Abstract
The farnesoid X receptor (FXR) has been recognized as a potential drug target for the treatment of non-alcoholic fatty liver disease (NAFLD). FXR agonists benefit NAFLD by modulating bile acid synthesis and transport, lipid metabolism, inflammation, and fibrosis pathways. However, there are still great challenges involved in developing safe and effective FXR agonists. To investigate the critical factors contributing to their activity on the FXR, 3D-QSAR molecular modeling was applied to a series of isoxazole derivatives, using comparative molecular field analysis (CoMFA (q2 = 0.664, r2 = 0.960, r2pred = 0.872)) and comparative molecular similarity indices analysis (CoMSIA (q2 = 0.706, r2 = 0.969, r2pred = 0.866)) models, which demonstrated strong predictive ability in our study. The contour maps generated from molecular modeling showed that the presence of hydrophobicity at the R2 group and electronegativity group at the R3 group in these compounds is crucial to their agonistic activity. A molecular dynamics (MD) simulation was carried out to further understand the binding modes and interactions between the FXR and its agonists in preclinical or clinical studies. The conformational motions of loops L: H1/H2 and L: H5/H6 in FXR–ligand binding domain (LBD) were crucial to the protein stability and agonistic activity of ligands. Hydrophobic interactions were formed between residues (such as LEU287, MET290, ALA291, HIS294, and VAL297) in helix H3 and ligands. In particular, our study found that residue ARG331 participated in salt bridges, and HIS447 participated in salt bridges and hydrogen bonds with ligands; these interactions were significant to protein–ligand binding. Eight new potent FXR agonists were designed according to our results, and their activities were predicted to be better than that of the first synthetic FXR agonist, GW4064.
1. Introduction
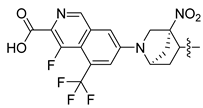
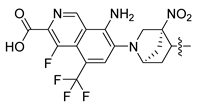
Non-alcoholic fatty liver disease (NAFLD) is becoming the most common liver disease in the world, with a global prevalence of about 25%. However, currently, there are no specific drugs available for the treatment of NAFLD [1,2]. The farnesoid X receptor (FXR) is a ligand-activated transcription factor, highly expressed in the liver and intestine, which has shown promising clinical results in the treatment of non-alcoholic steatohepatitis (NASH) [3,4,5]. Numerous FXR agonists that have been developed for the treatment of NAFLD are in preclinical and clinical trials at present. The most advanced FXR agonist in clinical development is obeticholic acid [6,7], which was approved by the US Food and Drug Administration (FDA) for the treatment primary biliary cholangitis (PBC) in 2016, and which is currently being investigated in clinical evaluation for the treatment of NAFLD. Obeticholic acid improves a variety of metabolic characteristics, such as liver steatosis, liver inflammation, and fibrosis [8]. However, the use of obeticholic acid was accompanied by some side effects during clinical trials, such as pruritus and drug-induced liver injury [9,10]. Various non-steroidal synthetic FXR agonists have been developed in order to avoid these adverse reactions, and a pose filter-based ensemble learning approach was also developed to discover promising lead compounds [11]; currently, the most abundant class of non-steroidal FXR agonists is that of trisubstituted isoxazole core compounds, derived from GW4064 [12,13]. Representative isoxazole derivatives, including cilofexor, tropifexor, PX20606, and LY2562175, are undergoing clinical trials for the treatment of NASH [14]. PX20606 from Phenex was the first chemical to reach Phase I testing, where the double-bond linker was replaced by a cyclopropyl moiety [15]. Gilead Sciences compound cilofexor and Lilly compound LY2562175 are clinical candidates for NASH [16,17]. These two candidates were obtained by replacing the terminal COOH-bearing aryl with a heteroaryl and by further changing the middle linker element. To date, many GW4064 derivatives have exhibited FXR-agonistic activity both in vivo and in vitro, while the structural requirements of these isoxazole derivatives, and the relationships between their structure and activities in detail for the development of efficient FXR agonists, are unclear [18,19].
The three-dimensional quantitative structure-activity relationship (3D-QSAR) is a computational method that involves constructing models (such as comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA)) that reflect the correlations between biological activities and molecular structures, as well as investigate the interaction characteristics between ligands and proteins using various statistical methods [20,21,22,23]. The advantages of the CoMFA model are that it can consider the internal interactions of molecules as well as the interactions between molecules and their environment, providing more accurate predictions of the structure–activity relationships of molecules. The advantage of the CoMSIA model is that it can consider molecular similarity, not only predicting the activity of molecules but also predicting properties such as hydrophilicity and lipophilicity. Both the CoMFA and CoMSIA models are important methods in the field of 3D-QSAR and can play a significant role in drug design and discovery.
Molecular dynamics (MD) simulations are always employed to investigate the dynamic structure and energetic information regarding protein and ligand interactions, which are important to understanding the structure–function relationship of the complex [24]. MD simulations can reveal the interactions, binding modes, and binding abilities between proteins and ligands, helping to understand the function of proteins and the activity of ligands. In addition, MD simulations can also predict the kinetic processes of protein–ligand binding, revealing their stability and dynamics.
The combination of 3D-QSAR and MD is regarded as an effective method through which to investigate the essence of the protein–ligand interaction and to guide the drug discovery and design processes. Hence, in the present work, we explored the SARs of the GW4064 derivatives with an isoxazole moiety by performing structure–agonistic activity relationship (SAR) analysis with an MD simulation, with the aim of identifying potential candidates and designing drugs with better activities through which to target the FXR.
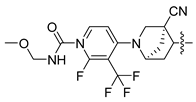
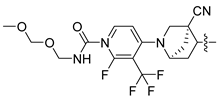
In this study, we demonstrate an effective molecular modeling of potent FXR agonists with an isoxazole moiety using 3D-QSAR and MD simulations. To investigate the structural requirements of GW4064 derivatives as FXR agonists, CoMFA and CoMSIA models were constructed to analyze the structures and experimental activities, and the activities of GW4064, trisubstituted isoxazole core agonists with different structures in clinical studies (cilofexor, LY2562175, and PX20606) (see Figure 1A below), and the novel designed compounds were predicted. Moreover, to reveal the dynamic binding process and find the crucial residues and conformational shifts involved in ligand binding, MD simulations were conducted and compared for the complex system activities of four agonists, and the tertiary structure of the FXR is shown in Figure 1B. Root mean square deviation (RMSD), root mean square fluctuation (RMSF), hydrogen bonding, and the radius of gyration of the proteins were evaluated, with the aim of determining the flexibility and stability of the FXR when bound to different ligands. To understand how isoxazole derivatives affect the overall dynamic characteristics of the FXR, the free energy landscape (FEL), principal component analysis (PCA), and porcupine plot methods were employed to demonstrate the movement of proteins toward stable conformations within the four complex systems. Finally, the binding free energy and per-residue energy decomposition calculations for these systems were systematically analyzed, with the aim of identifying the key residues of the interaction between the FXR and ligands. Altogether, our study explored the structure–activity relationship between the GW4064-derived agonists and the FXR through combining 3D-QSAR with MD simulation, providing new insights into FXR agonist development with better safety and efficiency.
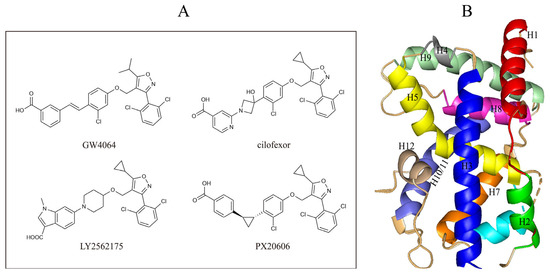
Figure 1.
(A) Clinical isoxazole-based farnesoid X receptor (FXR) agonists. (B) The tertiary structure of the FXR (PDB ID: 6HL1).
2. Results and Discussion
2.1. 3D-QSAR Model
To investigate the relationships between the structures and biological activities of the isoxazole compounds, a 3D-QSAR study was conducted using CoMFA and CoMSIA. The computational statistics of the CoMFA and CoMSIA models were constructed from a training set of 203 randomly selected compounds, with pEC50 values ranging from 6.0 to 10.0 (Supplementary Table S2). The training set superposition, based on the common skeleton of the template molecule 190, is shown in Figure 2, and the statistical parameters of the optimal models for the QSAR method are listed in Table 1. The best-constructed CoMFA model showed a cross-validation coefficient (q2) of 0.664, a non-cross-validation coefficient (r2) of 0.960, an F-test value of 581.48, a standard error of estimate (SEE) for cross-validation of 0.442, SEE for non-cross-validation of 0.152, SEE for the test set (SEEpred) of 0.261, and a prediction correlation coefficient (r2pred) of 0.872, demonstrating good predictive ability and statistical stability. The field contributions of the steric and electrostatic fields were 42.1% and 57.9%, respectively.
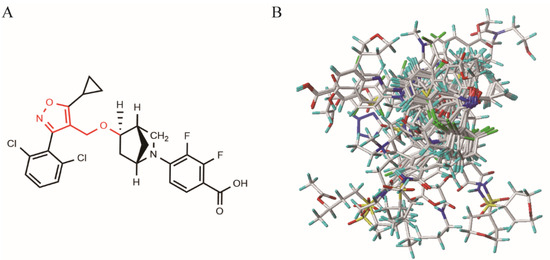
Figure 2.
(A) Structure of the template molecule 190. The common skeleton is displayed in red. (B) The superposition of ten high values of activity and ten low values of activity of the training set as representative compounds based on the common skeleton.

Table 1.
Statistical results of the CoMFA and CoMSIA models.
As compared to the CoMFA model, the CoMSIA model also calculates the hydrophobic (HF), hydrogen bond donor (HBD), and hydrogen bond acceptor (HBA) fields. For the CoMSIA study, the cross-validation coefficient was 0.706, the non-cross-validation coefficient was 0.969, the F-test value was 760.15, SEE for cross-validation was 0.413, SEE for non-cross-validation was 0.134, SEE for the test set was 0.266, and the prediction correlation coefficient was 0.866, suggesting good predictive abilities of the model (Table 1). The hydrogen bond donor field contributed the most to the molecular activity, with 30.3% (Table 1). The correlations between the predicted and experimental pEC50 values of the training and test sets for the CoMFA and CoMSIA models are shown in Figure 3. The activities of the four isoxazole FXR agonists under preclinical or clinical study were predicted using the QSAR model (Table 2), and they are ordered as follows: cilofexor > LY2562175 > PX20606 > GW4064. These observations are generally consistent with previous experimental results and need further verification [17,25,26,27].
