Peptide Self-Assembly Facilitating DNA Transfection and the Application in Inhibiting Cancer Cells
Abstract
1. Introduction
2. Results
2.1. Subsection
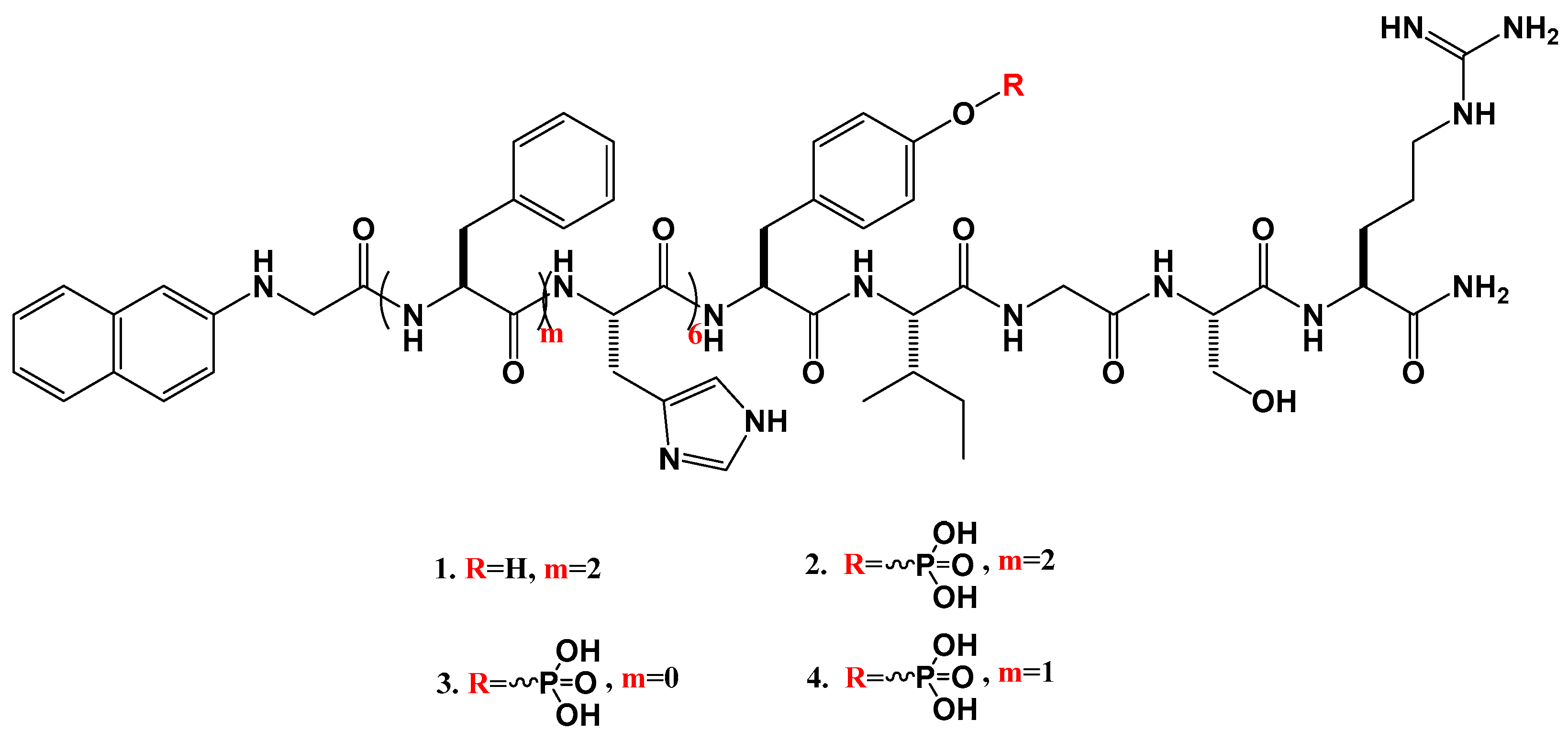
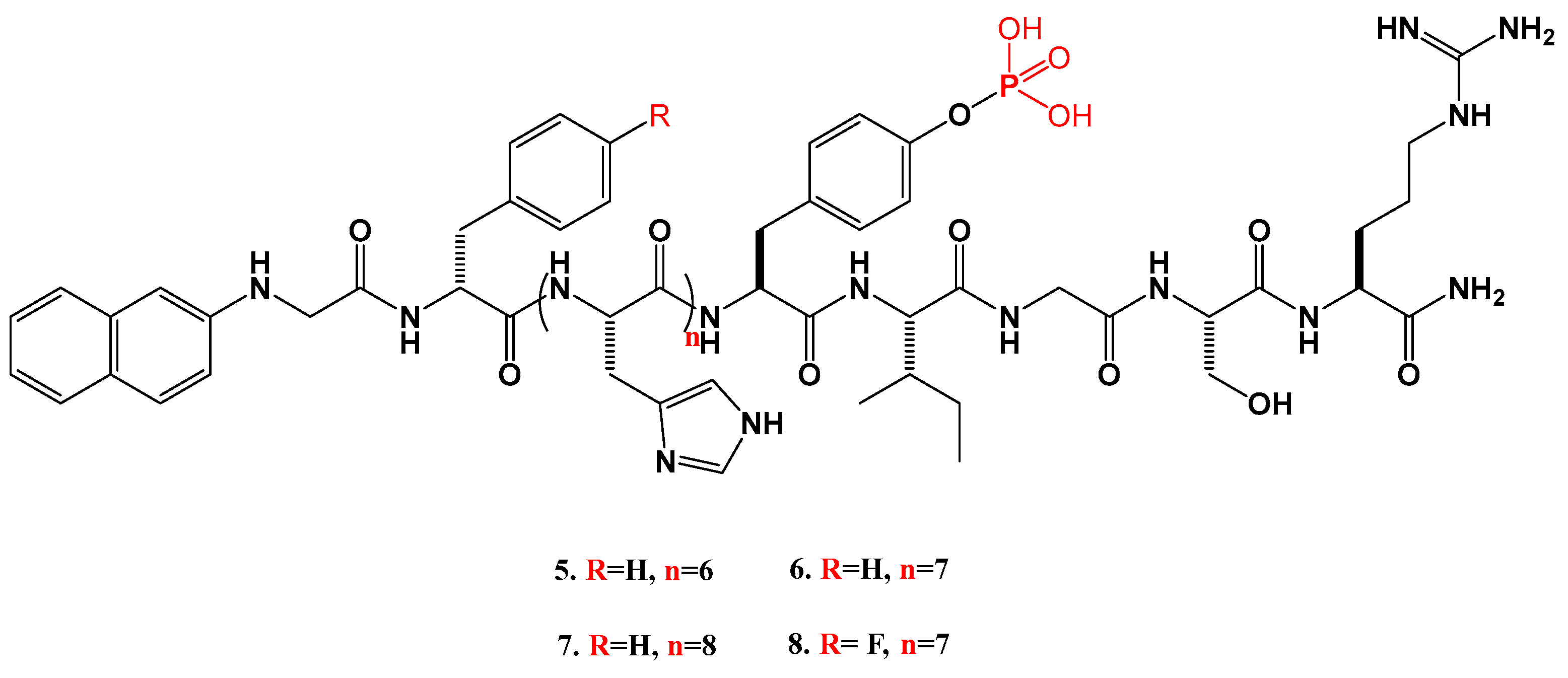
2.1.1. Synthesis of the Peptides
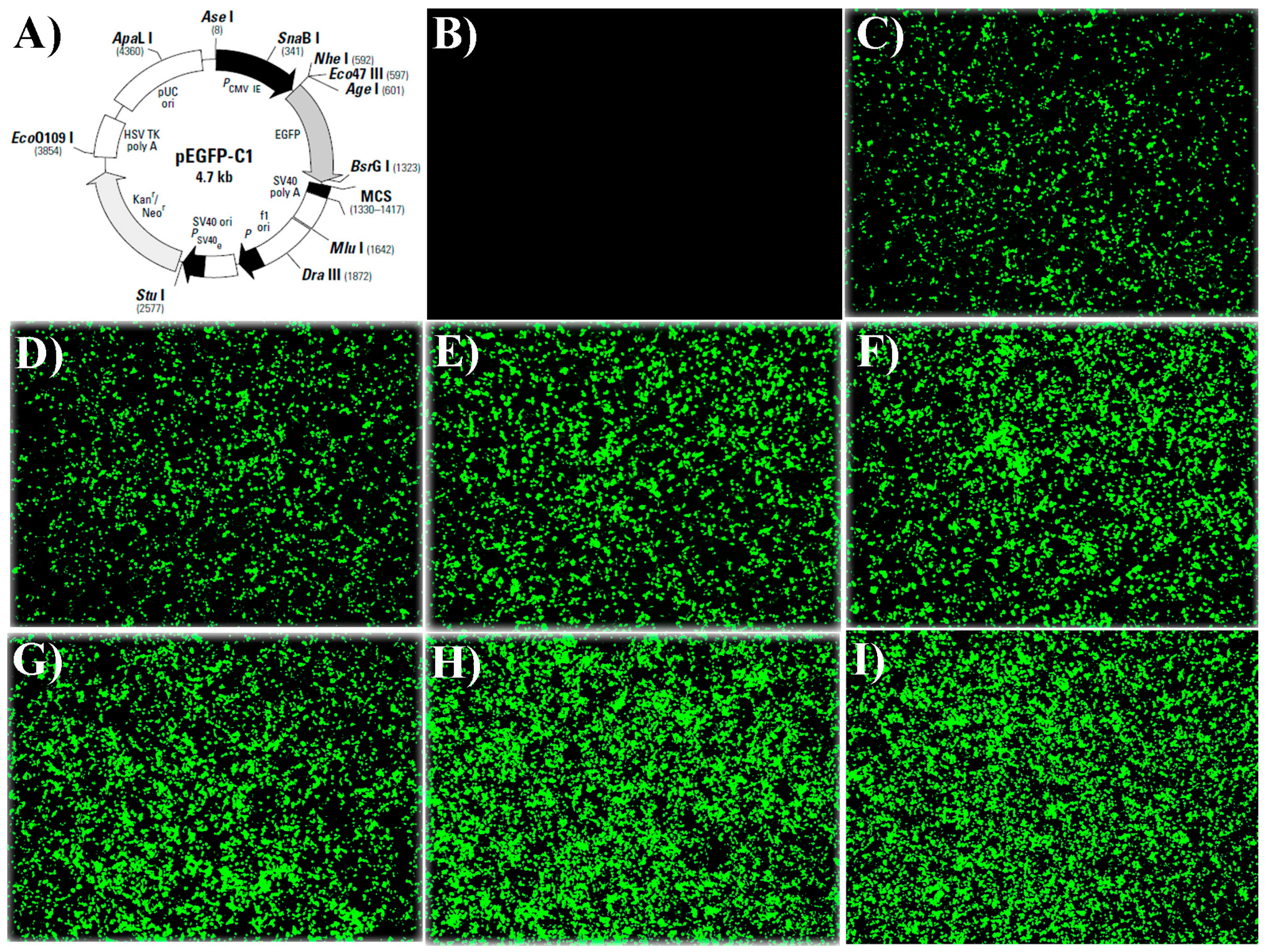
2.1.2. Fluorescence Microscope Results
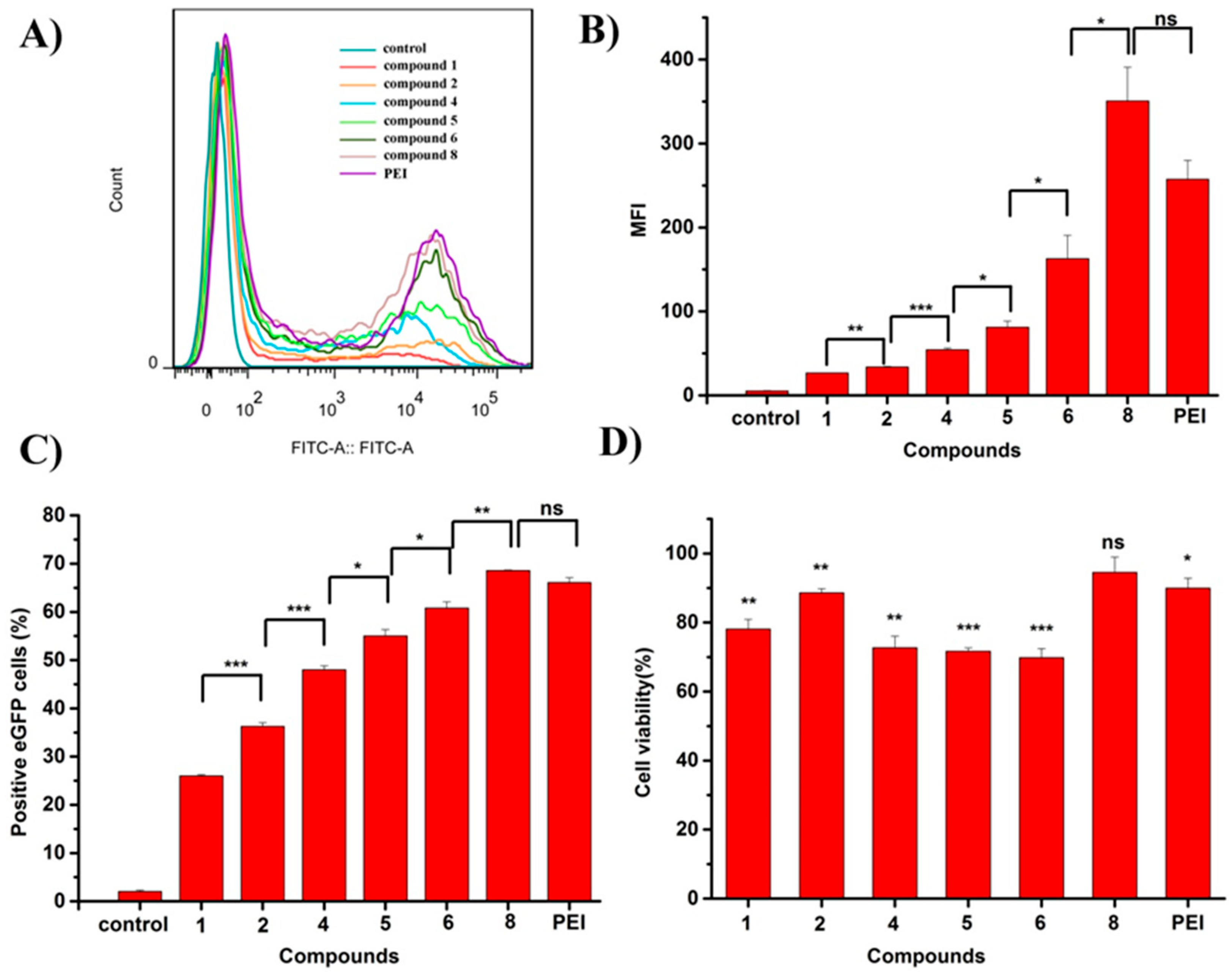
2.1.3. Flow Cytometry Results and Cytocompatibility
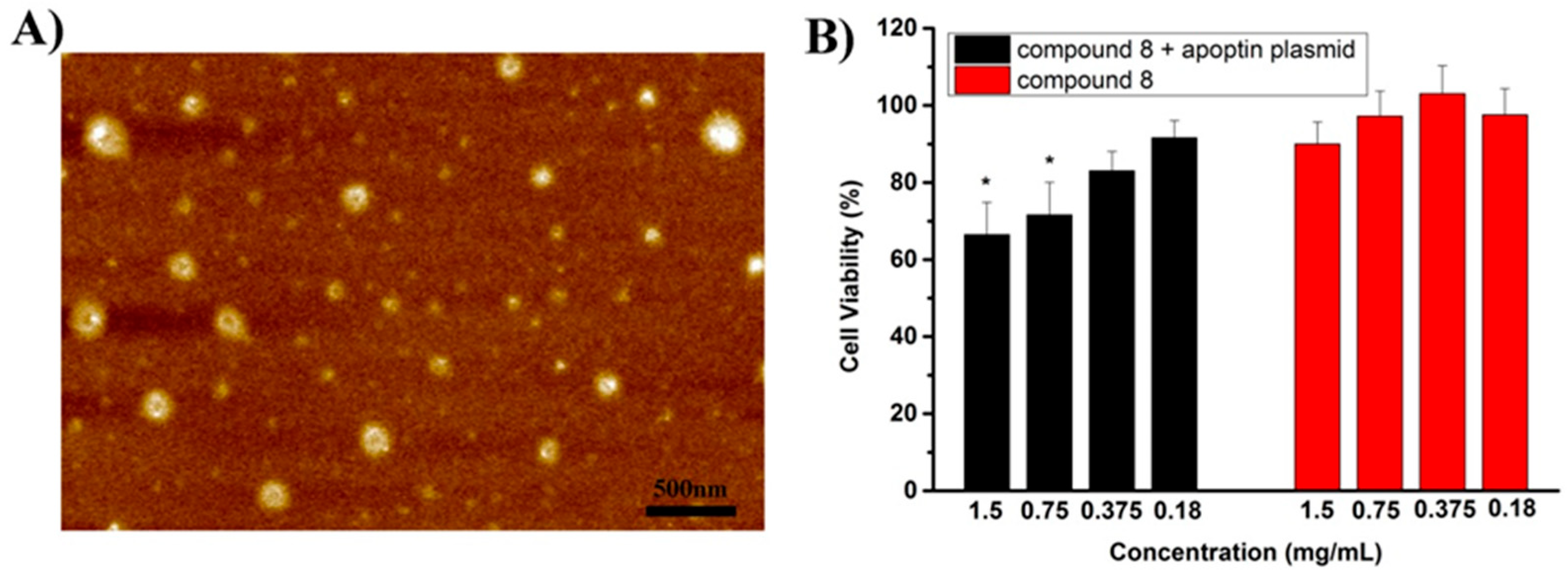
2.1.4. AFM Image and the Inhibitory Experiment
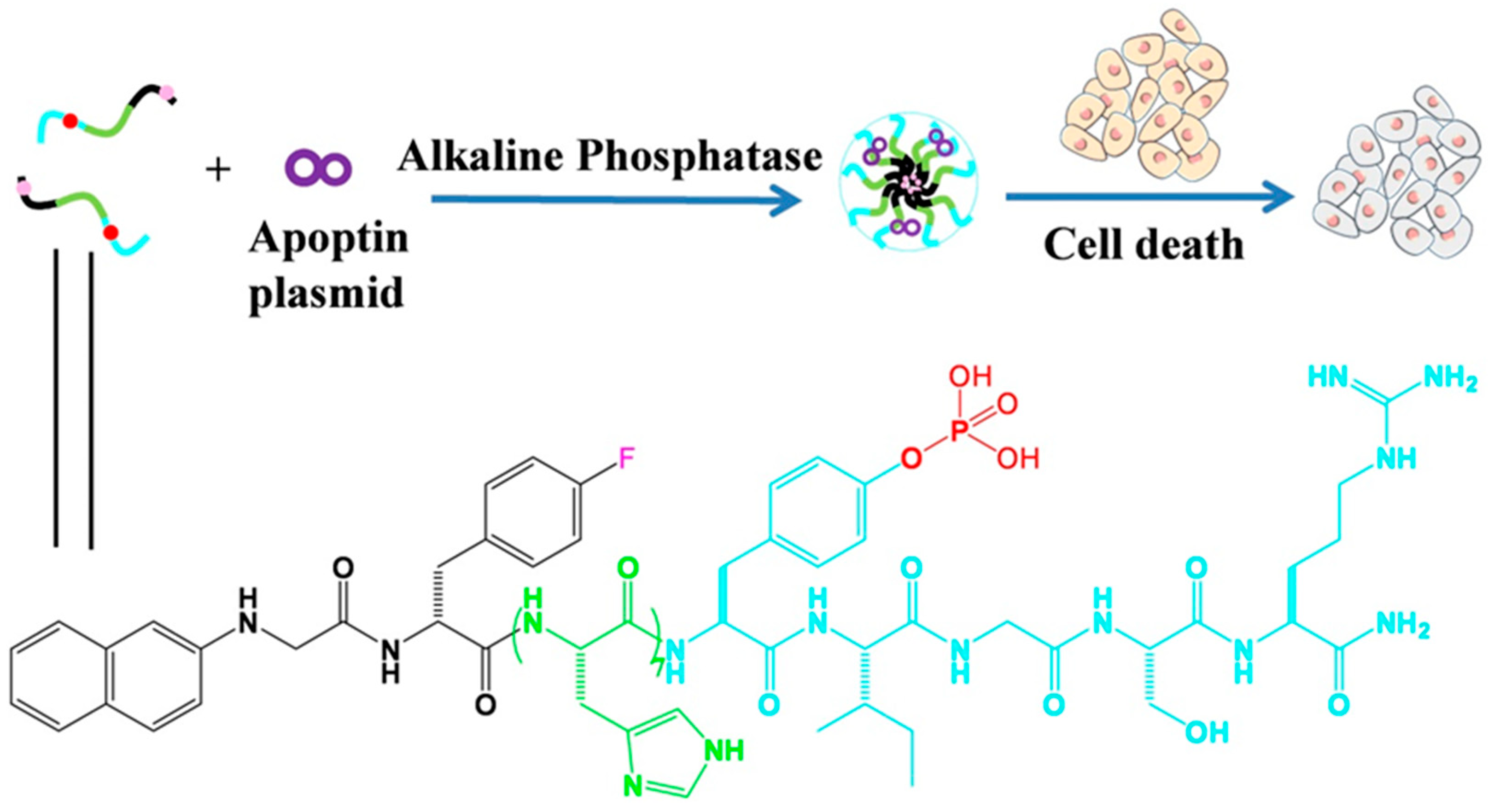
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. General Methods
4.3. Syntheses and Characterizations
4.3.1. Peptide Synthesis
4.3.2. Gene Transfection
4.3.3. Flow Cytometry
4.3.4. Cytotoxicity Assay after Transfection
4.3.5. AFM
4.3.6. Cell Viability after Inhibitory Experiment
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gao, X.; He, Z.; Ni, W.; Jian, X.; Hu, C.; Zhao, Y.; Yan, Y.; Wei, X. Layer-by-Layer Assembly of Functional Nanoparticles for Hepatocellular Carcinoma Therapy. Adv. Funct. Mater. 