Efficient Expression in Leishmania tarentolae (LEXSY) of the Receptor-Binding Domain of the SARS-CoV-2 S-Protein and the Acetylcholine-Binding Protein from Lymnaea stagnalis
Abstract
1. Introduction
2. Results
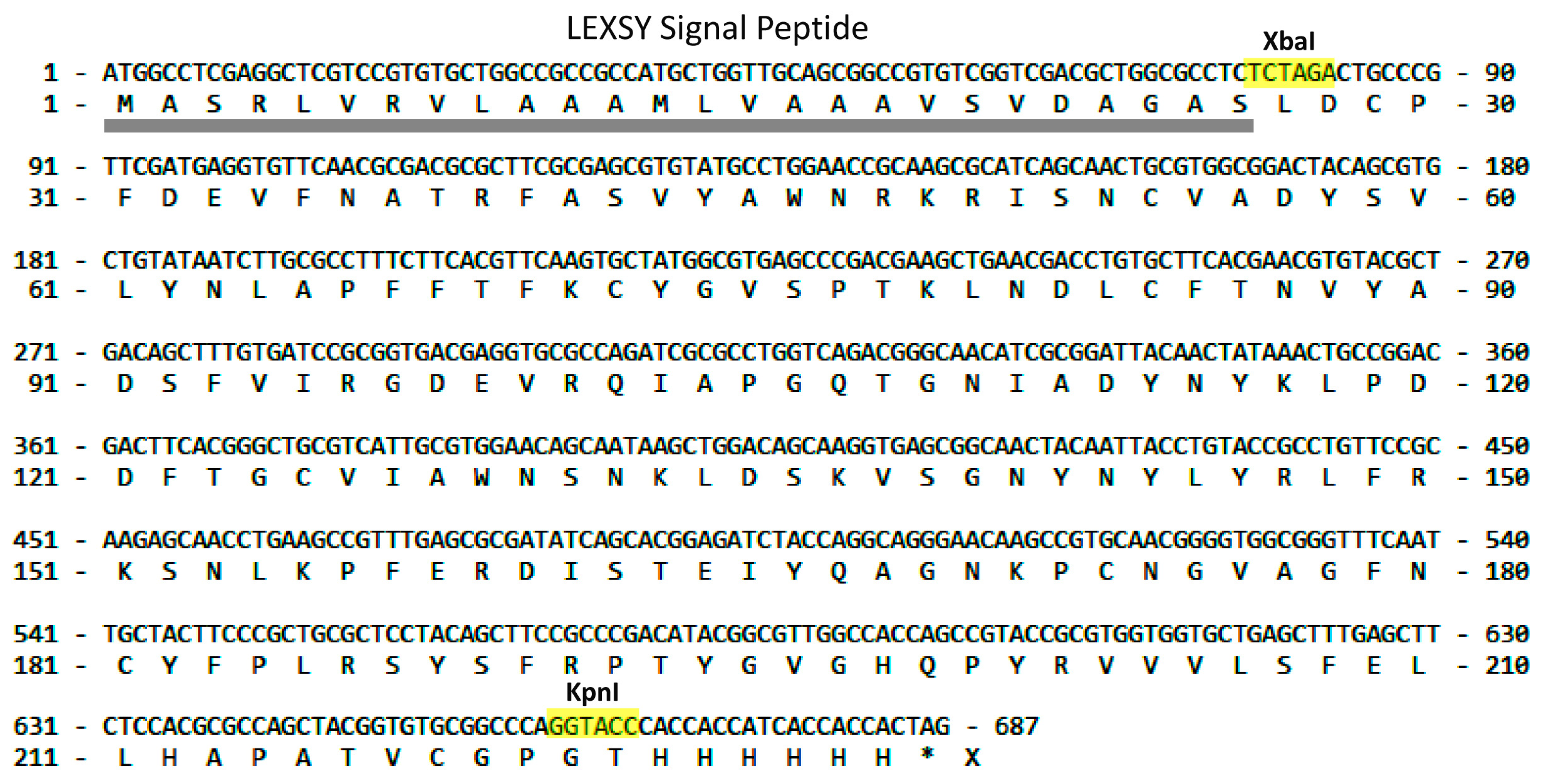
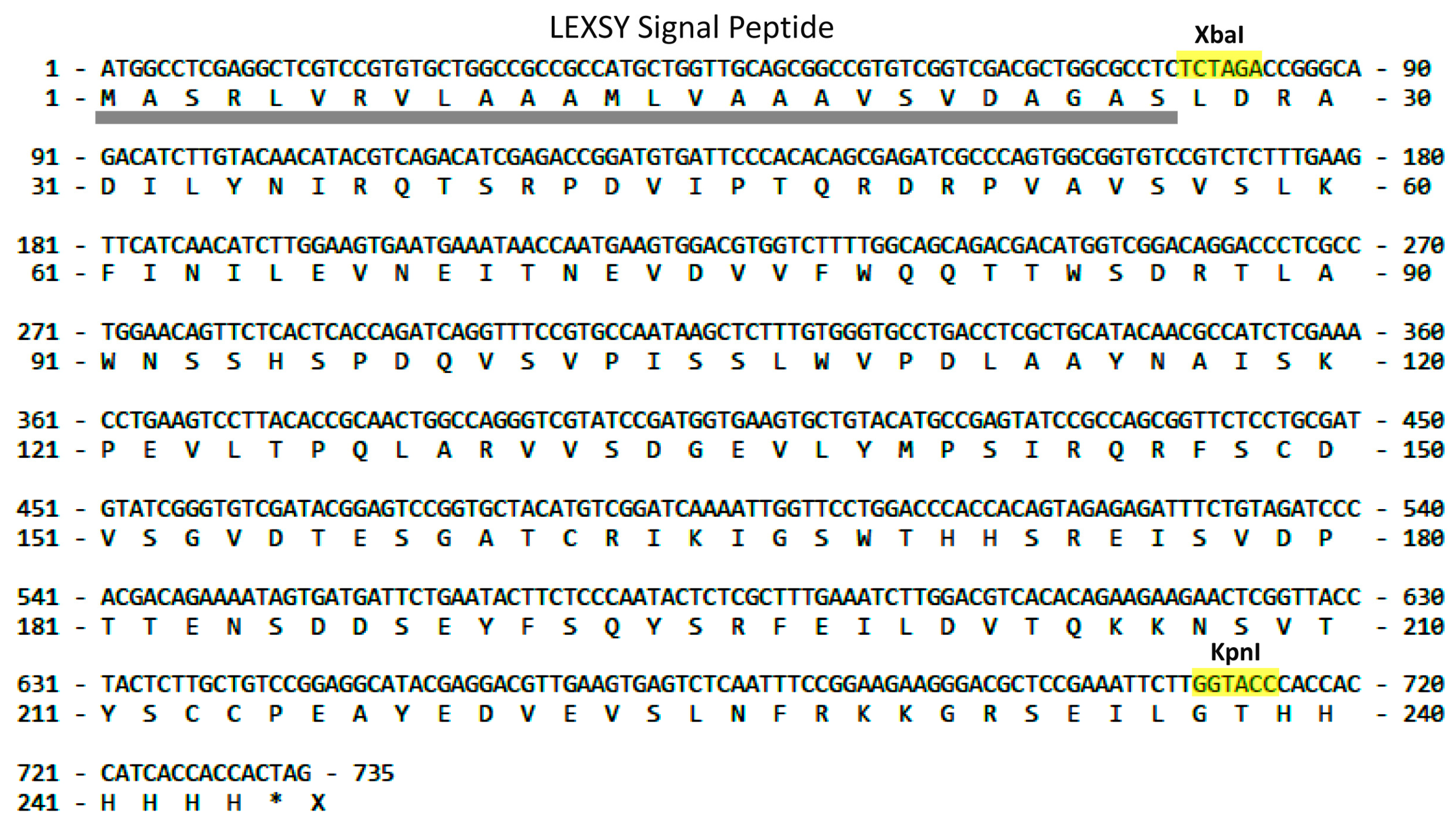
2.1. LEXSY Production of RBD and Lymnaea stagnalis AChBP
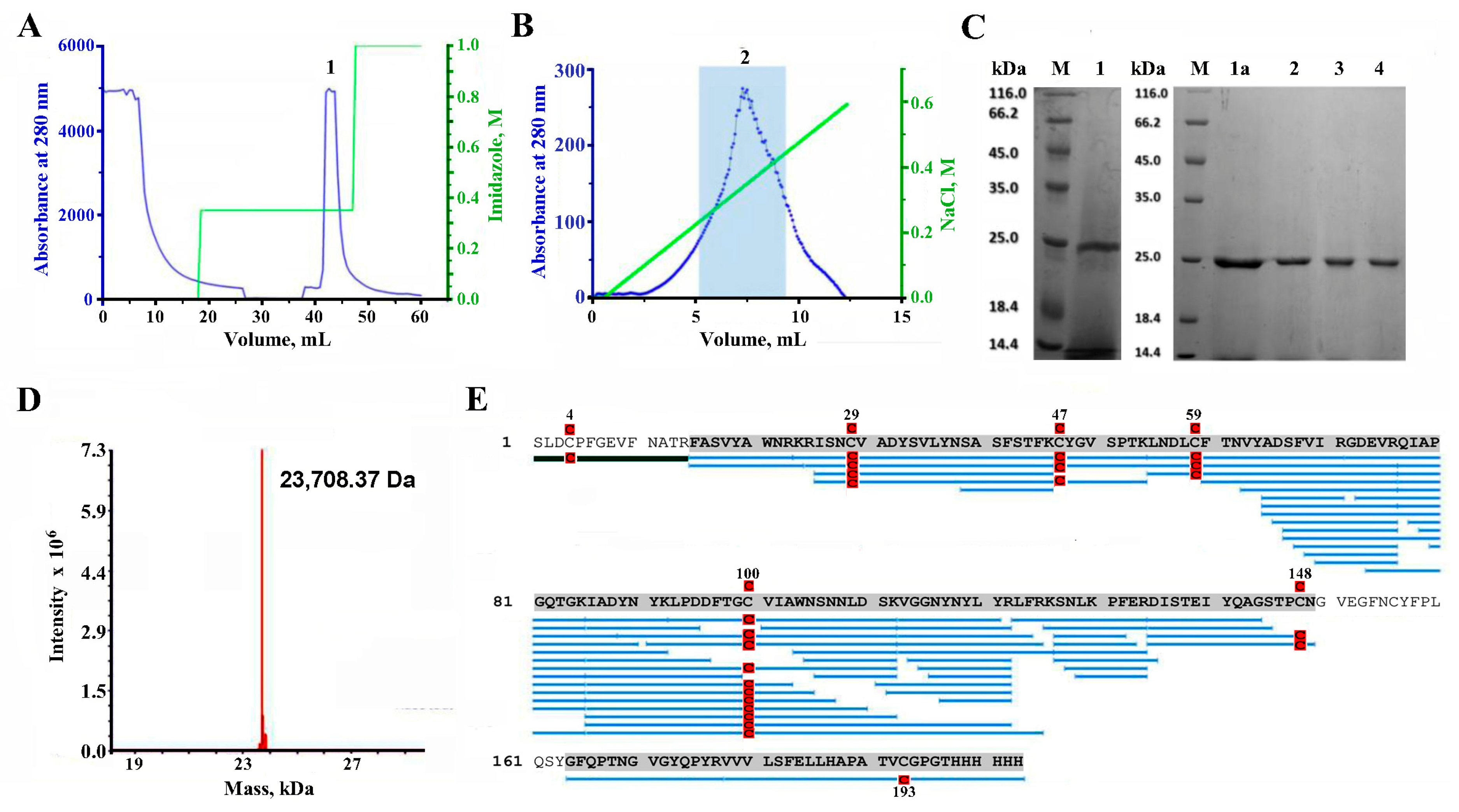
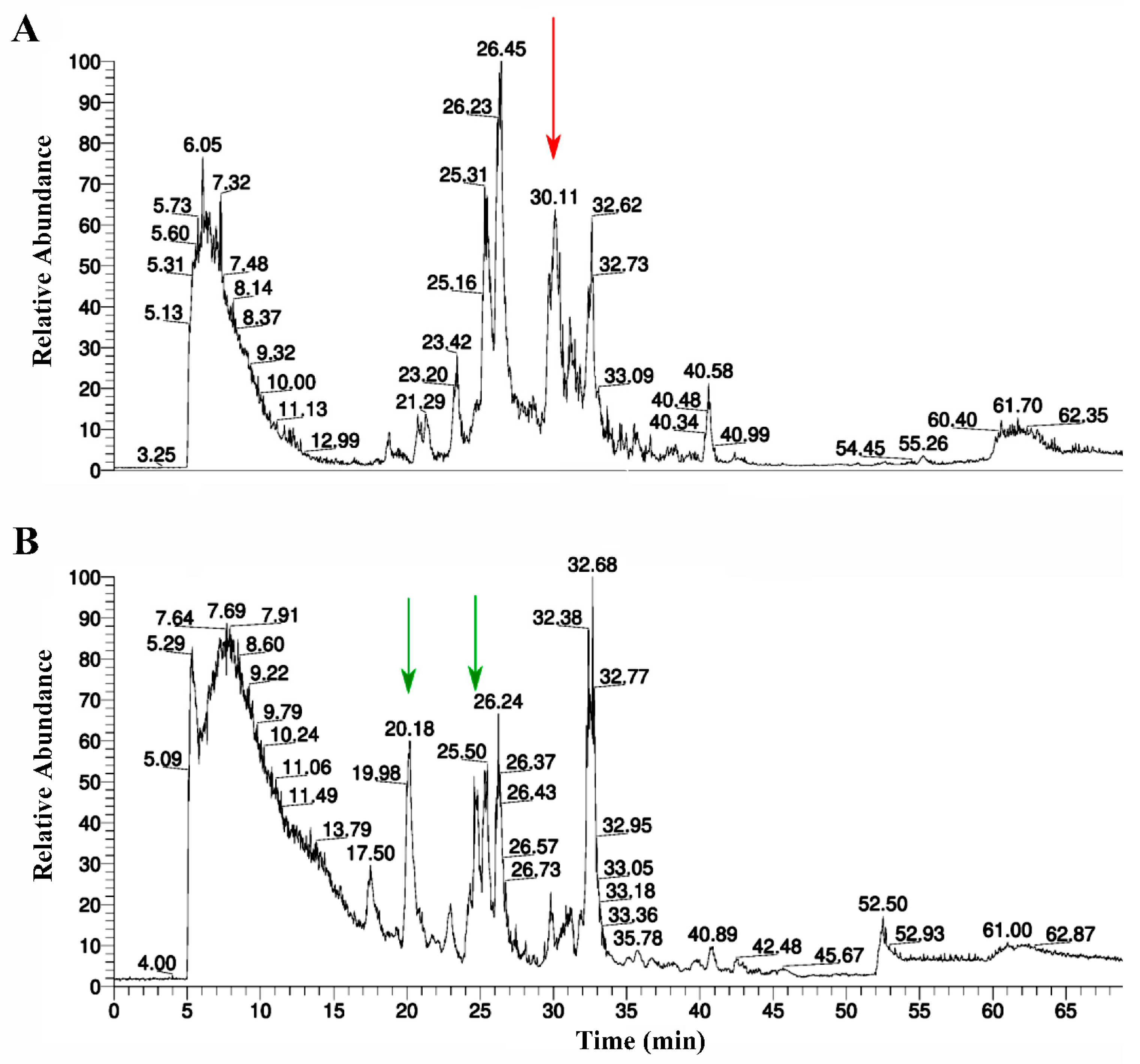
2.2. Purification of RBD
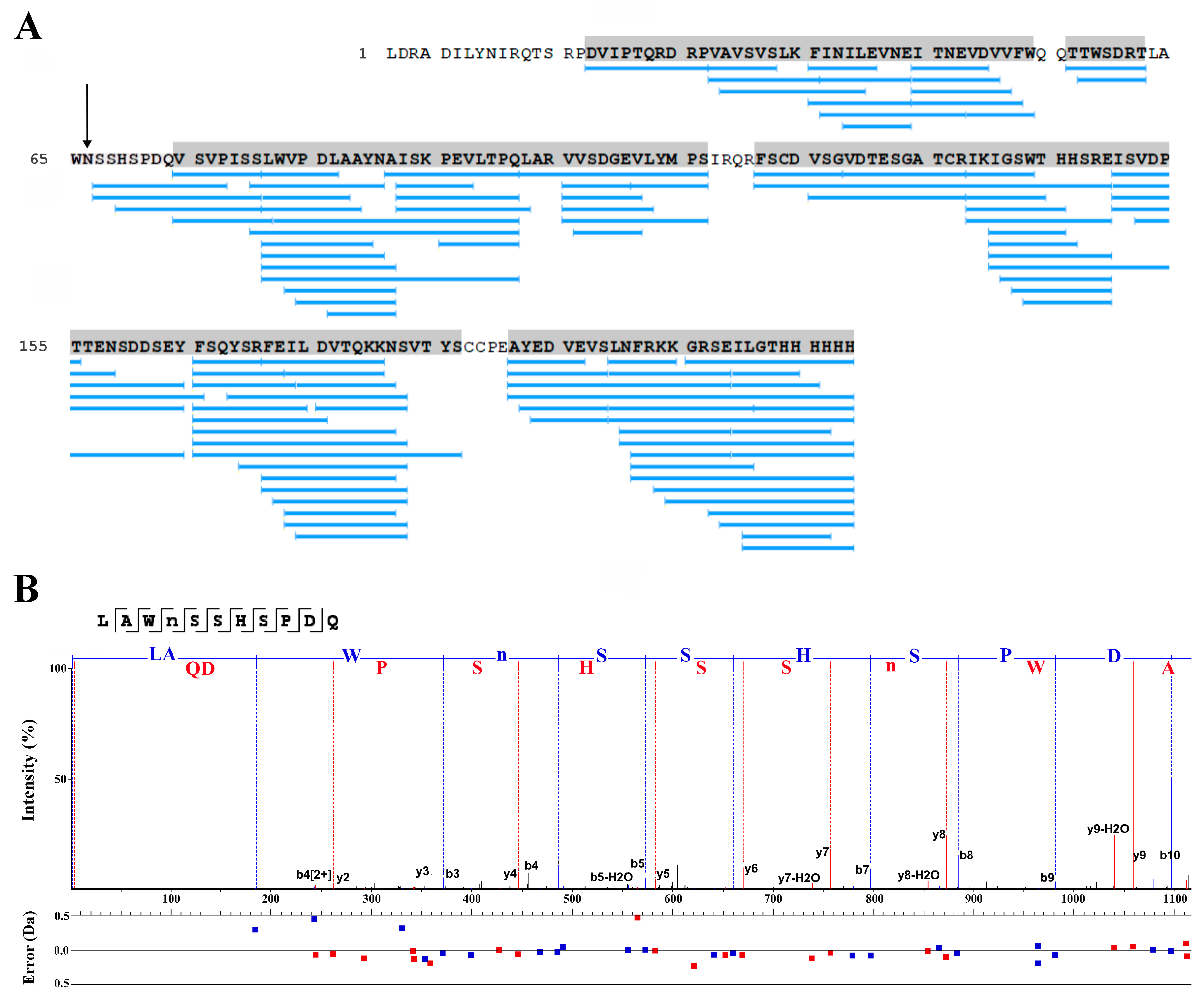
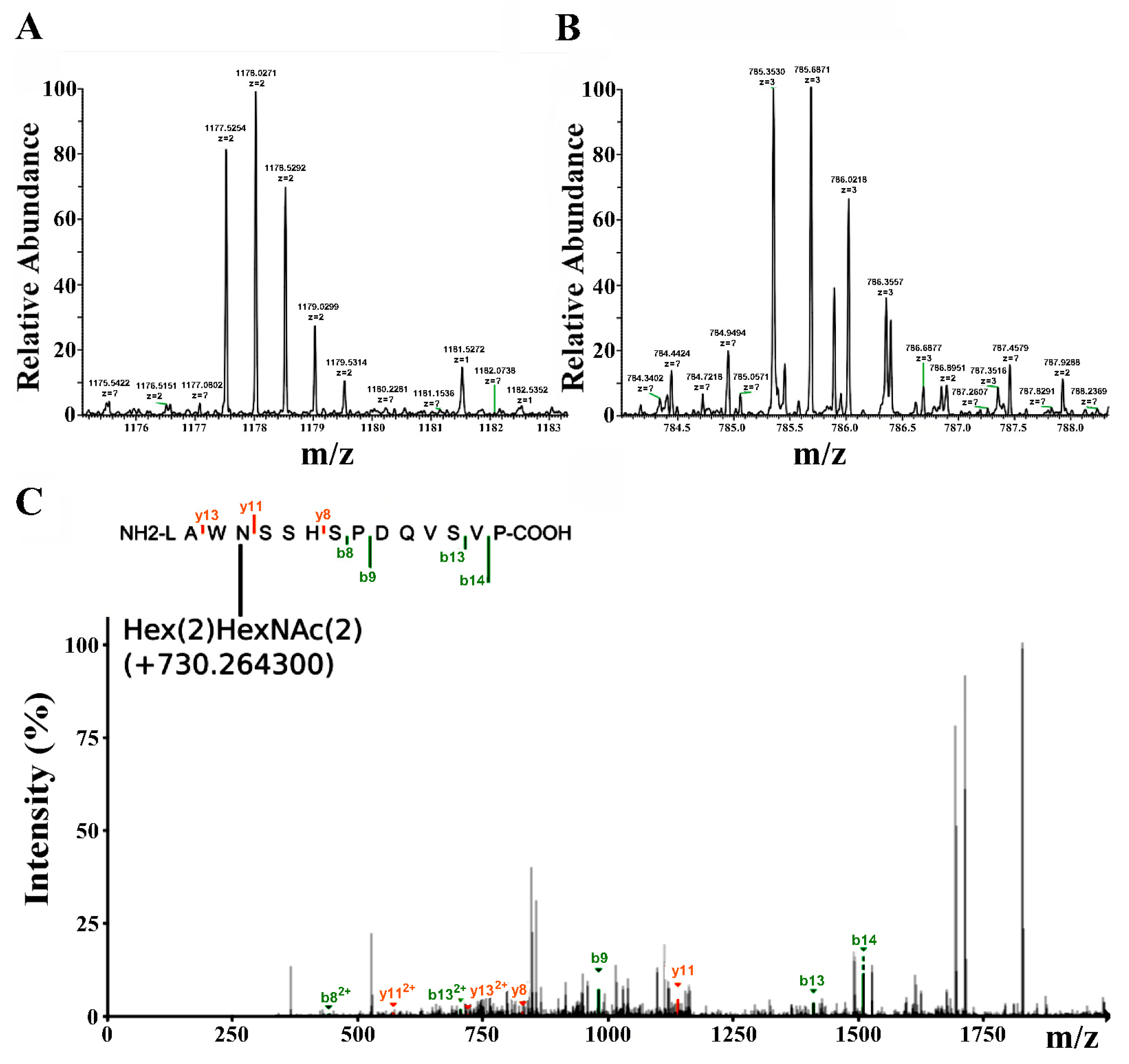
2.3. LC-MS Study of the Purified RBD
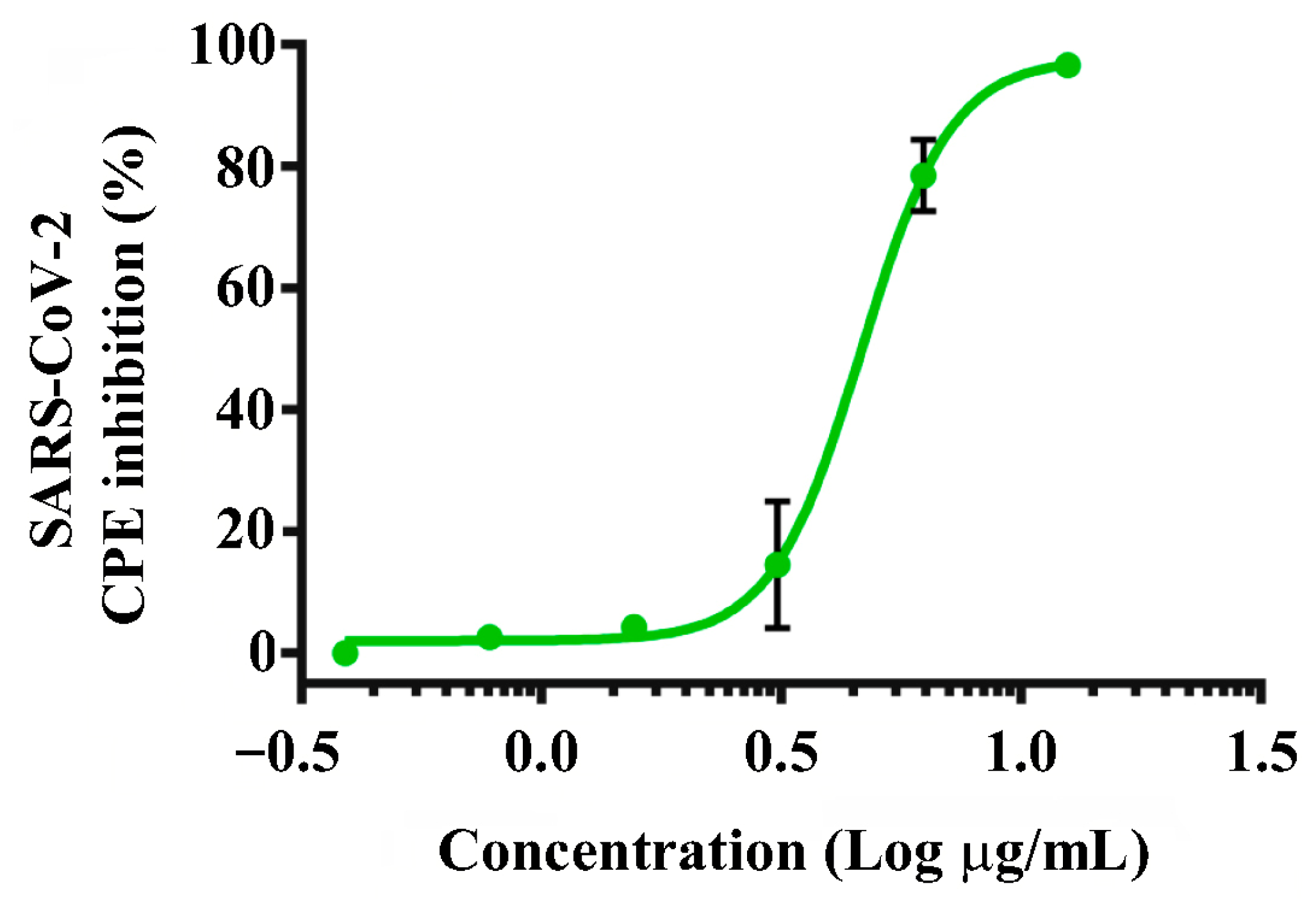
2.4. Testing the Recombinant RBD for SARS-CoV-2 Antiviral Effects
2.5. Purification of AChBP
2.6. LC-MS Study of the Purified Lymnaea stagnalis AChBP
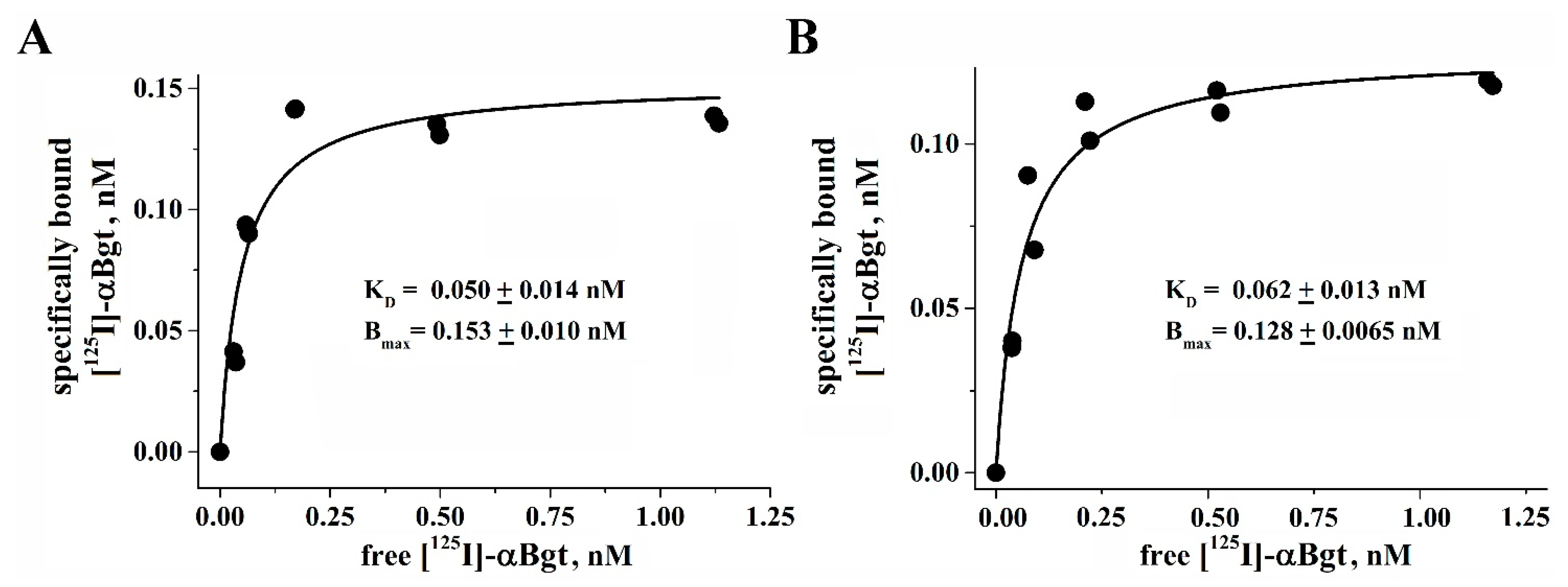
2.7. Radioligand Binding Assay
3. Discussion
4. Materials and Methods
4.1. LEXSY Production of RBD and Lymnaea stagnalis AChBP
4.2. RBD Purification
4.3. Enzymatic Digestion and Mass-Spectrometry of RBD
4.4. AChBP Purification
4.5. Enzymatic Digestion and Mass-Spectrometry of AChBP
4.6. SARS-CoV-2 Antiviral Assay
4.7. Radioligand Binding Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, J.; Wang, W.; Chen, Z.; Lu, S.; Yang, F.; Bi, Z.; Bao, L.; Mo, F.; Li, X.; Huang, Y.; et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 2020, 586, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Sun, Q. Antibodies and Vaccines Target RBD of SARS-CoV-2. Front. Mol. Biosci. 2021, 8, 671633. [Google Scholar] [CrossRef]
- Queirós-Reis, L.; Gomes da Silva, P.; Gonçalves, J.; Brancale, A.; Bassetto, M.; Mesquita, J.R. SARS-CoV-2 Virus-Host Interaction: Currently Available Structures and Implications of Variant Emergence on Infectivity and Immune Response. Int. J. Mol. Sci. 2021, 22, 10836. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Li, L.; Liu, S.; Wang, Q.; Zhang, D.; Xu, Z.; Han, P.; Li, X.; Peng, Q.; Su, C.; et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 2022, 185, 630–640.e10. [Google Scholar] [CrossRef]
- Borkotoky, S.; Dey, D.; Hazarika, Z. Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): A structural perspective. Mol. Biol. Rep. 2023, 50, 2713–2721. [Google Scholar] [CrossRef] [PubMed]
- Shahsavar, A.; Gajhede, M.; Kastrup, J.S.; Balle, T. Structural Studies of Nicotinic Acetylcholine Receptors: Using Acetylcholine-Binding Protein as a Structural Surrogate. Basic. Clin. Pharmacol. Toxicol. 2016, 118, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, E.X.; Pereira, E.F.; Alkondon, M.; Rogers, S.W. Mammalian nicotinic acetylcholine receptors: From structure to function. Physiol. Rev. 2009, 89, 73–120. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, D.; Terry, A.V., Jr. The wonderland of neuronal nicotinic acetylcholine receptors. Biochem. Pharmacol. 2018, 151, 214–225. [Google Scholar] [CrossRef]