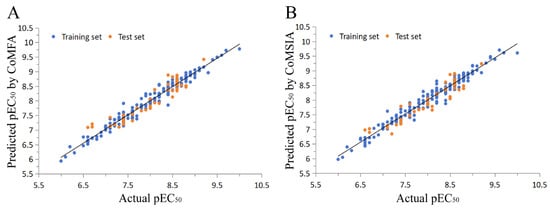
Figure 3.
Experimental and predicted pEC50 values for the training and test set compounds using (A) the CoMFA model and (B) the CoMSIA model.

Table 2.
Predicted pEC50 values of isoxazole-based FXR agonists under preclinical and clinical study.
2.2. 3D-QSAR Contour Map Analysis
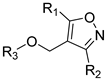
CoMFA and CoMSIA contour maps were generated in our study to explore the structural requirements of isoxazole-type FXR agonists. Compound 190 (Figure 4A) was chosen as the template molecule and is shown inside the field. The contours around the molecules present how the steric, electrostatic field, hydrogen bond acceptor, hydrogen bond donor, and hydrophobic interactions affect the agonistic activity. For the CoMFA model (Figure 4B), the yellow contours around the R1 group indicate that larger groups in this region may be detrimental to improving the agonistic activity of the compound. As can be seen from the figure, the large yellow outline around the R2 group suggests that the use of smaller groups in this region is beneficial for improving the agonist activity of the compound. These findings may explain the fact that the introduction of a larger group here of compounds 122 and 123 exhibited a lower pEC50 value. The CoMSIA steric contour map (Figure 4D) was similar to the CoMFA contour map, especially around the R3 group. The large green area surrounding the R3 group suggests that the introduction of bulky groups was favorable to FXR activity here, as in the cases of compound 87. Previous studies consistently reported that a bulky group at the R3 position could be a benefit to agonistic activity [16,17,28,29].
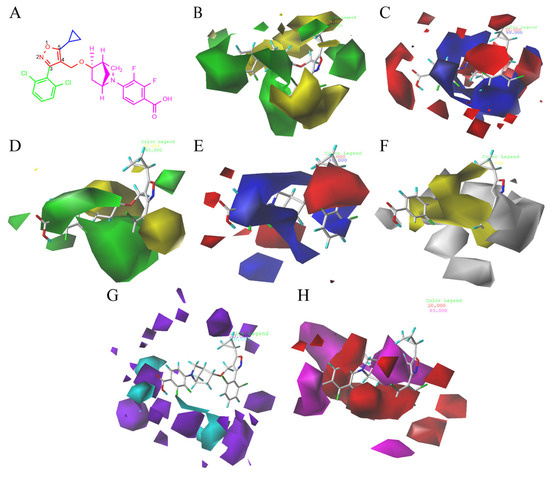
Figure 4.
CoMFA and CoMSIA contour maps of template compound 190. (A) The structure of compound 190. CoMFA contour maps: (B) favorable (green) and unfavorable (yellow) steric fields, (C) electropositive (blue) and electronegative (red) fields. CoMSIA contour maps: (D) favorable (green) and unfavorable (yellow) steric fields, (E) electropositive (blue) and electronegative (red) fields, (F) favorable (yellow) and unfavorable (white) hydrophobic fields, (G) favorable (cyan) and unfavorable (purple) hydrogen bond donor fields, and (H) favorable (magenta) and unfavorable (red) hydrogen bond acceptor fields.
In the 3D contour plots of the electrostatic fields in the CoMFA (Figure 4C) and CoMSIA (Figure 4E) models, the red coloring indicates that it is advantageous to introduce negatively charged functional groups, and the blue coloring indicates that it is advantageous to introduce positively charged functional groups at the indicated sites. A large blue contour was found around the benzene ring on the R2 group in both models, suggesting that the introduction of large electropositive substituents on the benzene ring will favor agonist activity and that the introduction of electronegativity groups is not conducive to agonist activity. As compared to compound 136, compound 135 bearing a fluorine atom which was electronegative at the R2 site exhibited weaker activity. In addition, the position of the azabicyclic group and the end of the R3 was occupied by the red contour map, suggesting that the addition of an electronegative group here may enhance agonist activity, as in compounds 86 and 87.
The 3D contour plot of the hydrophobic field in the CoMSIA model (Figure 4F) visualized the beneficial effects of hydrophobic groups (yellow) and hydrophilic groups (white) in the molecular structure. The large yellow contours around the R2 and R3 groups indicate that the introduction of hydrophobic groups in this position may improve biological activity. For instance, compound 126 exhibited better activity than compound 127 because of its hydrophobic group at the R2 site, halogen. At the same time, a white contour was located at the end of R3, suggesting that a hydrophilic substituent should be introduced in this position.
Figure 4G represents a contour map of the hydrogen bond donor field in the CoMSIA model, where cyan and purple contours refer to hydrogen bond donor-favored and -unfavored regions for activity, respectively. The purple contours located at the end of R3 indicate that the hydrogen bond donor group here may decrease biological activity. However, in the hydrogen bond donor field, the other color blocks are not explained due to their distance and small influence on the structure of the compounds.
In the contour plot of the hydrogen bond acceptor field in the CoMSIA model (Figure 4H), the favorable (magenta) and unfavorable (red) regions of the hydrogen bond acceptor field are visualized. The magenta contours at the end of R3 indicate that the hydrogen bond acceptor groups at these positions are important for the agonistic activity of the compound; this was exemplified by compounds 190 and 191. The presence of a red contour around the inter- and para-positions of the benzene ring at the R2 site indicates that the hydrogen bond receptor group may reduce biological activity.
2.3. Molecular Docking
To understand the differences in the mechanisms of FXR binding and agonistic activity of isoxazoles, the binding interactions of four isoxazole compounds with FXR were studied by molecular docking. The docking software is considered reliable when it is able to generate a pose that is very close to the native conformation in the crystal structure, with the root mean square deviation (RMSD) of less than 2 Å between the docked and crystal poses [30]. The overlap of the generated docked pose and the native conformation was as shown in Figure 5A; the RMSD between these two poses is 1.96 Å, proving that the docking protocol is well established.
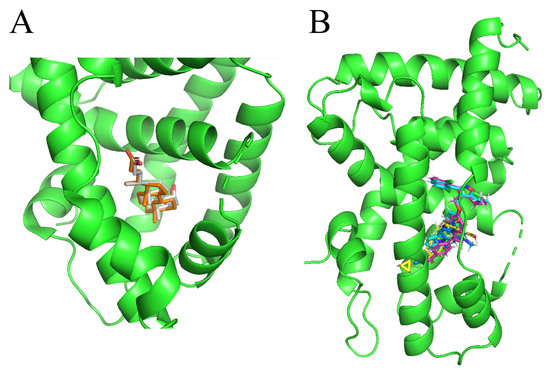
Figure 5.
(A) Overlap of the docked pose and native conformation from the crystal structure (PDB ID: 6HL1). The docked pose and native confirmation are colored orange and silver, respectively. (B) Overlay of the binding mode of FXR and four isoxazole ligands. The green indicates FXR, yellow is used for cilofexor, cyan for GW4064, blue for LY2562175, and magenta for PX20606.
As shown in Figure 5B, four compounds were docked to the active site of the protein using Discovery Studio 3.0 software. The conformational affinity of the molecule for binding to the protein at that point was determined by the docking score. The site and molecule conformation with the highest LibDockScore were determined for analysis, and the obtained results are presented in Table 3. As indicated in Table 3, all isoxazoles had higher docking scores with FXR-LBD in the range of 136.612 to 141.065, suggesting that all of them can bind to FXR-LBD as agonists. The docking scores of the ligands were in the following order: LY2562175 > cilofexor > GW4064 > PX20606.

Table 3.
Docking scores of FXR and isoxazole ligands.
2.4. Molecular Dynamic Simulations
To further investigate the mechanism of the binding between ligands and the FXR, analyze their global and local structural effects, and verify the stability of the docked pose of the four medicines, an MD simulation study was performed based on the four FXR agonists (GW4064, cilofexor, LY2562175, and PX20606) under preclinical or clinical study using GROMACS (University of Groningen). All selected protein-ligand complex structures were submitted to molecular dynamics analysis. Prior to MD simulations, the ligands were integrated into complexes with proteins based on the molecular docking using Discovery Studio 3.0.
2.4.1. Root Mean Square Deviation
The root mean square deviation (RMSD) of the protein backbone was tracked throughout the MD simulations (100 ns) to assess the stability of each binding system. In general, the RMSD values of all of the systems increased rapidly within 20 ns, and then reached states of equilibrium after 65 ns (Figure 6A). The average RMSD values for the FXR, FXR-GW4064, FXR-cilofexor, FXR-LY2562175, and FXR-PX20606 systems were 0.203 nm, 0.200 nm, 0.187 nm, 0.182 nm, and 0.195 nm, respectively (Figure 6B–E). The RMSD value of the FXR was higher than that of the FXR in complex systems, indicating a higher stability of the FXR with these ligands throughout the simulations. The tendency of stability in the four complex systems can be ranked as follows: FXR-cilofexor > FXR-LY2562175 > FXR-PX20606 > FXR-GW4064. The FXR-cilofexor system is the most stable one, showing the strongest binding interaction according to the comparison.

Figure 6.
Evolution of RMSDs during 100 ns MD simulations of complexes. (A) The RMSDs of five systems. (B) FXR-GW4064 (red), (C) FXR-cilofexor (green), (D) FXR-LY2562175 (blue), and (E) FXR-PX20606 (magenta) are compared with the RMSD of the FXR system (black).
2.4.2. Root Mean Square Fluctuation
Root mean square fluctuations (RMSFs) were measured in all systems throughout the 100 ns of MD simulation time and corresponded to the structural flexibilities of the FXR backbone atoms. The flexibilities of protein domains, and particularly loop structures close to the active site binding pocket, have earned significant attention regarding their impacts on ligand binding [31]. Binding to an FXR agonist increased the flexibility of the protein: loops L: H1/H2, L: H5/H6, L: H9/H10, and L: H11/H12, and helices H4, H7, and H11 were observed to fluctuate significantly in all systems (Figure 7A). Loop L: H1/H2 exhibited the highest flexibility in the FXR-GW4064 system, with an RMSF value reaching ~4.2 Å (Figure 7B); the other complex systems also showed high fluctuations in this region, with observations indicating that Loop L: H1/H2 significantly impacted protein flexibility. Loops L: H5/H6 and L: H9/H10 showed higher fluctuations in the FXR-LY2562175 system, with RMSF values of 3.1 Å and 2.9 Å, respectively (Figure 7D,F). Furthermore, upon comparing all of the FXR–ligand systems, helices H7 and H11 and loops L: H1/H2 and L: H9/H10 experienced lower fluctuations, but loop L: H11/H12 showed the highest fluctuation in the FXR-LY2562175 system (Figure 7B,E–H). These observations indicate that loop L: H11/H12 is an essential region in the FXR-LY2562175 system. In the FXR-PX20606 system, loops L: H5/H6 and L: H9/H10 and helices H4 and H7 showed high fluctuations. Particularly, loop L: H9/H10 showed the highest fluctuation, with an RMSF value of 4.5 Å, and helix H11 also showed a higher RMSF value, of ~2.5 Å (Figure 7C–G). It is worth noting that the complex with cilofexor showed a poor ability to increase FXR flexibility in loops L: H1/H2, L: H5/H6, and L: H9/H10 and helices H4, H7, and H11 (Figure 7B–G), which may indicate that the FXR is more dynamically stable with cilofexor. These observations are in accordance with our above-mentioned RMSD results. Consistently, ligands were reported to occupy the hydrophobic pocket of the FXR LBD and interact mainly with residues located on helices H3, H5, H7, H11, and H12 in a previous study [19].