2019, 29, 1–15. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z. Molecular cloning and characterization of LCRG1, a novel gene localized to the tumor suppressor locus D17S800–D17S930. Cancer Lett. 2004, 209, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Lehrman, S. Virus treatment questioned after gene therapy death. Nature 1999, 401, 517–518. [Google Scholar] [CrossRef] [PubMed]
- Ratner, M. Heart failure gene therapy disappoints but experts keep the faith. Nat. Biotechnol. 2015, 33, 573–574. [Google Scholar] [CrossRef] [PubMed]
- Monaco, L.; Faccio, L. Patient-driven search for rare disease therapies: The Fondazione Telethon success story and the strategy leading to Strimvelis. EMBO Mol. Med. 2017, 9, 289–292. [Google Scholar] [CrossRef]
- Lu, J.; Jiang, G. The journey of CAR-T therapy in hematological malignancies. Mol. Cancer 2022, 21, 1–15. [Google Scholar] [CrossRef]
- Keeler, A.M.; Flotte, T.R. Recombinant Adeno-Associated Virus Gene Therapy in Light of Luxturna (and Zolgensma and Glybera): Where Are We, and How Did We Get Here? Annu. Rev. Virol. 2019, 6, 601–621. [Google Scholar] [CrossRef]
- Hoy, S.M. Onasemnogene Abeparvovec: First Global Approval. Drugs 2019, 79, 1255–1262. [Google Scholar] [CrossRef]
- Lee, A. Nadofaragene Firadenovec: First Approval. Drugs 2023, 83, 353–357. [Google Scholar] [CrossRef]
- De Wolf, D.; Singh, K.; Chuah, M.K.; Vanden Driessche, T. Hemophilia Gene Therapy: The End of the Beginning? Hum. Gene Ther. 2023, 34, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Kamihira, M. Development of hybrid viral vectors for gene therapy. Biotechnol. Adv. 2013, 31, 208–223. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-C.; Tsai, Y.-H.; Chan, Y.-H.; Hu, C.-J.; Huang, C.-Y.; Xiao, R.; Hsu, C.-J.; Vandenberghe, L.H.; Wu, C.-C.; Cheng, Y.-F. Gene therapy with a synthetic adeno-associated viral vector improves audiovestibular phenotypes in Pjvk-mutant mice. J. Clin. Investig. 2022, 7, 152941. [Google Scholar] [CrossRef] [PubMed]
- Shirley, J.L.; de Jong, Y.P.; Terhorst, C.; Herzog, R.W. Immune Responses to Viral Gene Therapy Vectors. Mol. Ther. 2020, 28, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef]