- Shelukhina, I.V.; Zhmak, M.N.; Lobanov, A.V.; Ivanov, I.A.; Garifulina, A.I.; Kravchenko, I.N.; Rasskazova, E.A.; Salmova, M.A.; Tukhovskaya, E.A.; Rykov, V.A.; et al. Azemiopsin, a Selective Peptide Antagonist of Muscle Nicotinic Acetylcholine Receptor: Preclinical Evaluation as a Local Muscle Relaxant. Toxins 2018, 10, 34. [Google Scholar] [CrossRef]
- Siniavin, A.E.; Streltsova, M.A.; Kudryavtsev, D.S.; Shelukhina, I.V.; Utkin, Y.N.; Tsetlin, V.I. Activation of α7 Nicotinic Acetylcholine Receptor Upregulates HLA-DR and Macrophage Receptors: Potential Role in Adaptive Immunity and in Preventing Immunosuppression. Biomolecules 2020, 10, 507. [Google Scholar] [CrossRef]
- Li, X.; Tae, H.S.; Chu, Y.; Jiang, T.; Adams, D.J.; Yu, R. Medicinal chemistry, pharmacology, and therapeutic potential of α-conotoxins antagonizing the α9α10 nicotinic acetylcholine receptor. Pharmacol. Ther. 2021, 222, 107792. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.B.; Syed, N.I.; Schaap, D.; van Minnen, J.; Klumperman, J.; Kits, K.S.; Lodder, H.; van der Schors, R.C.; van Elk, R.; Sorgedrager, B.; et al. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature 2001, 411, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.B.; Talley, T.T.; Radic, Z.; Taylor, P. Structural and ligand recognition characteristics of an acetylcholine-binding protein from Aplysia californica. J. Biol. Chem. 2004, 279, 24197–24202. [Google Scholar] [CrossRef]
- Celie, P.H.; Klaassen, R.V.; van Rossum-Fikkert, S.E.; van Elk, R.; van Nierop, P.; Smit, A.B.; Sixma, T.K. Crystal structure of acetylcholine-binding protein from Bulinus truncatus reveals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors. J. Biol. Chem. 2005, 280, 26457–26466. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Meng, X.; Liu, Z. Spider acetylcholine binding proteins: An alternative model to study the interaction between insect nAChRs and neonicotinoids. Insect Biochem. Mol. Biol. 2017, 90, 82–89. [Google Scholar] [CrossRef]
- Brejc, K.K.; van Dijk, W.J.W.J.; Klaassen, R.V.; Schuurmans, M.; van der Oost, J.; Smit, A.B.; Sixma, T.K. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 2001, 411, 269–276. [Google Scholar] [CrossRef]
- Celie, P.H.N.N.; Kasheverov, I.E.; Mordvintsev, D.Y.; Hogg, R.C.; van Nierop, P.; Van Elk, R.; van Rossum-Fikkert, S.E.; Zhmak, M.N.; Bertrand, D.; Tsetlin, V.; et al. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an alpha-conotoxin PnIA variant. Nat. Struct. Mol. Biol. 2005, 12, 582–588. [Google Scholar] [CrossRef]
- Ho, T.N.T.; Abraham, N.; Lewis, R.J. Unique Pharmacological Properties of α-Conotoxin OmIA at α7 nAChRs. Front. Pharmacol. 2021, 12, 3605. [Google Scholar] [CrossRef]
- Huang, S.; Li, S.-X.; Bren, N.; Cheng, K.; Gomoto, R.; Chen, L.; Sine, S.M. Complex between α-bungarotoxin and an α7 nicotinic receptor ligand-binding domain chimaera. Biochem. J. 2013, 454, 303–310. [Google Scholar] [CrossRef]
