Hou, X.; Xiao, Z. Identification of Vernonia patula Merr. and Its Similar Varieties Based on a Combination of HPLC Fingerprinting and Chemical Pattern Recognition. Molecules 2024, 29, 1517. https://doi.org/10.3390/molecules29071517
Liu W, Huang L, Zhu J, Lu L, Su X, Hou X, Xiao Z. Identification of Vernonia patula Merr. and Its Similar Varieties Based on a Combination of HPLC Fingerprinting and Chemical Pattern Recognition. Molecules. 2024; 29(7):1517. https://doi.org/10.3390/molecules29071517
Chicago/Turabian StyleLiu, Wen, Liyuan Huang, Jiashan Zhu, Liwen Lu, Xiaoling Su, Xiaotao Hou, and Zeen Xiao. 2024. "Identification of Vernonia patula Merr. and Its Similar Varieties Based on a Combination of HPLC Fingerprinting and Chemical Pattern Recognition" Molecules 29, no. 7: 1517. https://doi.org/10.3390/molecules29071517
APA StyleLiu, W., Huang, L., Zhu, J., Lu, L., Su, X., Hou, X., & Xiao, Z. (2024). Identification of Vernonia patula Merr. and Its Similar Varieties Based on a Combination of HPLC Fingerprinting and Chemical Pattern Recognition. Molecules, 29(7), 1517. https://doi.org/10.3390/molecules29071517