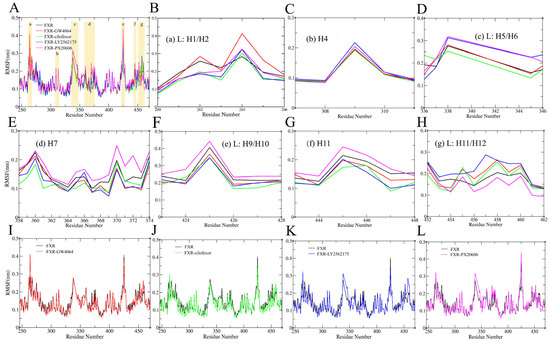
Figure 7.
RMSFs of the backbones in all FXR systems. (A) The RMSFs of five FXR systems. The region RMSF plots are shown for (B) loop L: H1/H2, (C) helix H4, (D) the loop between helices H5 and H6 (L: H5/H6), (E) helix H7, (F) the loop between H9 and H10 (L: H9/H10), (G) helix H11, and (H) the loop between helices H11 and H12 (L: H11/H12). FXR-GW4064 (I), FXR-cilofexor (J), FXR-LY2562175 (K), and FXR-PX20606 (L) were compared with the RMSF of the FXR system. FXR-GW4064, FXR-cilofexor, FXR-LY2562175 and FXR-PX20606 systems are represented with the colors red, green, blue, and magenta, respectively.
2.4.3. Hydrogen Bonding Analysis
The hydrogen bonding patterns of the complexes over the MD simulations were analyzed (Figure 8). Hydrogen bond interactions play a crucial role in the stability of proteins [32]. Our results showed that at least two hydrogen bonds were formed between the FXR and the ligands GW4064, cilofexor, and LY2562175 (Figure 8A–C); PX20606 and FXR could also form at least two hydrogen bonds in the later period of the simulation (Figure 8D). This stable interaction suggests a crucial role of hydrogen bonding in stabilizing these four complexes. Particularly, cilofexor exhibited a higher frequency of hydrogen bonding formation with the FXR protein compared to the other ligands, suggesting that it has a greater reliance on hydrogen bonding in the interaction with FXR. The FXR-cilofexor system was consistently found to be the most stable one among the complexes, according to RMSD analysis, in our study. These findings further validate the significance of hydrogen bonding in the stabilization of FXR–ligand complexes.
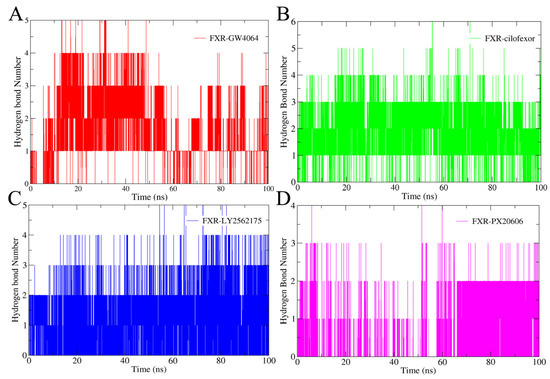
Figure 8.
The numbers of hydrogen bonds in the FXR-GW4064 system (A), the FXR-cilofexor system (B), the FXR-LY2562175 system (C), and the FXR-PX20606 system (D). The FXR-GW4064, FXR-cilofexor, FXR-LY2562175, and FXR-PX20606 systems are represented with the colors red, green, blue, and magenta, respectively.
2.4.4. Radius of Gyration Analysis
The radius of gyration (Rg) was employed to investigate protein structure compactness during MD simulations [33,34] (Figure 9). In our current study, the ligand–protein system was highly stable, with the Rg ranging between 1.77 and 1.83 nm. The Rg values of the four systems were observed to drop slightly, revealing more rigid complex structures during the simulation. The average Rg values of the FXR-GW4064, FXR-cilofexor, FXR-LY2562175, and FXR-PX20606 complexes were 1.788 nm, 1.798 nm, 1.792 nm, and 1.808 nm, respectively. Compared with the other complexes, the FXR-GW4064 complex showed a smaller Rg value, suggesting that it formed a more compact complex and a stronger interaction in the protein–ligand complexes. Meanwhile, the FXR system was the most compact, with an average Rg value of 1.780 nm, as compared to the other systems. This suggests that binding to the ligand increases the conformational changes to the protein and, thus, decreases protein compactness, which is consistent with the reported experimental results [35].
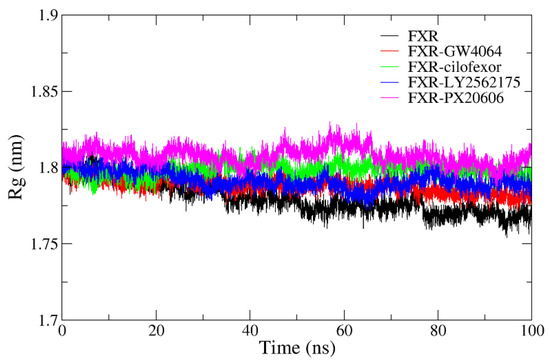
Figure 9.
Radius of gyration (Rg) values for the FXR (black), FXR-GW4064 (red), FXR-cilofexor (green), FXR-LY2562175 (blue) and FXR-PX20606 (magenta) systems.
2.4.5. Evaluation of Ligand Binding Affinities
Binding free energies, with neglected entropic contributions from the simulated systems, were calculated based on the molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method used in GROMACS (Table 4). The average values of binding free energies (∆Gbind) for the FXR with GW4064, cilofexor, LY2562175, and PX20606 were −36.71 kcal/mol, −49.83 kcal/mol, −43.46 kcal/mol, and −39.24 kcal/mol, respectively, suggesting that cilofexor possesses the strongest binding affinity with FXR, while GW4064 possesses the weakest. Smaller van der Waals force energy values represented larger contributions, and the van der Waals energies of the four complex systems (∆Evdw) were −64.69 kcal/mol, −78.87 kcal/mol, −71.11 kcal/mol, and −65.24 kcal/mol, respectively. Van der Waals energies showed much lower values than other energy terms, indicating it as the most significant contribution to the total binding free energy in our study. It is notable that the polar contributions (∆GPB) were positive in value, which means that they were not favorable for binding free energy.

Table 4.
MM-PBSA calculations of the simulated complex.
The obtained binding free energy was in the following order FXR-cilofexor > FXR-LY2562175 > FXR-PX20606 > FXR-GW4064, which is different from the order of molecular docking score: LY2562175 > cilofexor > GW4064 > PX20606. The inconsistent results may be because molecular docking misses the protein movement and shows activity in only one conformation [36,37]. Binding free energy was employed to assess the stabilities and driving forces of intermolecular interactions [31]. The analysis of the thermodynamic driving forces of ligand–protein binding suggests that they are key components to the selection and optimization of active compounds into drug candidates [38]. It is notable that the ordering of the values involved in MM-PBSA binding affinity and the predicted activity (pEC50) for the four agonists was consistent in our study, demonstrating that the more stable binding of the protein to the ligand leads to the higher FXR activity [39].
2.4.6. Per residue Energy Decomposition
In our study, an analysis of the free energy decomposition of the residues was carried out to investigate the individual energetic contributions of each residue involved in the stabilization of protein–ligand complexes (Figure 10). The residues showing contributions of −1 kcal/mol or above were considered hot-spot amino acids and were considered to contribute significantly to the stability of the complex. According to the cutoff of the residues, MET265, MET287, MET290, HIS294, VAL297, MET328, and ILE335 showed higher energy contributions in the FXR-GW4064 system. In the case of the FXR-cilofexor system, the residues MET265, LEU287, MET290, ALA291, HIS294, MET328, PHE329, ARG331, ILE335, and ILE352 showed the greatest contributions. MET265, LEU287, MET290, ALA291, HIS294, MET328, ILE335, and LEU348 showed high energy contributions in the FXR-LY2562175 system. Similarly, for the FXR-PX20606 system, the residues MET265, LEU287, MET290, ALA291, VAL297, MET328, and ILE335 exhibited high energy contributions. Among these, residues MET265, LEU287, MET290, MET328, and ILE335 all had contributions above −1 kcal/mol, indicating their important roles in the protein–ligand binding process. Notably, residues MET328 in the FXR-cilofexor system, MET265 and MET328 in the FXR-LY2562175 system, and MET265 in the FXR-PX20606 system showed energy contributions exceeding −2 kcal/mol. In addition, the largest number of amino acids contributed less than −1 kcal/mol was shown in the FXR-cilofexor system, an observation which is consistent with the most powerful binding free energy in the FXR-cilofexor system. The residue ARG331 showed an unfavorable contribution toward the total binding free energy in the FXR-GW4064 and FXR-PX20606 systems, while it contributed strongly to that in the FXR-cilofexor system; therefore, it may be a key residue for the effect on total binding free energy.
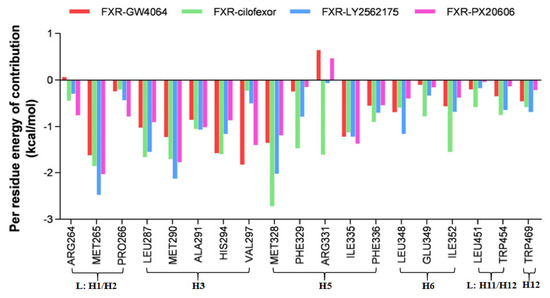
Figure 10.
Per residue energy contributions of the binding energy ΔGbind (kcal/mol) for the FXR-GW4064, FXR-cilofexor, FXR-LY2562175, and FXR-PX20606 systems. Amino acid residues with energy contributions < −1 kcal/mol are considered as key residues.