- Shahsavar, A.; Ahring, P.K.; Olsen, J.A.; Krintel, C.; Kastrup, J.S.; Balle, T.; Gajhede, M. Acetylcholine-Binding Protein Engineered to Mimic the α4-α4 Binding Pocket in α4β2 Nicotinic Acetylcholine Receptors Reveals Interface Specific Interactions Important for Binding and Activity. Mol. Pharmacol. 2015, 88, 697–707. [Google Scholar] [CrossRef]
- Montgomery, M.; Rendine, S.; Zimmer, C.T.; Elias, J.; Schaetzer, J.; Pitterna, T.; Benfatti, F.; Skaljac, M.; Bigot, A. Structural Biology-Guided Design, Synthesis, and Biological Evaluation of Novel Insect Nicotinic Acetylcholine Receptor Orthosteric Modulators. J. Med. Chem. 2022, 65, 2297–2312. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.S.; Aricescu, A.R. Crystal structure of a human GABAA receptor. Nature 2014, 512, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Morales-Perez, C.L.; Noviello, C.M.; Hibbs, R.E. X-ray structure of the human α4β2 nicotinic receptor. Nature 2016, 538, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.M.; Roh, S.H.; Gharpure, A.; Morales-Perez, C.L.; Teng, J.; Hibbs, R.E. Structural principles of distinct assemblies of the human α4β2 nicotinic receptor. Nature 2018, 557, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Gharpure, A.; Teng, J.; Zhuang, Y.; Noviello, C.M.; Walsh, R.M.; Cabuco, R.; Howard, R.J.; Zaveri, N.T.; Lindahl, E.; Hibbs, R.E. Agonist Selectivity and Ion Permeation in the α3β4 Ganglionic Nicotinic Receptor. Neuron 2019, 104, 501–511.e6. [Google Scholar] [CrossRef]
- Kasaragod, V.B.; Mortensen, M.; Hardwick, S.W.; Wahid, A.A.; Dorovykh, V.; Chirgadze, D.Y.; Smart, T.G.; Miller, P.S. Mechanisms of inhibition and activation of extrasynaptic αβ GABAA receptors. Nature 2022, 602, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Wei, J.; Kundu, R.T.; Adhikari, R.; Liu, Z.; Lee, J.; Versteeg, L.; Poveda, C.; Keegan, B.; Villar, M.J.; et al. Genetic modification to design a stable yeast-expressed recombinant SARS-CoV-2 receptor binding domain as a COVID-19 vaccine candidate. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129893. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhu, X.; Yu, J.; Yu, J.; Luo, S.; Wang, X. The crystal structure of Ac-AChBP in complex with α-conotoxin LvIA reveals the mechanism of its selectivity towards different nAChR subtypes. Protein Cell 2017, 8, 675–685. [Google Scholar] [CrossRef]
- García-Cordero, J.; Mendoza-Ramírez, J.; Fernández-Benavides, D.; Roa-Velazquez, D.; Filisola-Villaseñor, J.; Martínez-Frías, S.P.; Sanchez-Salguero, E.S.; Miguel-Rodríguez, C.E.; Maravillas Montero, J.L.; Torres-Ruiz, J.J.; et al. Recombinant Protein Expression and Purification of N, S1, and RBD of SARS-CoV-2 from Mammalian Cells and Their Potential Applications. Diagnostics 2021, 11, 1808. [Google Scholar] [CrossRef]
