SAR, Molecular Docking and Molecular Dynamic Simulation of Natural Inhibitors against SARS-CoV-2 Mpro Spike Protein
Abstract
1. Introduction
2. Results and Discussion
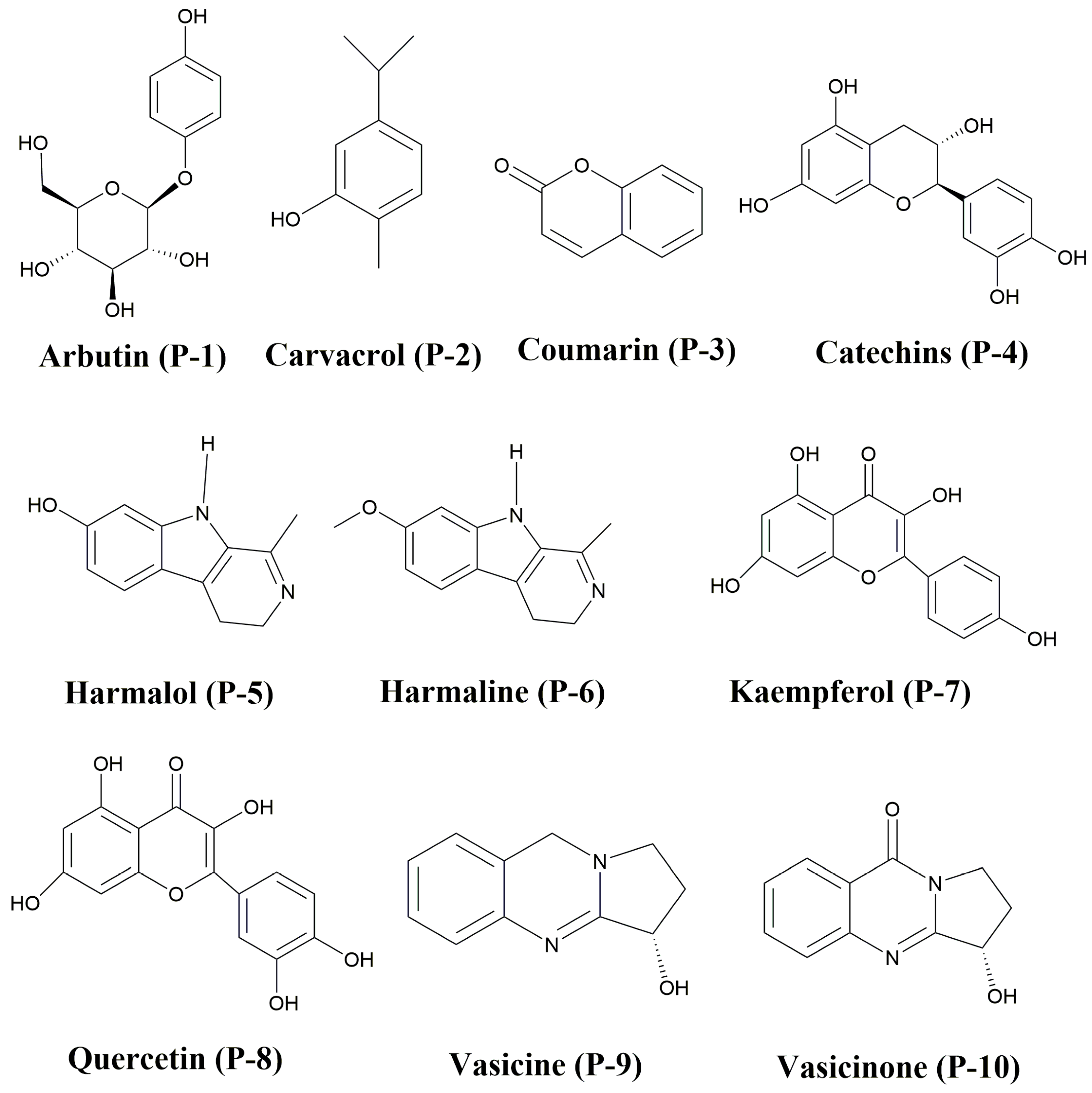
2.1. Ligand Modeling Studies (LMSs)
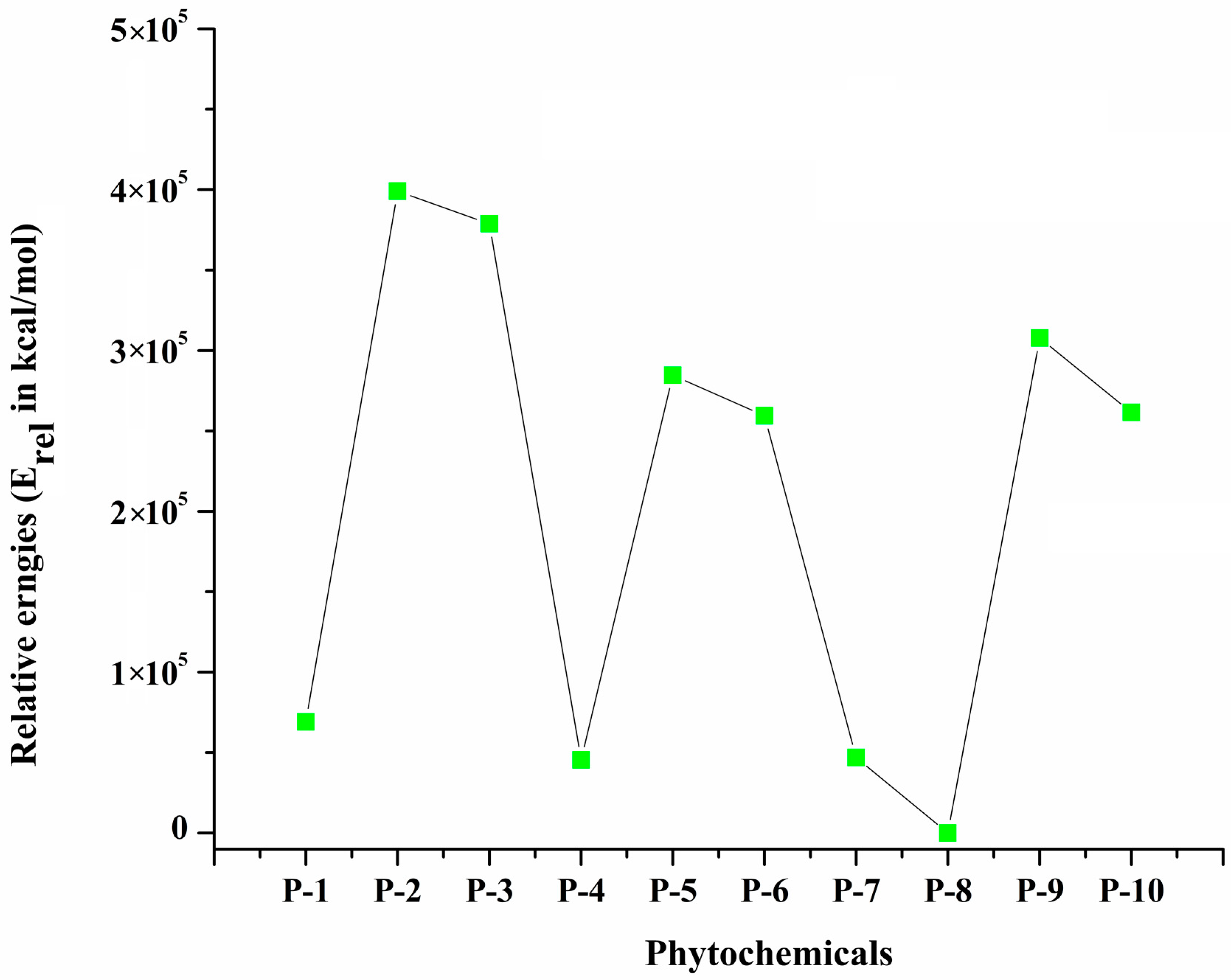
2.2. Frontiers Molecular Orbitals (FMOs) and Reactivity Descriptors of Selected Phytochemicals
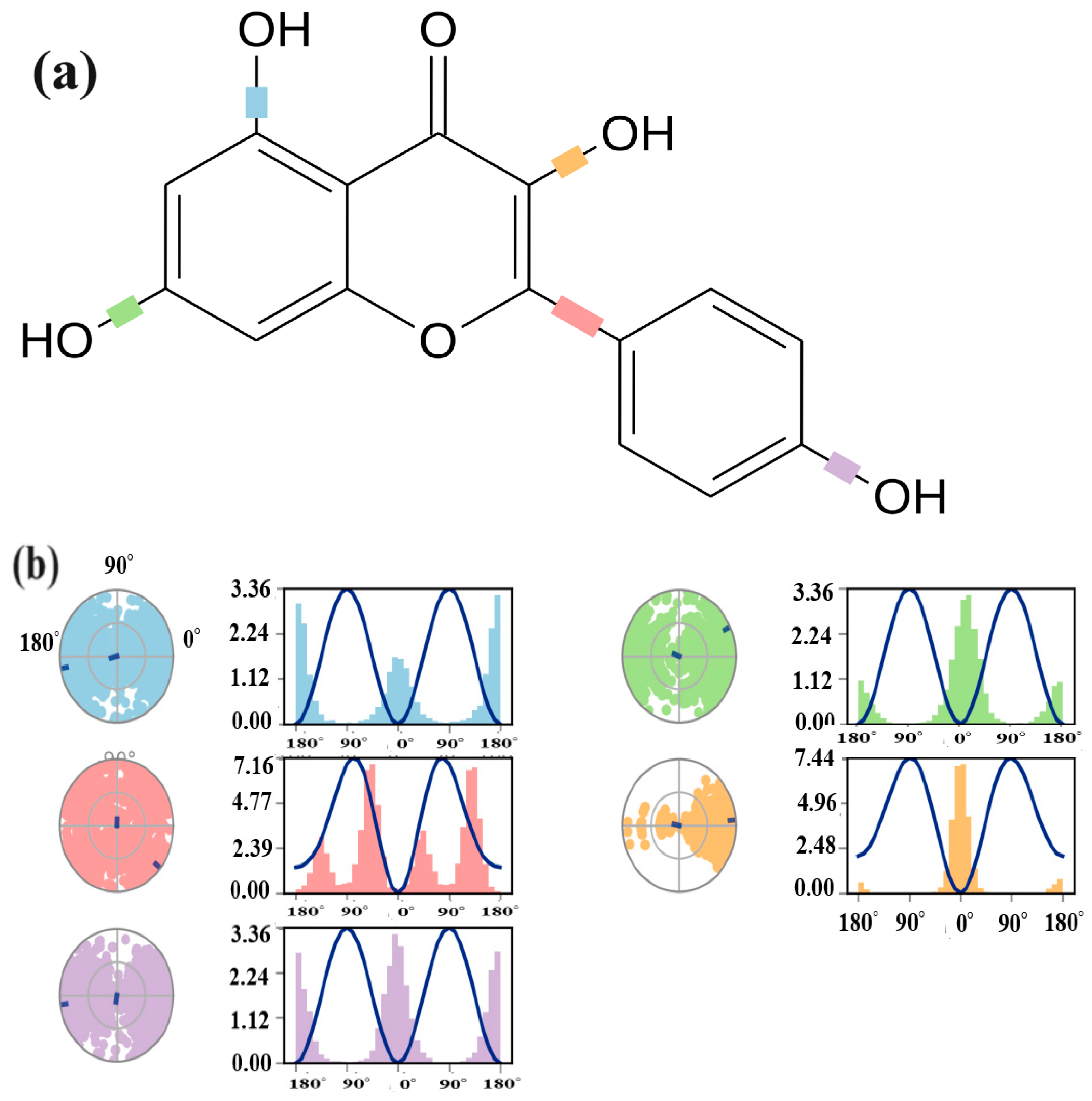
2.3. Structure and Activity Relationship (SAR) Analysis
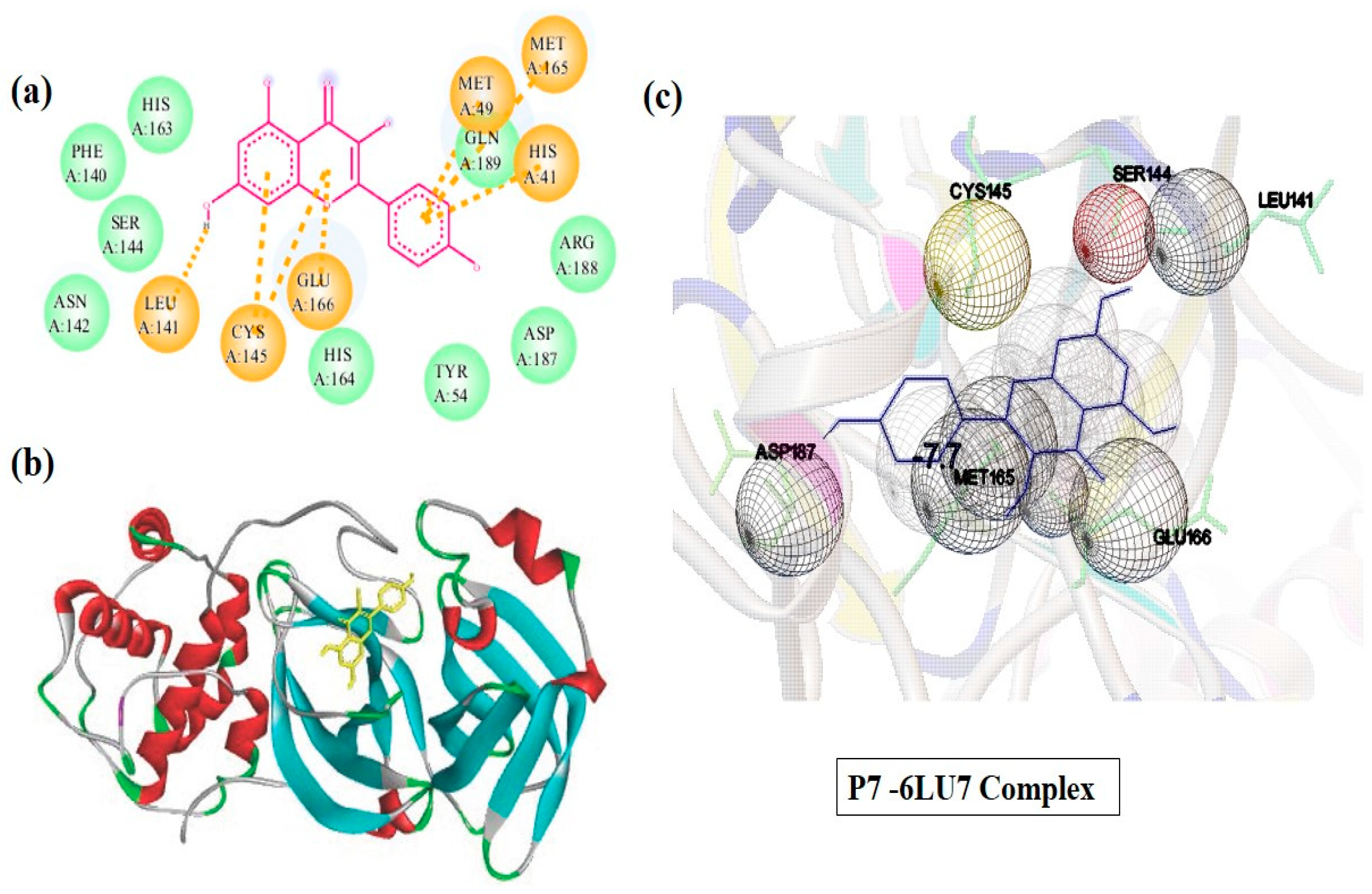
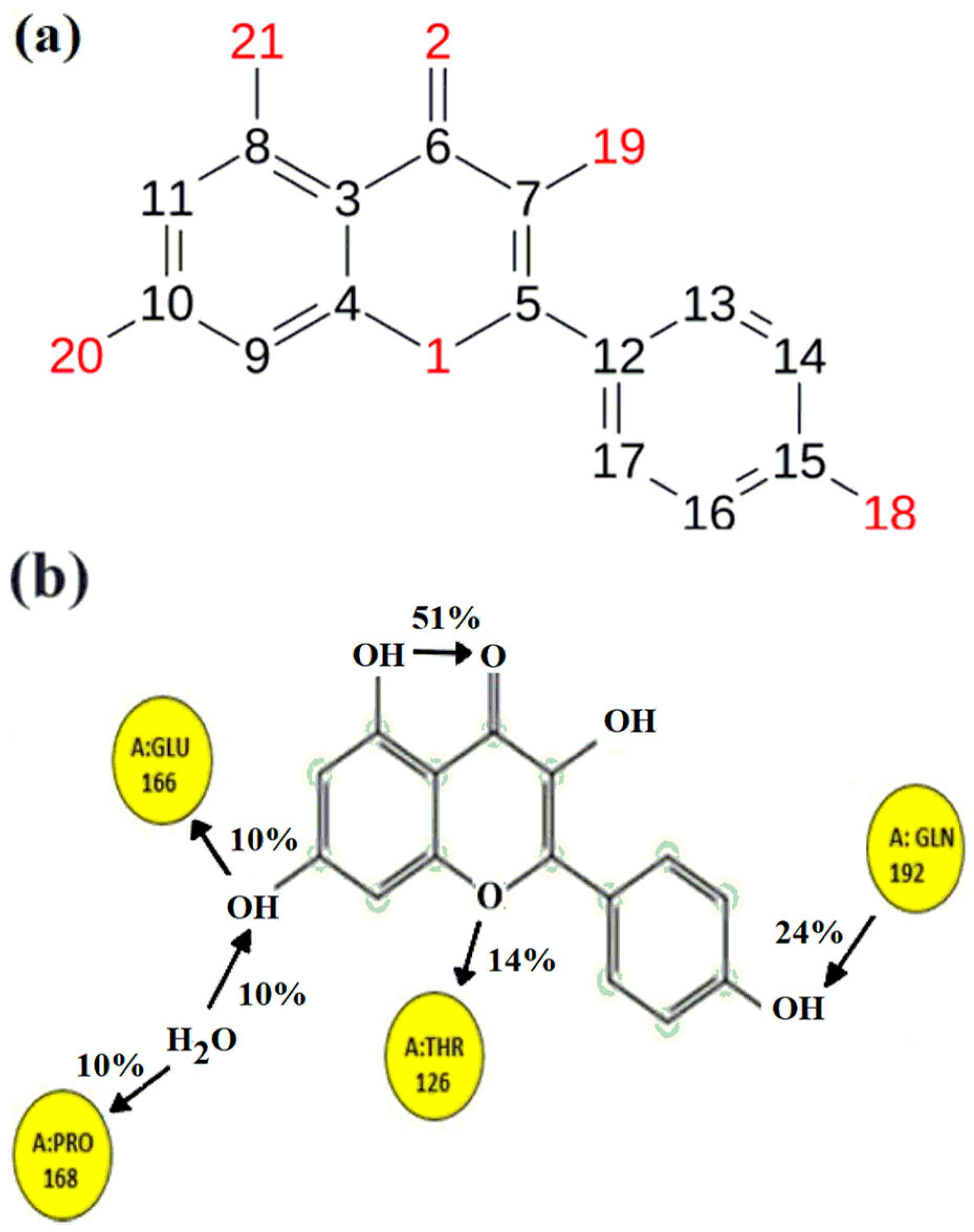