2.4.7. Protein–Ligand Interactions
To further explain ligand- and structure-based conformational relationships and to reveal the protein–ligand interactions, we visualized the interactions between residues and ligands at 95 ns and 100 ns in the simulations. Protein–ligand interactions were analyzed with PLIP online, and PyMOL 2.6.0 software was used for 3D visualization (Figure 11). As shown in Table 5, the binding of FXR to four ligands mainly occurred through hydrophobic interactions, which are considered to be the most primary forces that maintain the tertiary structure of proteins [40]. Residues LEU287, MET290, ALA291, HIS294, and VAL297, which are located on the flexible helix H3 site, showed high energy contributions in the four complexes. According to our per residue energy decomposition study, these observations demonstrate the important role of helix H3 in the LBD. At 100 ns in the simulation, hydrophobic interactions were observed between GW4064 and the residues ASN293, HIS294, and VAL297 in helix H3. Hydrophobic interactions were found between cilofexor and residues PHE284, LEU287, and MET290. Also, hydrophobic interactions were found between LY2562175 and the residues ALA291, HIS294, and VAL297, and there were hydrophobic interactions between PX20606 and residues LEU287, ASN293, and HIS294. These observations further demonstrated that helix H3 (SER279~LYS304) was essential for activating the FXR. Particularly, residue MET290 formed a hydrophobic interaction with the benzoic acid group of GW4064, as well as with the chlorophenyl groups of cilofexor and PX20606. The isopropyl group of residue VAL297 interacted hydrophobically with the pyridine group of cilofexor, as well as with the dichlorobenzene in GW4064 and LY2562175. These observations demonstrated the important roles of MET290 and VAL297 in protein–ligand interactions. In addition, we also found that helix H5 (ASP312~PHE336) played an important role in protein–ligand affinity, where residues MET328, ILE335, and PHE336 made remarkable contributions to binding free energies in the four systems (Figure 10). There were hydrogen bonding interactions and salt bridges between the proteins and ligands, in addition to the hydrophobic interactions. As shown in Figure 11, the carboxyl group (–COOH) of cilofexor formed a salt bridge with the terminal amino group (–NH2) of ARG331 and made strongly favorable contributions to binding free energy. However, the terminal amino group (–NH2) of residue ARG331 formed a hydrogen bond with the methoxy group of PX20606, revealing an unfavorable contribution towards the total binding free energy. The protein–ligand interaction analysis at 95 ns simulated conformations revealed that the hydrogen bonding between the imino group (=NH) of ARG331 and the oxygen atom in the isoxazole ring of GW4064 generated an unfavorable contribution to binding free energy. ARG331 may account for the loss in binding free energies in the FXR-GW4064 and FXR-PX20606 systems, but improved binding free energy in the FXR-cilofexor system. These results suggest that ARG331 may be a key residue on helix H5; the formation of the salt bridge with the carboxyl group of ligand generates a favorable contribution to binding free energy, while the formation of the hydrogen bond with the oxygen atom of the isoxazole ring and the methoxy group of the ligand is an unfavorable factor. Moreover, although without remarkable energy contribution, HIS447 may also be a crucial amino acid residue that affects activity, as HIS447 formed a hydrogen bond with the isoxazole nitrogen of cilofexor, and formed salt bridges with the carboxyl groups of LY2562175 and PX20606. It was consistent that the activation of HIS447~TRP469 has been reported to stabilize helix H12 in conformation [19]. Therefore, our results indicate that the salt bridges (with the terminal carboxyl groups) and the hydrogen bond (with the isoxazole nitrogen) formed by residue HIS447 would be significant to the activities of derivatives.
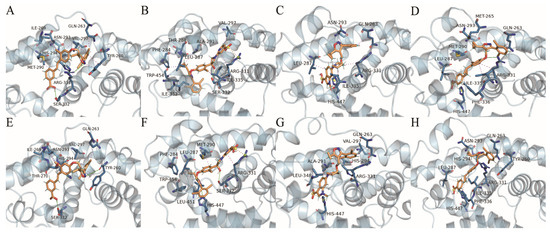
Figure 11.
(A–D) and (E–H) show protein–ligand interactions for GW4064, cilofexor, LY2562175, and PX20606 with the FXR (blue) at 95 ns and 100 ns in the simulation, respectively. The bonds of residues and ligands are displayed as blue and orange stick structures, respectively. Hydrogen bonds are displayed as solid green lines. Salt bridges and hydrophobic interactions are displayed as orange and magenta dashes, respectively.

Table 5.
Molecular interactions of FXR–ligand complexes.
2.4.8. Conformational Flexibility in the Ligand Binding Domain of the FXR
To identify protein backbone conformation shifts and monitor the effects of the four FXR agonists on the correlated motion of the FXR, an intercorrelation matrix analysis was conducted to evaluate the kinetic states of the FXR (Figure 12A–D) [41]. Protein residues were identified for correlated and anticorrelated motions using principal component analysis (PCA). The covariance matrix was diagonalized to obtain the principal components (PCs), and the eigenvalues and eigenvectors were solved to determine the directions and magnitudes of the motions. The eigenvectors show the directions of the motions, while the eigenvalues indicate their magnitudes along the direction. The output of this analysis is provided via a dynamic cross-correlation matrix (DCCM). DCCM analysis is a prominent method for analyzing the trajectory of MD simulations, with the aim of generating a matrix of cross-correlations, indicating correlations and anticorrelations between atoms during displacement or fluctuations. Correlations closer to +1 (inky blue) indicate that the residues were moving in the same direction, while correlations closer to −1 (yellowish) indicate that the residues were moving in opposite directions [42]. More yellowish blocks are shown in Figure 12B–D, suggesting more negative correlations in the motions of the residues in the FXR-cilofexor, FXR-LY2562175, and FXR-PX20606 systems and fewer negative correlations in the motions of the residues in the FXR-GW4064 system. The lowest anticorrelation in the FXR-GW4064 complex indicates less stability for the FXR-GW4064 system. These observations may help to explain why the FXR activation activity and binding free energy of GW4064 are lower than those of cilofexor, LY2562175, and PX20606.
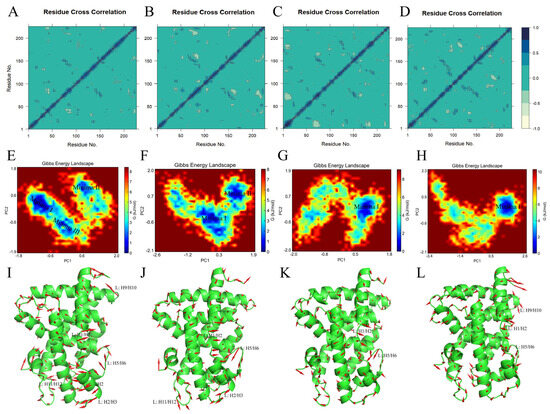
Figure 12.
Essential dynamics of FXR–LBD systems. (A–D) Dynamic cross–correlation matrices (DCCMs) for the FXR–GW4064, FXR–cilofexor, FXR–LY2562175, and FXR–PX20606 systems. In the DCCMs, inky blue coloring represents a positive correlation, and yellowish coloring represents a negative correlation; the closer the color is to inky blue, the closer the value is to 1; the closer the color is to yellowish, the closer the value is to −1. (E–H) Free energy landscapes (FELs) of each FXR–ligand system, respective to the aforementioned order. The blue areas identify the low-energy basins (minima). (I–L) Porcupine plots of the different FXR–ligand systems, respectively. The red arrows represent the trends of the protein from its initial conformation to its most stable one. The direction of motion is represented by the arrow, and its length indicates the movement strength.
To further investigate the effects of the four FXR agonists on the conformational space of the protein, their free energy landscapes (FELs) were analyzed. The FELs were calculated using the first two principal components, PC1 and PC2, as the coordinates with which to distinguish the conformational states of different complex systems. This method visualized the stability of the FXR and the conformational states of the four ligands by mapping out the probabilities of energy distribution for one or more variables of the protein system and identifying the low-energy basins (minima) [41,43]. The smaller and more concentrated blue areas in an FEL indicate a more stable ligand–protein complex system. Figure 12E–H depict the 2D plots of the FELs along the two primary components, PC1 versus PC2, of the FXR-GW4064, FXR-cilofexor, FXR-LY2562175, and FXR-PX20606 systems. The low- or high-energy conformations are specified with blue or red coloring in the 2D plots, respectively. Specifically, a single dominant minimum energy basin was observed for both the FXR-LY2562175 and the FXR-PX20606 complexes, while, upon the introduction of cilofexor, two major conformational regions were observed. In comparison, the conformational space of the FXR bound with GW4064 involved three energy minima, suggesting that GW4064 possesses the lowest ability to stabilize the FXR. To identify locations with high atomic fluctuations and their directionalities in the MD-simulated systems, we extracted the conformation of the deepest minima I, for each energy basin, and plotted porcupine plots using PyMOL 2.6.0 software. The basins in the FXR-GW4064 system correspond to the conformational changes in loops L: H1/H2, L: H2/H3, L: H5/H6, and L: H9/H10 and helix H2 (Figure 12I). According to the direction and magnitude of the porcupine vector, the highest fluctuation was found in loop L: H1/H2, which showed anticorrelated movement to loops L: H2/H3, L: H5/H6, and L: H9/H10 and helix H2. This indicates that the regions constituting the GW4064 binding pocket are flexible and move inward, resulting in reduced gyration and binding pocket volume, a result which is consistent with the findings previously discussed, regarding the drop in Rg. The basins in the FXR-cilofexor complex corresponded to the conformational changes in loops L: H1/H2, L: H2/H3, L: H5/H6, and L: H11/H12 (Figure 12J). In the porcupine plots, loop L: H1/H2 shows anticorrelated movement with loops L: H2/H3 and L: H5/H6, and loop L: H5/H6 showed the highest fluctuation. A slight inward movement in loop L: H11/H12 was observed in the FXR-LY2562175 system, where we observed the lowest basin (Minima I) (Figure 12K). Upon comparing the conformational changes, we found significant changes in loop L: H5/H6. The porcupine plot shows slight outward movements in loop L: H1/H2; L: H1/H2 shows anticorrelated motion with loop L: H5/H6. The FXR-PX20606 system captured slight fluctuation in loops L: H1/H2, L: H5/H6, and L: H9/H10 (Figure 12L). A slight inward movement in loop L: H1/H2 was observed. Loops L: H5/H6 and L: H9/H10 showed anticorrelated movement with L: H1/H2. In comparison, the FXR-GW4064 system showed a larger conformational deviation than the FXR-cilofexor, FXR-LY2562175, and FXR-PX20606 systems did. The region with the greatest flexibility in binding with GW4064 was L: H1/H2, which exhibited an outward movement trend, enlarging the cavity of the FXR-LBD complex and leading to a decrease in its stability. These observations may indicate that stability between loop L: H1/H2 and loop L: H5/H6 in the LBD were essential for its activation.