- Abraham, N.; Paul, B.; Ragnarsson, L.; Lewis, R.J. Escherichia coli Protein Expression System for Acetylcholine Binding Proteins (AChBPs). PLoS ONE 2016, 11, e0157363. [Google Scholar] [CrossRef]
- Antipova, N.V.; Larionova, T.D.; Siniavin, A.E.; Nikiforova, M.A.; Gushchin, V.A.; Babichenko, I.I.; Volkov, A.V.; Shakhparonov, M.I.; Pavlyukov, M.S. Establishment of Murine Hybridoma Cells Producing Antibodies against Spike Protein of SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 9167. [Google Scholar] [CrossRef]
- Dellisanti, C.D.; Yao, Y.; Stroud, J.C.; Wang, Z.-Z.; Chen, L. Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution. Nat. Neurosci. 2007, 10, 953–962. [Google Scholar] [CrossRef]
- Rahman, M.M.; Teng, J.; Worrell, B.T.; Noviello, C.M.; Lee, M.; Karlin, A.; Stowell, M.H.B.; Hibbs, R.E. Structure of the native muscle-type nicotinic receptor and inhibition by snake venom toxins. Neuron 2020, 106, 952–962.e5. [Google Scholar] [CrossRef]
- Breitling, R.; Klingner, S.; Callewaert, N.; Pietrucha, R.; Geyer, A.; Ehrlich, G.; Hartung, R.; Müller, A.; Contreras, R.; Beverley, S.M.; et al. Non-pathogenic trypanosomatid protozoa as a platform for protein research and production. Protein Expr. Purif. 2002, 25, 209–218. [Google Scholar] [CrossRef]
- Khan, A.H.; Bayat, H.; Rajabibazl, M.; Sabri, S.; Rahimpour, A. Humanizing glycosylation pathways in eukaryotic expression systems. World J. Microbiol. Biotechnol. 2016, 33, 4. [Google Scholar] [CrossRef] [PubMed]
- Celie, P.H.N.; van Rossum-Fikkert, S.E.; van Dijk, W.J.; Brejc, K.; Smit, A.B.; Sixma, T.K. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 2004, 41, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Ihara, M.; Okajima, T.; Yamashita, A.; Oda, T.; Hirata, K.; Nishiwaki, H.; Morimoto, T.; Akamatsu, M.; Ashikawa, Y.; Kuroda, S.; et al. Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin. Invert. Neurosci. 2008, 8, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Shahsavar, A.; Kastrup, J.S.; Nielsen, E.; Kristensen, J.L.; Gajhede, M.; Balle, T. Crystal Structure of Lymnaea stagnalis AChBP Complexed with the Potent nAChR Antagonist DHβE Suggests a Unique Mode of Antagonism. PLoS ONE 2012, 7, e40757. [Google Scholar] [CrossRef] [PubMed]
- Niculae, A.; Bayer, P.; Cirstea, I.; Bergbrede, T.; Pietrucha, R.; Gruen, M.; Breitling, R.; Alexandrov, K. Isotopic labeling of recombinant proteins expressed in the protozoan host Leishmania tarentolae. Protein Expr. Purif. 2006, 48, 167–172. [Google Scholar] [CrossRef]
- Foldynová-Trantírková, S.; Matulová, J.; Dötsch, V.; Löhr, F.; Cirstea, I.; Alexandov, K.; Breitling, R.; Lukeš, J.; Trantírek, L. A Cost-effective Amino-acid-type Selective Isotope Labeling of Proteins Expressed in Leishmania tarentolae. J. Biomol. Struct. Dyn. 2009, 26, 755–761. [Google Scholar] [CrossRef]