- Yonezawa, S.; Koide, H.; Asai, T. Recent advances in siRNA delivery mediated by lipid-based nanoparticles. Adv. Drug Deliv. Rev. 2020, 154–155, 64–78. [Google Scholar] [CrossRef]
- Lamb, Y.N. Inclisiran: First Approval. Drugs 2021, 81, 389–395. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Zhan, J.; Cai, Y.; He, S.; Wang, L.; Yang, Z. Tandem Molecular Self-Assembly in Liver Cancer Cells. Angew. Chem. Int. Ed. Engl. 2018, 57, 1813–1816. [Google Scholar] [CrossRef]
- He, H.; Guo, J.; Lin, X.; Xu, B. Enzyme-Instructed Assemblies Enable Mitochondria Localization of Histone H2B in Cancer Cells. Angew. Chem. Int. Ed. 2020, 59, 9330–9334. [Google Scholar] [CrossRef]
- He, H.; Lin, X.; Guo, J.; Wang, J.; Xu, B. Perimitochondrial Enzymatic Self-Assembly for Selective Targeting the Mitochondria of Cancer Cells. ACS Nano 2020, 14, 6947–6955. [Google Scholar] [CrossRef]
- Dong, Y.; Chen, Y.; Zhu, D.; Shi, K.; Ma, C.; Zhang, W.; Rocchi, P.; Jiang, L.; Liu, X. Self-assembly of amphiphilic phospholipid peptide dendrimer-based nanovectors for effective delivery of siRNA therapeutics in prostate cancer therapy. J. Control Release 2020, 322, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Lei, Q.; Jia, H.; Wang, S.; Yin, W.; Chen, W.; Cheng, S.; Zhang, X. A Tumor Targeted Chimeric Peptide for Synergistic Endosomal Escape and Therapy by Dual-Stage Light Manipulation. Adv. Funct. Mater. 2015, 25, 1248–1257. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, H.; Liu, Y.; Mao, L.; Chen, W.; Zhu, Z.; Liu, W.; Zheng, W.; Zhao, Y.; Kong, D.; et al. A peptide-based nanofibrous hydrogel as a promising DNA nanovector for optimizing the efficacy of HIV vaccine. Nano Lett. 2014, 14, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Munder, A.; Hedtfeld, S.; Braubach, P.; Glage, S.; Zhang, L.; Lienenklaus, S.; Schultze, A.; Hasenpusch, G.; Garrels, W.; et al. Self-assembled peptide–poloxamine nanoparticles enable in vitro and in vivo genome restoration for cystic fibrosis. Nat. Nanotechnol. 