2.5. Summary of Structure–Activity Relationships
In the present work, 3D-QSAR analysis was performed using CoMFA and CoMSIA models and MD simulations, which provided a theoretical basis for the interactions between ligands and agonistic activities (Figure 13). The introduction of a hydrophobic group at the terminal position of the R2 site would enhance the activities of such agonists. However, the terminus of the R2 site was an unfavorable region for a hydrogen bond acceptor group, and the introduction of bulky and electronegative groups at the R2 position would result in a loss of bioactivity. A bulky group at the R3 site was required for improving the agonistic activity of isoxazole-type FXR agonists. Meanwhile, the introduction of a hydrogen bond acceptor group and enhancement of electronegativity at the terminal carboxyl position of R3 would increase the bioactivity. Alternatively, the hydrophobic group in the middle of R3 is also beneficial to bioactivity. Moreover, according to the MD analysis, hydrophobic interactions that formed between the ligand and the residues LEU287, MET290, ALA291, HIS294, and VAL297 were crucial for agonist activity. A protein–ligand interaction analysis demonstrated that residue MET290 formed hydrophobic interactions with benzoic acid groups, and the isopropyl group of residue VAL297 interacted hydrophobically with benzoic acid groups and with dichlorobenzene. In particular, residue HIS447, which formed salt bridges with the terminal carboxyl group and a hydrogen bond with the isoxazole nitrogen, would benefit agonistic activity. Additionally, ARG331 was a key residue of the LBD that contributed favorably to the salt bridge bound to the carboxyl group of the ligand. However, ARG331 had an unfavorable effect on the formation of hydrogen bonds with isoxazole. These results provided clues for the further modification of isoxazole compounds for better FXR agonists.
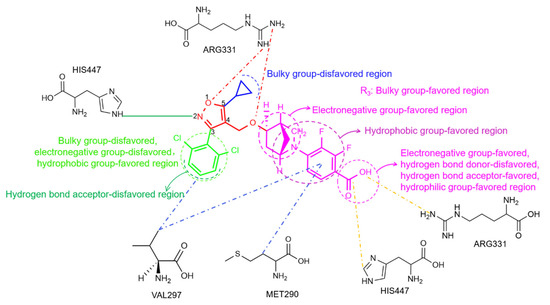
Figure 13.
Structure–activity relationship results from the present study. The common skeleton, and its R1, R2, and R3 sites are shown in red, blue, green, and magenta, respectively. Unfavorable interactions, salt bridges, and hydrophobic interactions are shown as red, orange, and blue dashes, respectively. Hydrogen bonds are shown as solid green lines.
2.6. Design of Novel Compound
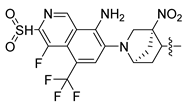
The SAR analysis provided useful information on structural features for the improvement in FXR agonist activity. Hence, we designed eight new compounds containing isoxazole cores as novel FXR agonists (compounds 246–257) by modifying the structure of the template molecule 190. When designing drugs, we avoided making significant changes to R1 and R2, as both of them are not suitable locations for introducing bulky groups. Instead, we focused on modifying R3. Compounds 247–253 were designed to increase the electronegativity and hydrophobicity of R3 by replacing a fluorine atom (–F) with trifluoromethyl (–CF3). We designed compounds 246–251 by introducing bulky groups (quinoline and naphthalene) at the R3 site. We also altered the hydrogen bond acceptor group–carboxyl group, as shown in compounds 249, 252, and 253. Furthermore, we added strong electron-withdrawing groups at the azabicyclic group to increase the electronegativity of the compounds. Meanwhile, cyano (–CN) and nitro groups (–NO2) were added to compounds 246–253. In addition, we added hydrogen bond acceptor compounds at the R3 group to form hydrophobic interactions and salt bridges with ARG331. The pEC50 values of the newly designed compounds were predicted using the CoMFA and CoMSIA models (Table 6). All of the designed compounds showed better predicted activity than compound 190 did, and these predictions are consistent with the conclusions of the SAR analysis in this study.

Table 6.
Chemical structures and predicted pEC50 values (Pred.) of the newly designed molecules.
3. Materials and Methods
3.1. Database Selection and Biological Activity
To construct the pharmacophore model, a series of isoxazole compounds acting as FXR agonists were chosen from previously published studies (US11718619, US10793568). The experimental data obtained were all measured under the same experimental conditions, and the experimental data (EC50) were measured using the fluorescent resonance energy transfer (FRET) method [44]. The FRET signal is measured as the ratio of 520 nm/495 nm emission following excitation at 340 nm. We transformed the EC50 values into –logEC50 (pEC50) to reduce the skewed data distribution’s adverse effects and stabilize the variance [45]. Based on their structural diversity and activities, 203 compounds were randomly selected as the training set, and 42 molecules as the test set, from the 245 molecules (all sets contained template molecule) to limit the error caused by confounding factors [45]. Supplementary Table S1 lists their structures and bioactivity values, and the test set is indicated with a superscript asterisk (“*”). All compounds were obtained from PubChem database and prepared using SYBYL-X 2.0 software.
3.2. Molecular Optimization and Alignment
The structures of the isoxazole compounds were optimized through energy minimization by incorporating Gasteiger–Hückel charges and a Tripos force field [46,47]. The maximum iteration parameter was set to 10,000 times, the energy convergence was limited to 0.05 kcal/mol, and the other values were set to default [48]. Molecular energy optimization was performed on the molecules in the training set to obtain the lowest energy conformation for each molecule, as the quality of molecular alignment is an important factor in 3D-QSAR modeling [49]. For 3D-QSAR studies, the common skeleton was used for compound alignment. In order to maximize the comparison, the most active compound 190 was selected as the template molecule for comparison (as shown in Figure 2A).
3.3. Construction of the 3D-QSAR Model
The aligned training set molecules were placed in a 3D lattice box with a grid spacing of 2 Å. Then, CoMFA steric (S) and electrostatic (E) fields between the aligned compounds, and a carbon atom probe with sp3 hybridization and a charge of +1.0, were computed using Coulombic and Lennard–Jones potentials, respectively [50]. The CoMSIA models were created by using the same molecular alignments that were used for the CoMFA ones. The aligned compounds were placed in a 3D lattice box, with a grid spacing of 2.0 Å. For the CoMSIA model, in addition to steric and electrostatic fields, hydrophobic (H), as well as hydrogen bond donor (D) and acceptor (A), descriptors were calculated by using the same lattice box as for the CoMFA model and a sp3 carbon probe with a +1.0 charge [51]. Partial least squares (PLS) analysis was used to construct a linear correlation between the CoMFA/CoMSIA descriptors as independent variables and the biological activity (pEC50) as dependent variables [52]. The training set was cross-validated using the leave-one-out (LOO) procedure to obtain the cross-validation coefficient (q2) and the optimal component number (N). A regression analysis was performed with non-cross-validation to obtain the non-cross-validation coefficients (r2), the significance test value (F), and the standard error of estimate (SEE) [53]. The parameters q2, F, r2, and SEE are the main indices used to determine the quality of 3D-QSAR models [54,55].To ensure that the results of the 3D-QSAR model were more informative, the following requirements for these evaluation parameters must be met: q2 > 0.5, r2 > 0.9, and F > 100 [48,56].
3.4. External Validation of the CoMFA and CoMSIA Models
The predictive capabilities of the developed models were then evaluated using the test set of 42 compounds. After these compounds were aligned to the template, the pEC50 values of all compounds were predicted via the developed CoMFA and CoMSIA models. Thus, the predictive correlation coefficients (r2pred) between the predicted and experimental activities of the test set were obtained. In general, r2pred > 0.5 is necessary for a good model [57]. The predictive correlation coefficients for the predictive performance of the PLS model were measured via the following equation [47,58,59,60]:
where SD is the sum of squared deviations from the mean of the observed activities, and Press is the predicted sum of squares, which is the sum of the squares of the difference between predicted and experimental activity.
r2pred = (SD − Press)/SD
3.5. Molecular Docking
Molecular docking was performed in Discovery Studio 3.0 software. The crystal structure of the FXR (PDB ID: 6HL1) with a high resolution at 1.60 Å was acquired from the Protein Data Bank (PDB) database [61]. The 3D structures of compounds were obtained from the PubChem database. Prior to molecular docking, the FXR crystal structure was cleaned, and water and original ligands were removed. The protein was pre-processed by CHARMm force field using Discovery Studio 3.0, and the ligands GW4064, cilofexor, LY2562175, and PX20606 were optimized using the MMFF force field. Then, the optimized ligands were docked into the corresponding active pockets where the original ligands were located using LibDock module, respectively. The poses obtained from docking were ranked according to the LibDock score, and the complexes with the highest LibDock score were selected as starting poses to perform MD simulation [62].
3.6. Molecular Dynamic Simulations
3.6.1. System Preparation
GROMACS 2020.4 software was used to perform 100 ns molecular dynamics (MD) simulations for each complex.
Here, we explored five systems—no ligand in the FXR system, the FXR-GW4064 system, the FXR-cilofexor system, the FXR-LY2562175 system, and the FXR-PX20606 system—to identify the motion mechanisms of the FXR with its binding partners and determine their binding sites. A GAFF force field was employed to parameterize the ligands, while an AMBER ff99SB force field was employed for the FXR structure [63,64]. Each complex was then solvated by a dodecahedral box of TIP3P waters, with the solute 10 Å away from the water box boundary [65]. After the solvation, each system was neutralized with the addition of counter ions (NaCl), followed by the steepest descent method, in order to minimize the system every 1000 steps. Afterward, each system was heated from 0 to 300 K in 100 ps in the NVT ensemble, and then the systems were equilibrated for 100 ps at 300 K and 1 atm pressure in the NPT ensemble. Moreover, the particle mesh Ewald (PME) was utilized to derive long-range electrostatic interactions [66]. The bonds involving hydrogen were constrained by applying the SHAKE algorithm [67], allowing for a time step of about 2 fs, and periodic boundary conditions were applied in all directions. The systems were subjected to a 100 ns MD simulation at constant pressure and temperature. To assess the stability and availability of the MD, triplicate all-atom MD simulations were performed using GROMACS 2020.4 [35].
3.6.2. MD Trajectory Analysis
The root mean square deviation (RMSD), root mean square fluctuation (RMSF), hydrogen bonding analysis, and radius of gyration (Rg) were used to analyze the MD trajectory. The RMSD was used to analyze global conformational dynamics and the stabilities of proteins [42]. RMSF was used to describe the fluctuation of each amino acid residue [68]. Rg was used to characterize the tightness of each system [69]. Hydrogen bonds are determined by an acceptor–donor distance of <3.5 Å and an acceptor–H–donor angle of >135° [70]. The xmgrace program (version 5.1.22) was used to generate two-dimensional (2D) plots.