- Nayak, A.; Akpunarlieva, S.; Barrett, M.; Burchmore, R. A defined medium for Leishmania culture allows definition of essential amino acids. Exp. Parasitol. 2018, 185, 39–52. [Google Scholar] [CrossRef]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, W.; Mao, Y.; Chen, Y.; Wang, S.; Zhong, Y.; Su, T.; Gong, M.; Du, D.; Lu, X.; et al. Site-specific N-glycosylation Characterization of Recombinant SARS-CoV-2 Spike Proteins. Mol. Cell. Proteom. 2021, 20, 100058. [Google Scholar] [CrossRef]
- Selcuk Unal, E.; Zhao, R.; Qiu, A.; Goldman, I.D. N-linked glycosylation and its impact on the electrophoretic mobility and function of the human proton-coupled folate transporter (HsPCFT). Biochim. Biophys. Acta Biomembr. 2008, 1778, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.B.; Radic’, Z.; Talley, T.T.; Molles, B.E.; Deerinck, T.; Tsigelny, I.; Taylor, P. Tryptophan fluorescence reveals conformational changes in the acetylcholine binding protein. J. Biol. Chem. 2002, 277, 41299–41302. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.B.; Sulzenbacher, G.; Huxford, T.; Marchot, P.; Taylor, P.; Bourne, Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005, 24, 3635–3646. [Google Scholar] [CrossRef]
- Basavarajappa, S.C.; Liu, A.R.; Bruchez, A.; Li, Z.; Suzart, V.G.; Liu, Z.; Chen, Y.; Xiao, T.S.; Buck, M.; Ramakrishnan, P. Trimeric receptor-binding domain of SARS-CoV-2 acts as a potent inhibitor of ACE2 receptor-mediated viral entry. iScience 2022, 25, 104716. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Liu, M.; Wang, C.; Xu, W.; Lan, Q.; Feng, S.; Qi, F.; Bao, L.; Du, L.; Liu, S.; et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell. Res. 2020, 30, 343–355. [Google Scholar] [CrossRef]
- Shin, H.J.; Ku, K.B.; Kim, H.S.; Moon, H.W.; Jeong, G.U.; Hwang, I.; Yoon, G.Y.; Lee, S.; Lee, S.; Ahn, D.G.; et al. Receptor-binding domain of SARS-CoV-2 spike protein efficiently inhibits SARS-CoV-2 infection and attachment to mouse lung. Int. J. Biol. Sci. 2021, 17, 3786–3794. [Google Scholar] [CrossRef]
- Gao, X.; Peng, S.; Mei, S.; Liang, K.; Khan, M.S.I.; Vong, E.G.; Zhan, J. Expression and functional identification of recombinant SARS-CoV-2 receptor binding domain (RBD) from E. coli system. Prep. Biochem. Biotechnol. 2022, 52, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Brindha, S.; Yoshizue, T.; Wongnak, R.; Takemae, H.; Oba, M.; Mizutani, T.; Kuroda, Y. An Escherichia coli Expressed Multi-Disulfide Bonded SARS-CoV-2 RBD Shows Native-like Biophysical Properties and Elicits Neutralizing Antisera in a Mouse Model. Int. J. Mol. Sci. 2022, 23, 15744. [Google Scholar] [CrossRef]
- Gstöttner, C.; Zhang, T.; Resemann, A.; Ruben, S.; Pengelley, S.; Suckau, D.; Welsink, T.; Wuhrer, M.; Domínguez-Vega, E. Structural and Functional Characterization of SARS-CoV-2 RBD Domains Produced in Mammalian Cells. Anal. Chem. 2021, 93, 6839–6847. [Google Scholar] [CrossRef]