2.4. Molecular Docking Study
2.5. Pharmacokinetics and Pharmacodynamics Predictions of Selected Phytochemicals
2.6. Toxicity Analysis by ProTox
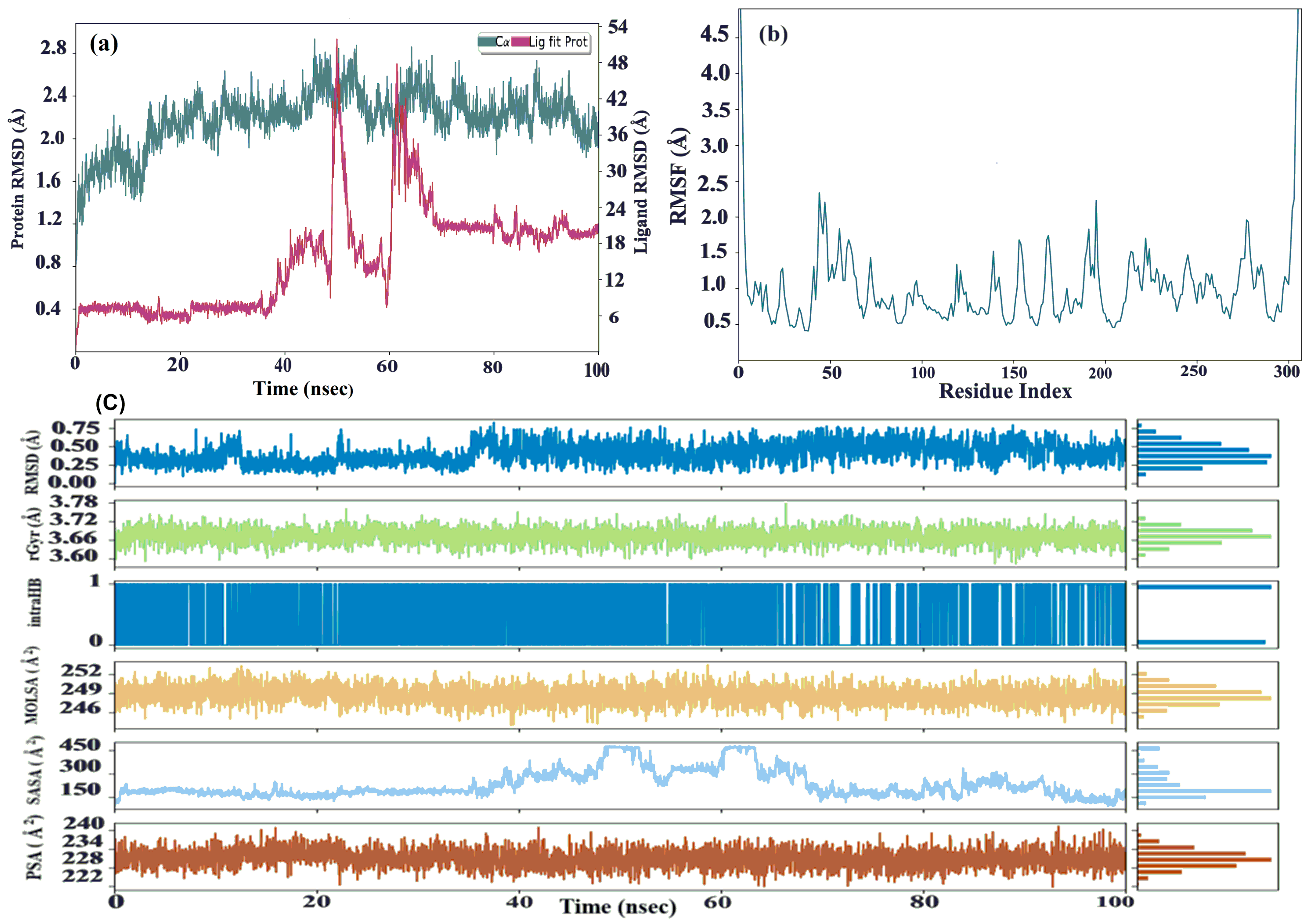
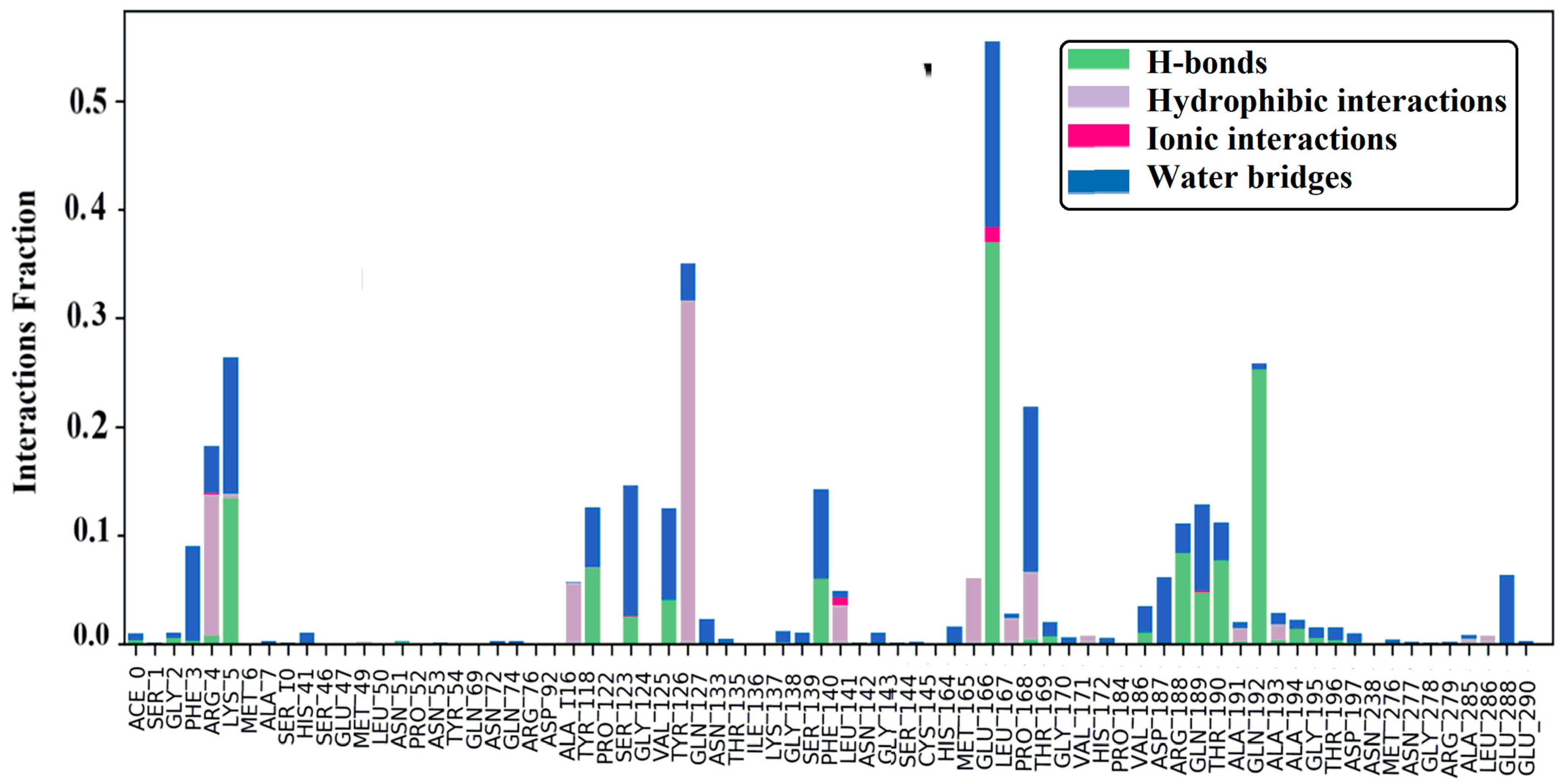
2.7. MD Simulation
3. Materials and Methods
3.1. Density Functional Theory (DFT) Methods
3.2. Molecular Docking Methods
3.3. Pharmacokinetic and Pharmacodynamics Investigations
3.4. Molecular Dynamic Simulation Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chojnacka, K.; Witek-Krowiak, A.; Skrzypczak, D.; Mikula, K.; Młynarz, P. Phytochemicals