2019, 14, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Zhan, J.; Xu, G.; Chen, Y.; Qin, Q.; Liao, X.; Ma, S.; Yang, Z.; Cai, Y. Enzyme-Instructed Self-Assembly Enabled Monomer–Excimer Transition to Construct Higher Ordered Luminescent Supramolecular Assembly for Activity-based Bioimaging. Angew. Chem. Int. Ed. 2021, 60, 8121–8129. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Kang, Z.; Sun, C.; Yang, S.; Zhao, B.; Feng, S.; Meng, Q.; Liu, K. Enhanced gene transfection efficiency by use of peptide vectors containing laminin receptor-targeting sequence YIGSR. Nanoscale 2017, 10, 1215–1227. [Google Scholar] [CrossRef]
- Liang, C.; Zheng, D.; Shi, F.; Xu, T.; Yang, C.; Liu, J.; Wang, L.; Yang, Z. Enzyme-assisted peptide folding, assembly and anti-cancer properties. Nanoscale 2017, 9, 11987–11993. [Google Scholar] [CrossRef]
- Wang, H.; Yang, C.; Tan, M.; Wang, L.; Kong, D.; Yang, Z. A structure–gelation ability study in a short peptide-based ‘Super Hydrogelator’ system. Soft Matter 2011, 7, 3897–3905. [Google Scholar] [CrossRef]