- Merkuleva, I.A.; Shcherbakov, D.N.; Borgoyakova, M.B.; Shanshin, D.V.; Rudometov, A.P.; Karpenko, L.I.; Belenkaya, S.V.; Isaeva, A.A.; Nesmeyanova, V.S.; Kazachinskaia, E.I.; et al. Comparative Immunogenicity of the Recombinant Receptor-Binding Domain of Protein S SARS-CoV-2 Obtained in Prokaryotic and Mammalian Expression Systems. Vaccines 2022, 10, 96. [Google Scholar] [CrossRef]
- Shi, J.; Zheng, J.; Tai, W.; Verma, A.K.; Zhang, X.; Geng, Q.; Wang, G.; Guan, X.; Malisheni, M.M.; Odle, A.E.; et al. A Glycosylated RBD Protein Induces Enhanced Neutralizing Antibodies against Omicron and Other Variants with Improved Protection against SARS-CoV-2 Infection. J. Virol. 2022, 96, e0011822. [Google Scholar] [CrossRef] [PubMed]
- Rahnama, S.; Azimzadeh Irani, M.; Amininasab, M.; Ejtehadi, M.R. S494 O-glycosylation site on the SARS-CoV-2 RBD affects the virus affinity to ACE2 and its infectivity; a molecular dynamics study. Sci. Rep. 2021, 11, 15162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yang, Y.; Lan, J.; Wang, Z.; Gao, Y.; Li, X.; Mao, W.; Xie, J.; Mi, L.Z.; Zhang, X.; et al. Inducing enhanced neutralizing antibodies against broad SARS-CoV-2 variants through glycan-shielding multiple non-neutralizing epitopes of RBD. Front. Immunol. 2023, 14, 1259386. [Google Scholar] [CrossRef]
- Carlson, E.C.; Macsai, M.; Bertrand, S.; Bertrand, D.; Nau, J. The SARS-CoV-2 Virus and the Cholinergic System: Spike Protein Interaction with Human Nicotinic Acetylcholine Receptors and the Nicotinic Agonist Varenicline. Int. J. Mol. Sci. 2023, 24, 5597. [Google Scholar] [CrossRef]
- Bourne, Y.; Talley, T.T.; Hansen, S.B.; Taylor, P.; Marchot, P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. EMBO J. 2005, 24, 1512–1522. [Google Scholar] [CrossRef]
- Olsen, J.A.; Ahring, P.K.; Kastrup, J.S.; Gajhede, M.; Balle, T. Structural and functional studies of the modulator NS9283 reveal agonist-like mechanism of action at α4β2 nicotinic acetylcholine receptors. J. Biol. Chem. 2014, 289, 24911–24921. [Google Scholar] [CrossRef]
- McCormack, T.; Petrovich, R.M.; Mercier, K.A.; DeRose, E.F.; Cuneo, M.J.; Williams, J.; Johnson, K.L.; Lamb, P.W.; London, R.E.; Yakel, J.L. Identification and functional characterization of a novel acetylcholine-binding protein from the marine annelid Capitella teleta. Biochemistry 2010, 49, 2279–2287. [Google Scholar] [CrossRef][Green Version]
- Hansen, S.B.; Taylor, P. Galanthamine and non-competitive inhibitor binding to ACh-binding protein: Evidence for a binding site on non-alpha-subunit interfaces of heteromeric neuronal nicotinic receptors. J. Mol. Biol. 2007, 369, 895–901. [Google Scholar] [CrossRef]
- Hibbs, R.E.; Sulzenbacher, G.; Shi, J.; Talley, T.T.; Conrod, S.; Kem, W.R.; Taylor, P.; Marchot, P.; Bourne, Y. Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal α7 nicotinic acetylcholine receptor. EMBO J. 2009, 28, 3040–3051. [Google Scholar] [CrossRef]