Containing Biologically Active Polyphenols as an Effective Agent against COVID-19-Inducing Coronavirus. J. Funct. Foods 2020, 73, 104146. [Google Scholar] [CrossRef] [PubMed]
- Ciotti, M.; Ciccozzi, M.; Terrinoni, A.; Jiang, W.-C.; Wang, C.-B.; Bernardini, S. The COVID-19 Pandemic. Crit. Rev. Clin. Lab. Sci. 2020, 57, 365–388. [Google Scholar] [CrossRef] [PubMed]
- Baj, J.; Karakuła-Juchnowicz, H.; Teresiński, G.; Buszewicz, G.; Ciesielka, M.; Sitarz, R.; Forma, A.; Karakuła, K.; Flieger, W.; Portincasa, P.; et al. COVID-19: Specific and Non-Specific Clinical Manifestations and Symptoms: The Current State of Knowledge. J. Clin. Med. 2020, 9, 1753. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, X.; Zhu, B.; Liang, H.; Fang, C.; Gong, Y.; Guo, Q.; Sun, X.; Zhao, D.; Shen, J.; et al. Characteristics of Pediatric SARS-CoV-2 Infection and Potential Evidence for Persistent Fecal Viral Shedding. Nat. Med. 2020, 26, 502–505. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, S.; Liu, J.; Zhang, Z.; Wan, X.; Huang, B.; Chen, Y.; Zhang, Y. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020, 5, 128. [Google Scholar] [CrossRef] [PubMed]