3.6.3. Binding Free Energy Calculations
The molecular mechanics Poisson–Boltzmann Surface Area (MM-PBSA) method was used to calculate the binding free energy of protein–ligand complexes in each system. In this study, calculations using MM-PBSA were performed to explore the binding affinity of protein–ligand complexes and systematically compare the binding free energies of each system [71]. Meanwhile, the MM-PBSA method can decompose the total binding free energy to confirm key residues that contribute significantly to binding. The last 10 ns of the MD trajectories were selected for the calculation of binding free energy (G). The specific calculation formulas are as follows [72,73]:
where G is the total binding free energy, and Gcomplex, Gprotein, and Gligand are the free energies of the complex, protein, and ligand, respectively. Egas, the gas phase interaction energy, is evaluated as the sum of the van der Waals (Evdw) and electrostatic energies (Eele). The solvation free energy (Gsol) is divided into polar (EPB) and nonpolar (ESA) components, in which the polar solvation energy was calculated using the Poisson–Boltzmann equation, and the nonpolar solvation energy was computed based on the solvent-accessible surface area (SASA), with γ set to the default value [74]. The configuration entropy (TΔS) was attributed to the contributions of entropy temperature and S entropy [75].
ΔG = Gcomplex − (Gprotein + Gligand)
G = Egas + Gsol − TΔS
Egas = Evdw + Eele
Gsol = EPB + ESA
GSA = γ·SASA
3.6.4. Protein–Ligand Interaction Analysis
Ligand binding to protein is a dynamic process involving numerous structural changes in proteins and dynamic processes of ligand binding. Thus, the conformations at 95 ns and 100 ns were captured in order to explore common crucial residues, which was performed using the module gmx trjconv command. Contact residues of the complexes’ structures were analyzed using the Protein–Ligand Interaction Profiler (PLIP), and then the results of image plotting were processed using PyMOL software.
3.6.5. Principal Component Analysis and Dynamic Cross-Correlation Matrix
The PCA method utilizes the covariance matrix to extract the most important functional motions from the protein dynamics coordinates. The covariance matrix includes the atomic fluctuations of each residue’s C-alpha and provides the orthogonal eigenvectors and their corresponding eigenvalues [75]. The eigenvalues represent the amplitudes of the motions, while eigenvectors describe the directions of the motions, which are then used to calculate principal components (PCs). The covariance matrix shows the displacements of the C-alpha atoms relative to their average positions; the following equation was used to calculate the covariance matrix element (Cij):
where xi represents the position of the ith carbon alpha atom in Cartesian coordinates, and the brackets represent the average of all structures that were sampled throughout the MD trajectory. Positive Cij values show correlated motion between residues i and j. Negative values of Cij show anticorrelated motion between residues i and j. The parentheses show the average of all structures sampled on the MD trajectory. The dynamic cross-correlation matrix (DCCM) was used to identify the cross-correlation shifts of the backbone atoms and to show protein backbone conformation shifts, and was performed using RStudio software (version 4.3.1) and the Bio3d application [75,76,77]. The cross-correlation coefficient value fluctuated from −1 to 1, and the resulting dynamic cross-correlation matrices displayed amino acids that have both positive and negative effects.
Cij = <(xi − <xi>)(xj − <xj>)>
3.6.6. Free Energy Landscape
The PCA method was employed to calculate eigenvectors and eigenvalues and their projection, along with the first two principal components. The FEL was plotted based on the first two principal components (PC1 and PC2), which were attained using the gmx sham module of GROMACS software and the xpm2txt.py tool [78]. Porcupine plotting allowed visualization of the directions and extents of primary motions exhibited by the backbone residues of different ligands and the FXR. The trajectory of each system, from its initial motion towards the lowest-energy conformation of the FEL, was analyzed in a porcupine plot analysis. All possible atomic motions were visualized as mode vector graphs. The porcupine plot was constructed using a Python script and PyMOL software.
4. Conclusions
We carried out a molecular modeling study on isoxazole derivatives as FXR agonists using 3D-QSAR and MD simulations. The CoMFA and CoMSIA models, with good predictive ability, were constructed for the 3D-QSAR study; the obtained contour maps depicted the structural requirements of isoxazole derivatives serving as FXR agonists, thus promoting the development of derivatives with improved agonistic activities. MD simulations of GW4064 and three isoxazole-derived FXR agonists that have entered clinical studies were investigated in our study. GW4064 exhibited the least stability in the LBD of the FXR, showing the fewest hydrogen bond interactions and the lowest binding free energy, while cilofexor demonstrated the opposite. The stability of the L: H1/H2 and L: H5/H6 loops seemed to correlate with agonistic activity in our study. A free energy decomposition analysis revealed that the residues MET265, LEU287, MET290, MET328, and ILE335 made crucial contributions to ligand binding. In addition, according to the analysis of conformational relationships between ligands and structures, the interactions in which ligands connected with residues MET290 and VAL297 in helix H3 via hydrophobic interactions would be favorable to protein–ligand affinity. Residue ARG331 of helix H5 engaged in salt bridges, and residue HIS447 participated in salt bridges and hydrogen bonding; these two residues were determined as the key residues for agonistic activity in our study. Eight new potential FXR agonists, with better predicted agonistic activity, were designed based on a structure–activity relationship analysis. In summary, these structural features contribute to the understanding of drug hotspots for the FXR and provide promising avenues for the discovery of novel FXR agonists, thus providing potential therapeutic strategies for NASH.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29061210/s1, Table S1: The structures of dataset compounds; Table S2: The actual pEC50, predicted pEC50 (Pred.), and the residuals (res.) of dataset compounds.
Author Contributions
Conceptualization, Q.L. and D.Y.; methodology, Y.Y. and H.S.; software, K.Y. and Z.L.; validation, Q.L. and D.Y.; formal analysis, K.Y. and Q.L.; investigation, Y.Y., H.S. and K.Y.; resources, Q.L., Z.L. and D.Y.; data curation, Q.L., D.Y., Y.Y. and K.Y.; writing—original draft preparation, Q.L. and Y.Y.; writing—review and editing, Q.L., D.Y., Z.L. and Y.Y.; visualization, Y.Y. and H.S.; supervision, Q.L.; project administration, Q.L.; funding acquisition, Q.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Hubei Province (2023AFB102 (Q.L.)), the Hubei Province Key Laboratory of Occupational Hazard Identification and Control, and the Wuhan University of Science and Technology (OHIC2021Y02 (Q.L.)).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and supplementary materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rom, O.; Liu, Y.; Liu, Z.; Zhao, Y.; Wu, J.; Ghrayeb, A.; Villacorta, L.; Fan, Y.; Chang, L.; Wang, L.; et al. Glycine-based treatment ameliorates NAFLD by modulating fatty acid oxidation, glutathione synthesis, and the gut microbiome. Sci. Transl. Med. 2020, 12, eaaz2841. [Google Scholar] [CrossRef]
- Tomasiewicz, K.; Flisiak, R.; Halota, W.; Jaroszewicz, J.; Lebensztejn, D.; Lisik, W.; Małkowski, P.; Pawłowska, M.; Piekarska, A.; Simon, K.; et al. Recommendations for the management of non-alcoholic fatty liver disease (NAFLD). Clin. Exp. Hepatol. 2018, 4, 153–157. [Google Scholar] [CrossRef]
- Eloranta, J.J.; Kullak-Ublick, G.A. The role of FXR in disorders of bile acid homeostasis. Physiology 2008, 23, 286–295. [Google Scholar] [CrossRef]
- Sepe, V.; Distrutti, E.; Fiorucci, S.; Zampella, A. Farnesoid X receptor modulators 2014-present: A patent review. Expert Opin. Ther. Pat. 2018, 28, 351–364. [Google Scholar] [CrossRef]
- Taylor, R.; Armstrong, L.; Bhattacharya, A.; Henry, Z.; Brinker, A.; Buckley, B.; Kong, B.; Guo, G. Myclobutanil-mediated alteration of liver-gut FXR signaling in mice. Toxicol. Sci. 2023, 191, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Woo, D.C.; Kang, J.; Ra, M.; Kim, K.H.; Lee, S.R.; Choi, D.K.; Lee, H.; Hong, K.B.; Min, S.H.; et al. The Role of the Histone Methyltransferase EZH2 in Liver Inflammation and Fibrosis in STAM NASH Mice. Biology 2020, 9, 93. [Google Scholar] [CrossRef] [PubMed]
- Gay, M.D.; Cao, H.; Shivapurkar, N.; Dakshanamurthy, S.; Kallakury, B.; Tucker, R.D.; Kwagyan, J.; Smith, J.P. Proglumide Reverses Nonalcoholic Steatohepatitis by Interaction with the Farnesoid X Receptor and Altering the Microbiome. Int. J. Mol. Sci. 2022, 23, 1899. [Google Scholar] [CrossRef]