- Delbart, F.; Brams, M.; Gruss, F.; Noppen, S.; Peigneur, S.; Boland, S.; Chaltin, P.; Brandao-Neto, J.; von Delft, F.; Touw, W.G.; et al. An allosteric binding site of the α7 nicotinic acetylcholine receptor revealed in a humanized acetylcholine-binding protein. J. Biol. Chem. 2018, 293, 2534–2545. [Google Scholar] [CrossRef] [PubMed]
- Dawson, A.; Trumper, P.; de Souza, J.O.; Parker, H.; Jones, M.J.; Hales, T.G.; Hunter, W.N. Engineering a surrogate human heteromeric α/β glycine receptor orthosteric site exploiting the structural homology and stability of acetylcholine-binding protein. IUCr J. 2019, 6 Pt 6, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Meng, H.; Bing, H.; Zhangsun, D.; Luo, S. Efficient expression of acetylcholine-binding protein from Aplysia californica in Bac-to-Bac system. Biomed. Res. Int. 2014, 2014, 691480. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Xu, M.; Zhu, X.; Wu, Y.; Liu, X.; Zhangsun, D.; Hu, Y.; Xiang, S.H.; Kasheverov, I.E.; Tsetlin, V.I.; et al. From crystal structure of α-conotoxin GIC in complex with Ac-AChBP to molecular determinants of its high selectivity for α3β2 nAChR. Sci. Rep. 2016, 6, 22349. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Pan, S.; Xu, M.; Zhang, L.; Yu, J.; Yu, J.; Wu, Y.; Fan, Y.; Li, H.; Kasheverov, I.E.; et al. High Selectivity of an α-Conotoxin LvIA Analogue for α3β2 Nicotinic Acetylcholine Receptors Is Mediated by β2 Functionally Important Residues. J. Med. Chem. 2020, 63, 13656–13668. [Google Scholar] [CrossRef] [PubMed]
- Nirthanan, S. Snake three-finger α-neurotoxins and nicotinic acetylcholine receptors: Molecules, mechanisms and medicine. Biochem. Pharmacol. 2020, 181, 114168. [Google Scholar] [CrossRef] [PubMed]
- Tsetlin, V.I.; Kasheverov, I.E.; Utkin, Y.N. Three-finger proteins from snakes and humans acting on nicotinic receptors: Old and new. J. Neurochem. 2021, 158, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Shoji, H.; Takahashi, N.; Nomoto, H.; Ishikawa, M.; Shimada, I.; Arata, Y.; Hayashi, K. Detailed structural analysis of asparagine-linked oligosaccharides of the nicotinic acetylcholine receptor from Torpedo californica. Eur. J. Biochem. 1992, 207, 631–641. [Google Scholar] [CrossRef]
- Chen, L. In pursuit of the high-resolution structure of nicotinic acetylcholine receptors. J. Physiol. 2010, 588 Pt 4, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Metzger, S.; Lochner, M.; Ruepp, M.D. The binding orientation of epibatidine at α7 nACh receptors. Neuropharmacology 2017, 116, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Li, S.X.; Huang, S.; Bren, N.; Noridomi, K.; Dellisanti, C.D.; Sine, S.M.; Chen, L. Ligand-binding domain of an α7-nicotinic receptor chimera and its complex with agonist. Nat. Neurosci. 2011, 14, 1253–1259. [Google Scholar] [CrossRef]