- Gangadaran, P.; Padinjarathil, H.; Rajendran, S.H.S.; Jogalekar, M.P.; Hong, C.M.; Aruchamy, B.; Rajendran, U.M.; Gurunagarajan, S.; Krishnan, A.; Ramani, P.; et al. COVID-19 and Diabetes: What Do We Know so Far? Exp. Biol. Med. 2022, 247, 1330–1334. [Google Scholar] [CrossRef] [PubMed]
- Gavriatopoulou, M.; Korompoki, E.; Fotiou, D.; Ntanasis-Stathopoulos, I.; Psaltopoulou, T.; Kastritis, E.; Terpos, E.; Dimopoulos, M.A. Organ-Specific Manifestations of COVID-19 Infection. Clin. Exp. Med. 2020, 20, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Krishnan, A.; Gangadaran, P.; Chavda, V.P.; Jogalekar, M.P.; Muthusamy, R.; Valu, D.; Vadivalagan, C.; Ramani, P.; Laishevtcev, A.; Katari, N.K.; et al. Convalescent Serum-Derived Exosomes: Attractive Niche as COVID-19 Diagnostic Tool and Vehicle for MRNA Delivery. Exp. Biol. Med. 2022, 247, 1244–1252. [Google Scholar] [CrossRef]
- Guljaš, S.; Bosnić, Z.; Salha, T.; Berecki, M.; Krivdić Dupan, Z.; Rudan, S.; Majnarić Trtica, L. Lack of Informations about COVID-19 Vaccine: From Implications to