- Roy, P.P.; Mahtab, M.A.; Rahim, M.A.; Yesmin, S.S.; Islam, S.B.; Akbar, S.M.F. Treatment of Nonalcoholic Steatohepatitis by Obeticholic Acid: Current Status. Euroasian J. Hepato-Gastroenterol. 2022, 12 (Suppl. S1), S46–S50. [Google Scholar]
- Neuschwander-Tetri, B.A.; Loomba, R.; Sanyal, A.J.; Lavine, J.E.; Van Natta, M.L.; Abdelmalek, M.F.; Chalasani, N.; Dasarathy, S.; Diehl, A.M.; Hameed, B.; et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): A multicentre, randomised, placebo-controlled trial. Lancet 2015, 385, 956–965. [Google Scholar] [CrossRef]
- Xiao, J.; Ng, C.H.; Chin, Y.H.; Tan, D.J.H.; Lim, W.H.; Lim, G.; Quek, J.; Tang, A.S.P.; Chan, K.E.; Soong, R.Y.; et al. A Class Effect Network Meta-analysis of Lipid Modulation in Non-alcoholic Steatohepatitis for Dyslipidemia. J. Clin. Transl. Hepatol. 2022, 10, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wang, Z.; Huan, Y.; Xue, W.; Wang, X.; Wang, Y.; Liu, Z.; Hsieh, J.-H.; Zhang, L.; Wu, S.; et al. Pose Filter-Based Ensemble Learning Enables Discovery of Orally Active, Nonsteroidal Farnesoid X Receptor Agonists. J. Chem. Inf. Model. 2020, 60, 1202–1214. [Google Scholar] [CrossRef]
- Maloney, P.R.; Parks, D.J.; Haffner, C.D.; Fivush, A.M.; Chandra, G.; Plunket, K.D.; Creech, K.L.; Moore, L.B.; Wilson, J.G.; Lewis, M.C.; et al. Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 2000, 43, 2971–2974. [Google Scholar] [CrossRef] [PubMed]
- Abel, U.; Schlüter, T.; Schulz, A.; Hambruch, E.; Steeneck, C.; Hornberger, M.; Hoffmann, T.; Perović-Ottstadt, S.; Kinzel, O.; Burnet, M.; et al. Synthesis and pharmacological validation of a novel series of non-steroidal FXR agonists. Bioorg. Med. Chem. Lett. 2010, 20, 4911–4917. [Google Scholar] [CrossRef]
- Fiorucci, S.; Biagioli, M.; Sepe, V.; Zampella, A.; Distrutti, E. Bile acid modulators for the treatment of nonalcoholic steatohepatitis (NASH). Expert. Opin. Investig. Drugs 2020, 29, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Gege, C.; Kinzel, O.; Steeneck, C.; Schulz, A.; Kremoser, C. Knocking on FXR’s door: The “hammerhead”-structure series of FXR agonists—Amphiphilic isoxazoles with potent in vitro and in vivo activities. Curr. Top. Med. Chem. 2014, 14, 2143–2158. [Google Scholar] [CrossRef] [PubMed]
- Kinzel, O.; Steeneck, C.; Schlüter, T.; Schulz, A.; Gege, C.; Hahn, U.; Hambruch, E.; Hornberger, M.; Spalwisz, A.; Frick, K.; et al. Novel substituted isoxazole FXR agonists with cyclopropyl, hydroxycyclobutyl and hydroxyazetidinyl linkers: Understanding and improving key determinants of pharmacological properties. Bioorg. Med. Chem. Lett. 2016, 26, 3746–3753. [Google Scholar] [CrossRef] [PubMed]
- Genin, M.J.; Bueno, A.B.; Agejas Francisco, J.; Manninen, P.R.; Bocchinfuso, W.P.; Montrose-Rafizadeh, C.; Cannady, E.A.; Jones, T.M.; Stille, J.R.; Raddad, E.; et al. Discovery of 6-(4-{[5-Cyclopropyl-3-(2,6-dichlorophenyl)isoxazol-4-yl]methoxy}piperidin-1-yl)-1-methyl-1H-indole-3-carboxylic Acid: A Novel FXR Agonist for the Treatment of Dyslipidemia. J. Med. Chem. 2015, 58, 9768–9772. [Google Scholar] [CrossRef] [PubMed]
- Sepe, V.; Marchianò, S.; Finamore, C.; Baronissi, G.; Di Leva, F.S.; Carino, A.; Biagioli, M.; Fiorucci, C.; Cassiano, C.; Monti, M.C.; et al. Novel Isoxazole Derivatives with Potent FXR Agonistic Activity Prevent Acetaminophen-Induced Liver Injury. ACS Med. Chem. Lett. 2019, 10, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Hegazy, L.; Finck, B.N.; Elgendy, B. Recent Advances in the Medicinal Chemistry of Farnesoid X Receptor. J. Med. Chem. 2021, 64, 17545–17571. [Google Scholar] [CrossRef]
- Xie, H.; Chen, L.; Zhang, J.; Xie, X.; Qiu, K.; Fu, J. A Combined Pharmacophore Modeling, 3D QSAR and Virtual Screening Studies on Imidazopyridines as B-Raf Inhibitors. Int. J. Mol. Sci. 2015, 16, 12307–12323. [Google Scholar] [CrossRef]
- Żołnowska, B.; Sławiński, J.; Brzozowski, Z.; Kawiak, A.; Belka, M.; Zielińska, J.; Bączek, T.; Chojnacki, J. Synthesis, Molecular Structure, Anticancer Activity, and QSAR Study of N-(aryl/heteroaryl)-4-(1H-pyrrol-1-yl)Benzenesulfonamide Derivatives. Int. J. Mol. Sci. 2018, 19, 1482. [Google Scholar] [CrossRef]
- Guidotti, I.L.; Neis, A.; Martinez, D.P.; Seixas, F.K.; Machado, K.; Kremer, F.S. Bambu and its applications in the discovery of active molecules against melanoma. J. Mol. Graph. Model. 2023, 124, 108564. [Google Scholar] [CrossRef]
- Abdullahi, S.H.; Uzairu, A.; Shallangwa, G.A.; Uba, S.; Umar, A.B. Pharmacokinetic profiling of quinazoline-4(3H)-one analogs as EGFR inhibitors: 3D-QSAR modeling, molecular docking studies and the design of therapeutic agents. J. Taibah Univ. Med. Sci. 2023, 18, 1018–1029. [Google Scholar] [CrossRef]
- Liu, X.; Shi, D.; Zhou, S.; Liu, H.; Liu, H.; Yao, X. Molecular dynamics simulations and novel drug discovery. Expert. Opin. Drug Discov. 2018, 13, 23–37. [Google Scholar] [CrossRef]
- Panzitt, K.; Zollner, G.; Marschall, H.U.; Wagner, M. Recent advances on FXR-targeting therapeutics. Mol. Cell. Endocrinol. 2022, 552, 111678. [Google Scholar] [CrossRef] [PubMed]
- Trauner, M.; Gulamhusein, A.; Hameed, B.; Caldwell, S.; Shiffman, M.L.; Landis, C.; Eksteen, B.; Agarwal, K.; Muir, A.; Rushbrook, S.; et al. The Nonsteroidal Farnesoid X Receptor Agonist Cilofexor (GS-9674) Improves Markers of Cholestasis and Liver Injury in Patients With Primary Sclerosing Cholangitis. Hepatology 2019, 70, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Akwabi-Ameyaw, A.; Bass, J.Y.; Caldwell, R.D.; Caravella, J.A.; Chen, L.; Creech, K.L.; Deaton, D.N.; Jones, S.A.; Kaldor, I.; Liu, Y.; et al. Conformationally constrained farnesoid X receptor (FXR) agonists: Naphthoic acid-based analogs of GW 4064. Bioorg. Med. Chem. Lett. 2008, 18, 4339–4343. [Google Scholar] [CrossRef] [PubMed]
- Tully, D.C.; Rucker, P.V.; Chianelli, D.; Williams, J.; Vidal, A.; Alper, P.B.; Mutnick, D.; Bursulaya, B.; Schmeits, J.; Wu, X.; et al. Discovery of Tropifexor (LJN452), a Highly Potent Non-bile Acid FXR Agonist for the Treatment of Cholestatic Liver Diseases and Nonalcoholic Steatohepatitis (NASH). J. Med. Chem. 2017, 60, 9960–9973. [Google Scholar] [CrossRef] [PubMed]
- Bass, J.Y.; Caravella, J.A.; Chen, L.; Creech, K.L.; Deaton, D.N.; Madauss, K.P.; Marr, H.B.; McFadyen, R.B.; Miller, A.B.; Mills, W.Y.; et al. Conformationally constrained farnesoid X receptor (FXR) agonists: Heteroaryl replacements of the naphthalene. Bioorg. Med. Chem. Lett. 2011, 21, 1206–1213. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, B.V.; Babu, T.M.C.; Rammohan, A.; Zheng, G.Y.; Zyryanov, G.V.; Gu, W. Structure-Based Virtual Screening of Pseudomonas aeruginosa LpxA Inhibitors Using Pharmacophore-Based Approach. Biomolecules 2020, 10, 266. [Google Scholar] [CrossRef] [PubMed]
- Meyners, C.; Krämer, A.; Yildiz, Ö.; Meyer-Almes, F.J. The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1855–1863. [Google Scholar] [CrossRef] [PubMed]
- Mehranfar, A.; Khavani, M.; Izadyar, M. A molecular dynamic study on the ability of phosphorene for designing new sensor for SARS-CoV-2 detection. J. Mol. Liq. 2022, 345, 117852. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Gupta, S.K.; Naaz, F.; Sen Gupta, P.S.; Yadav, M.; Singh, V.K.; Panda, S.K.; Biswal, S.; Rana, M.K.; Gupta, S.K.; et al. Exploring antiviral potency of N-1 substituted pyrimidines against HIV-1 and other DNA/RNA viruses: Design, synthesis, characterization, ADMET analysis, docking, molecular dynamics and biological activity. Comput. Biol. Chem. 2023, 106, 107910. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Mathur, K.; Verma, R.K.; Jayadev Magani, S.K.; Vyas, D.K.; Singh, A. Molecular dynamics study of tropical calcific pancreatitis (TCP) associated calcium-sensing receptor single nucleotide variation. Front. Mol. Biosci. 2022, 9, 982831. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Mittal, L.; Srivastava, M.; Pathak, D.P.; Asthana, S. Conformational Characterization of the Co-Activator Binding Site Revealed the Mechanism to Achieve the Bioactive State of FXR. Front. Mol. Biosci. 2021, 8, 658312. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, X.; Qi, Y.; Lin, Y.; Zhao, K.; Jin, Y.; Luo, J.; Xu, L.; Yu, D.; Li, C. Binding, activity and risk assessment of bisphenols toward farnesoid X receptor pathway: In vitro and in silico study. Sci. Total Environ. 2023, 869, 161701. [Google Scholar] [CrossRef]
- Elkwafi, G.; Mohamed, N.; Elabbar, F.; Alnajjar, R. Flavonoid content of the Libyan Onosma Cyrenaicum: Isolation, identification, electronic chemical reactivity, drug likeness, docking, and MD study. J. Biomol. Struct. Dyn. 2022, 40, 7351–7366. [Google Scholar] [CrossRef]
- Dong, Y.; Fu, L.; Song, J.; Zhang, S.; Li, X.; Fang, W.; Cui, Q.; Gao, L. Thermodynamic Driving Forces for Divalent Cations Binding to Zwitterionic Phospholipid Membranes. J. Phys. Chem. Lett. 2022, 13, 11237–11244. [Google Scholar] [CrossRef]