- Lazaridis, K.; Zisimopoulou, P.; Giastas, P.; Bitzopoulou, K.; Evangelakou, P.; Sideri, A.; Tzartos, S.J. Expression of human AChR extracellular domain mutants with improved characteristics. Int. J. Biol. Macromol. 2014, 63, 210–217. [Google Scholar] [CrossRef]
- Siniavin, A.E.; Streltsova, M.A.; Nikiforova, M.A.; Kudryavtsev, D.S.; Grinkina, S.D.; Gushchin, V.A.; Mozhaeva, V.A.; Starkov, V.G.; Osipov, A.V.; Lummis, S.C.R.; et al. Snake venom phospholipase A2s exhibit strong virucidal activity against SARS-CoV-2 and inhibit the viral spike glycoprotein interaction with ACE2. Cell Mol. Life Sci. 2021, 78, 7777–7794. [Google Scholar] [CrossRef]

| Stage | Fraction Volume, mL | Concentration (SDS-PAGE Evaluation), μg/mL | Total Protein Amount, μg |
|---|---|---|---|
| crude supernatant | 150 | ND | ND |
| affinity chromatography | 5 | 100 | 500 |
| ion-exchange chromatography | 4 | 85 | 340 |
| desalting and concentration | 1 | 270 1 | 270 1 |
| Stage | Fraction Volume, mL | Concentration (SDS-PAGE Evaluation), μg/mL | Total Protein Amount, μg |
|---|---|---|---|
| crude supernatant | 300 | ND | ND |
| affinity chromatography | 5 | 200 | 1000 |
| size-exclusion chromatography | 10 | 70 | 700 |
| concentration | 1 | 400 1 | 400 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, L.; Kost, V.; Maiorov, V.; Sukhov, D.; Arkhangelskaya, P.; Ivanov, I.; Kudryavtsev, D.; Siniavin, A.; Utkin, Y.; Kasheverov, I. Efficient Expression in Leishmania tarentolae (LEXSY) of the Receptor-Binding Domain of the SARS-CoV-2 S-Protein and the Acetylcholine-Binding Protein from Lymnaea stagnalis. Molecules 2024, 29, 943. https://doi.org/10.3390/molecules29050943
Son L, Kost V, Maiorov V, Sukhov D, Arkhangelskaya P, Ivanov I, Kudryavtsev D, Siniavin A, Utkin Y, Kasheverov I. Efficient Expression in Leishmania tarentolae (LEXSY) of the Receptor-Binding Domain of the SARS-CoV-2 S-Protein and the Acetylcholine-Binding Protein from Lymnaea stagnalis. Molecules. 2024; 29(5):943. https://doi.org/10.3390/molecules29050943
Chicago/Turabian StyleSon, Lina, Vladimir Kost, Valery Maiorov, Dmitry Sukhov, Polina Arkhangelskaya, Igor Ivanov, Denis Kudryavtsev, Andrei Siniavin, Yuri Utkin, and Igor Kasheverov. 2024. "Efficient Expression in Leishmania tarentolae (LEXSY) of the Receptor-Binding Domain of the SARS-CoV-2 S-Protein and the Acetylcholine-Binding Protein from Lymnaea stagnalis" Molecules 29, no. 5: 943. https://doi.org/10.3390/molecules29050943
APA StyleSon, L., Kost, V., Maiorov, V., Sukhov, D., Arkhangelskaya, P., Ivanov, I., Kudryavtsev, D., Siniavin, A., Utkin, Y., & Kasheverov, I. (2024). Efficient Expression in Leishmania tarentolae (LEXSY) of the Receptor-Binding Domain of the SARS-CoV-2 S-Protein and the Acetylcholine-Binding Protein from Lymnaea stagnalis. Molecules, 29(5), 943. https://doi.org/10.3390/molecules29050943