- Yang, C.; Chu, L.; Zhang, Y.; Shi, Y.; Liu, J.; Liu, Q.; Fan, S.; Yang, Z.; Ding, D.; Kong, D.; et al. Dynamic biostability, biodistribution, and toxicity of l/d-peptide-based supramolecular nanofibers. ACS Appl. Mater. Interfaces 2015, 7, 2735–2744. [Google Scholar] [CrossRef] [PubMed]
- Porosk, L.; Arukuusk, P.; Põhako, K.; Kurrikoff, K.; Kiisholts, K.; Padari, K.; Pooga, M.; Langel, U. Enhancement of siRNA transfection by the optimization of fatty acid length and histidine content in the CPP. Biomater. Sci. 2019, 7, 4363–4374. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, H.; Li, L.; Cheng, Y. A fluorinated dendrimer achieves excellent gene transfection efficacy at extremely low nitrogen to phosphorus ratios. Nat. Commun. 2014, 5, 3053. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, Y.; Wang, Y.; Hu, J.; Li, T.; Liu, H.; Zhang, Q.; Cheng, Y. Self-Assembled Fluorodendrimers Combine the Features of Lipid and Polymeric Vectors in Gene Delivery. Angew. Chem. Int. Ed. Engl. 2015, 54, 11647–11651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Guo, T.; Liu, X.; Guo, S.; Wang, Y.; Zhu, B.; Zhang, M.; Gao, X.; Wang, J. Apoptin and apoptotic protease-activating factor 1 plasmid-assisted multi-functional nanoparticles in hepatocellular carcinoma therapy. Int. J. Biol. Macromol. 2023, 253, 126870. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Ye, M.; Zhu, B. Peptide Self-Assembly Facilitating DNA Transfection and the Application in Inhibiting Cancer Cells. Molecules 2024, 29, 932. https://doi.org/10.3390/molecules29050932
Wang J, Ye M, Zhu B. Peptide Self-Assembly Facilitating DNA Transfection and the Application in Inhibiting Cancer Cells. Molecules. 2024; 29(5):932. https://doi.org/10.3390/molecules29050932
Chicago/Turabian StyleWang, Jingyu, Min Ye, and Baokuan Zhu. 2024. "Peptide Self-Assembly Facilitating DNA Transfection and the Application in Inhibiting Cancer Cells" Molecules 29, no. 5: 932. https://doi.org/10.3390/molecules29050932
APA StyleWang, J., Ye, M., & Zhu, B. (2024). Peptide Self-Assembly Facilitating DNA Transfection and the Application in Inhibiting Cancer Cells. Molecules, 29(5), 932. https://doi.org/10.3390/molecules29050932