- Lee, K.; Nada, H.; Byun, H.J.; Lee, C.H.; Elkamhawy, A. Hit Identification of a Novel Quinazoline Sulfonamide as a Promising EphB3 Inhibitor: Design, Virtual Combinatorial Library, Synthesis, Biological Evaluation, and Docking Simulation Studies. Pharmaceuticals 2021, 14, 1247. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, M.; Su, J.; Hu, H.; Yang, M.; Huang, Z.; Chen, D.; Wu, J.; Feng, Z. Overcoming biomass recalcitrance by synergistic pretreatment of mechanical activation and metal salt for enhancing enzymatic conversion of lignocellulose. Biotechnol. Biofuels 2019, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Shafiee, Z.; Karami, L.; Akbari, M.; Rezaee, E.; Maaza, M.; Tabatabai, S.A.; Karimi, G. Insights into the molecular mechanism of triazolopyrimidinone derivatives effects on the modulation of α(1)β(2)γ(2) subtype of GABA(A) receptor: An in silico approach. Arch. Biochem. Biophys. 2022, 729, 109380. [Google Scholar] [CrossRef]
- Akash, S.; Bayıl, I.; Rahman, M.A.; Mukerjee, N.; Maitra, S.; Islam, M.R.; Rajkhowa, S.; Ghosh, A.; Al-Hussain, S.A.; Zaki, M.E.A.; et al. Target specific inhibition of West Nile virus envelope glycoprotein and methyltransferase using phytocompounds: An in silico strategy leveraging molecular docking and dynamics simulation. Front. Microbiol. 2023, 14, 1189786. [Google Scholar] [CrossRef] [PubMed]
- Kumari, G.; Nigam, V.K.; Pandey, D.M. Mutational analysis of flavonol synthase of M. pinnata towards enhancement of binding affinity: A computational approach. J. Biomol. Struct. Dyn. 2023, 1–14. [Google Scholar] [CrossRef]
- Ma, C.; Liu, W.-G.; Liu, W.-D.; Xi, C.-C.; Xiong, F.; Zhang, S.-P. Molecular Docking and 3D-QSAR Studies on a Series of Benzenesulfonamide Derivatives as a Hepatitis B Virus Capsid Assembly Inhibitor. Polycycl. Aromat. Compd. 2021, 42, 3622–3637. [Google Scholar] [CrossRef]
- Ren, R.; Gao, L.; Li, G.; Wang, S.; Zhao, Y.; Wang, H.; Liu, J. 2D, 3D-QSAR study and docking of vascular endothelial growth factor receptor 3 (VEGFR3) inhibitors for potential treatment of retinoblastoma. Front. Pharmacol. 2023, 14, 1177282. [Google Scholar] [CrossRef] [PubMed]
- Ershov, P.V.; Veselovsky, A.V.; Mezentsev, Y.V.; Yablokov, E.O.; Kaluzhskiy, L.A.; Tumilovich, A.M.; Kavaleuski, A.A.; Gilep, A.A.; Moskovkina, T.V.; Medvedev, A.E.; et al. Mechanism of the Affinity-Enhancing Effect of Isatin on Human Ferrochelatase and Adrenodoxin Reductase Complex Formation: Implication for Protein Interactome Regulation. Int. J. Mol. Sci. 2020, 21, 7605. [Google Scholar] [CrossRef]
- Thillainayagam, M.; Malathi, K.; Anbarasu, A.; Singh, H.; Bahadur, R.; Ramaiah, S. Insights on inhibition of Plasmodium falciparum plasmepsin I by novel epoxyazadiradione derivatives—Molecular docking and comparative molecular field analysis. J. Biomol. Struct. Dyn. 2019, 37, 3168–3182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, L.; Wang, Z.; Zhu, Y.; Jiang, H.; Xu, J.; Xiong, F. Design of novel DABO derivatives as HIV-1 RT inhibitors using molecular docking, molecular dynamics simulations and ADMET properties. J. Biomol. Struct. Dyn. 2023, 1–18, Online ahead of print. [Google Scholar] [CrossRef]
- Zięba, A.; Żuk, J.; Bartuzi, D.; Matosiuk, D.; Poso, A.; Kaczor, A.A. The Universal 3D QSAR Model for Dopamine D(2) Receptor Antagonists. Int. J. Mol. Sci. 2019, 20, 4555. [Google Scholar] [CrossRef]
- Medeiros, A.R.; Ferreira, L.L.G.; de Souza, M.L.; de Oliveira Rezende Junior, C.; Espinoza-Chávez, R.M.; Dias, L.C.; Andricopulo, A.D. Chemoinformatics Studies on a Series of Imidazoles as Cruzain Inhibitors. Biomolecules 2021, 11, 579. [Google Scholar] [CrossRef]
- Liu, G.; Wang, W.; Wan, Y.; Ju, X.; Gu, S. Application of 3D-QSAR, Pharmacophore, and Molecular Docking in the Molecular Design of Diarylpyrimidine Derivatives as HIV-1 Nonnucleoside Reverse Transcriptase Inhibitors. Int. J. Mol. Sci. 2018, 19, 1436. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.L.; Luo, J.X.; Zhang, B.C.; Jiang, G.F.; Ding, W.; Zhang, Y.Q. 3D-QSAR and Molecular Docking Studies on the TcPMCA1-Mediated Detoxification of Scopoletin and Coumarin Derivatives. Int. J. Mol. Sci. 2017, 18, 1380. [Google Scholar] [CrossRef]
- Gao, J.; Li, X.; Fu, R.; Li, Y. Mechanism analysis and improved molecular modification: Design of high efficiency and environmentally friendly triazole fungicide substitutes. Chemosphere 2023, 336, 139150. [Google Scholar] [CrossRef]
- Abdizadeh, R.; Hadizadeh, F.; Abdizadeh, T. Molecular Modeling Studies of Anti-Alzheimer Agents by QSAR, Molecular Docking and Molecular Dynamics Simulations Techniques. Med. Chem. 2020, 16, 903–927. [Google Scholar] [CrossRef]
- Clark, M.; Cramer, R.D.; Van Opdenbosch, N. Validation of the general purpose tripos 5.2 force field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar] [CrossRef]
- Roy, K. On some aspects of validation of predictive quantitative structure-activity relationship models. Expert. Opin. Drug Discov. 2007, 2, 1567–1577. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, Y.; Qiang, S.; Jin, R.; Li, Z.; Tang, Y.; Leung, E.L.H.; Guo, H.; Yao, X. 3D-QSAR, Molecular Docking, and MD Simulations of Anthraquinone Derivatives as PGAM1 Inhibitors. Front. Pharmacol. 2021, 12, 764351. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Liu, P.; Liu, H.; Wei, Q.; Yang, J.; Cao, P.; Lai, Y. 3D-QSAR, molecular docking and molecular dynamics simulations study of 3-pyrimidin-4-yl-oxazolidin-2-one derivatives to explore the structure requirements of mutant IDH1 inhibitors. J. Mol. Struct. 2019, 1189, 187–202. [Google Scholar] [CrossRef]
- Oprea, T.I.; Waller, C.L.; Marshall, G.R. Three-dimensional quantitative structure-activity relationship of human immunodeficiency virus (I) protease inhibitors. 2. Predictive power using limited exploration of alternate binding modes. J. Med. Chem. 1994, 37, 2206–2215. [Google Scholar] [CrossRef]
- Gajjar, K.A.; Gajjar, A.K. CoMFA, CoMSIA and HQSAR Analysis of 3-aryl-3-ethoxypropanoic Acid Derivatives as GPR40 Modulators. Curr. Drug Discov. Technol. 2020, 17, 100–118. [Google Scholar] [CrossRef]
- Merk, D.; Sreeramulu, S.; Kudlinzki, D.; Saxena, K.; Linhard, V.; Gande, S.L.; Hiller, F.; Lamers, C.; Nilsson, E.; Aagaard, A.; et al. Molecular tuning of farnesoid X receptor partial agonism. Nat. Commun. 2019, 10, 2915. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, Y.; Teng, L.; Luo, E.; Liu, D.; Zhou, F.; Wang, K.; Chen, W.; Bi, L. Zi Shen Decoction Inhibits Growth and Metastasis of Lung Cancer via Regulating the AKT/GSK-3β/β-Catenin Pathway. Oxidative Med. Cell. Longev. 2021, 2021, 6685282. [Google Scholar] [CrossRef]
- Li, W.; Fu, J.; Cheng, F.; Zheng, M.; Zhang, J.; Liu, G.; Tang, Y. Unbinding pathways of GW4064 from human farnesoid X receptor as revealed by molecular dynamics simulations. J. Chem. Inf. Model. 2012, 52, 3043–3052. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jiao, Y.; Yang, M.; Wu, L.; Long, G.; Hu, W. Network Pharmacology, Molecular Docking and Molecular Dynamics to Explore the Potential Immunomodulatory Mechanisms of Deer Antler. Int. J. Mol. Sci. 2023, 24, 10370. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Berendsen, J.-P.R.G.C.H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar]
- Farid, N.; Bux, K.; Ali, K.; Bashir, A.; Tahir, R. Repurposing Amphotericin B: Anti-microbial, molecular docking and molecular dynamics simulation studies suggest inhibition potential of Amphotericin B against MRSA. BMC Chem. 2023, 17, 67. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, S.; Sun, Y.; Zhou, W. Transcriptomics Changes in the Peritoneum of Mice with Lipopolysaccharide-Induced Peritonitis. Int. J. Mol. Sci. 2021, 22, 13008. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, X.; Pang, L.; Zhang, J.Z.H.; Zhu, T. Effect of mutations on binding of ligands to guanine riboswitch probed by free energy perturbation and molecular dynamics simulations. Nucleic Acids Res. 2019, 47, 6618–6631. [Google Scholar] [CrossRef] [PubMed]
- Enayatkhani, M.; Hasaniazad, M.; Faezi, S.; Gouklani, H.; Davoodian, P.; Ahmadi, N.; Einakian, M.A.; Karmostaji, A.; Ahmadi, K. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. J. Biomol. Struct. Dyn. 2021, 39, 2857–2872. [Google Scholar] [CrossRef]
- Ahmad, S.; Waheed, Y.; Ismail, S.; Abbasi, S.W.; Najmi, M.H. A computational study to disclose potential drugs and vaccine ensemble for COVID-19 conundrum. J. Mol. Liq. 2021, 324, 114734. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Jin, Y.; Wang, B.; Liu, B. Molecular Mechanism of Small-Molecule Inhibitors in Blocking the PD-1/PD-L1 Pathway through PD-L1 Dimerization. Int. J. Mol. Sci. 2021, 22, 4766. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liang, J.; Liu, B.; Jin, Y. Molecular Mechanism of Food-Derived Polyphenols on PD-L1 Dimerization: A Molecular Dynamics Simulation Study. Int. J. Mol. Sci. 2021, 22, 10924. [Google Scholar] [CrossRef] [PubMed]
- Prajapat, M.; Singh, H.; Chaudhary, G.; Sarma, P.; Kaur, G.; Prakash Patel, A.; Medhi, B. A Novel Inhibitor of DKK1/LRP6 Interactions Against the Alzheimer Disease: An Insilco Approach. Bioinform. Biol. Insights 2023, 17, 11779322231183762. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xia, Z.; Zhao, Q.; Zheng, B.; Zhang, M.; Ying, Y. Insights Into the Resistance Mechanisms of Inhibitors to FLT3 F691L Mutation via an Integrated Computational Approach. Front. Pharmacol. 2019, 10, 1050. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Dalby, P.A. A beginner’s guide to molecular dynamics simulations and the identification of cross-correlation networks for enzyme engineering. Methods Enzym. 2020, 643, 15–49. [Google Scholar]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).