Intervention for Supporting Public Health Communications in COVID-19 Pandemic. Int. J. Environ. Res. Public Health 2021, 18, 6141. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, N.; Yang, L.; Song, X. Bioactive Natural Products in COVID-19 Therapy. Front. Pharmacol. 2022, 13, 926507. [Google Scholar] [CrossRef] [PubMed]
- Attah, A.F.; Fagbemi, A.A.; Olubiyi, O.; Dada-Adegbola, H.; Oluwadotun, A.; Elujoba, A.; Babalola, C.P. Therapeutic Potentials of Antiviral Plants Used in Traditional African Medicine with COVID-19 in Focus: A Nigerian Perspective. Front. Pharmacol. 2021, 12, 596855. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.A.; Urban, S.; Roessner, U. A Historical Overview of Natural Products in Drug Discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef] [PubMed]
- Petkou, D.; Diamantidis, G.; Vasilakakis, M. Arbutin Oxidation by Pear (Pyrus communis L.) Peroxidases. Plant Sci. 2002, 162, 115–119. [Google Scholar] [CrossRef]
- Maltas, E.; Yildiz, S. Evaluation of Phytochemicals and Antioxidant Activity of Ginkgo Biloba from Turkey. Pharmacologia 2012, 3, 113–120. [Google Scholar] [CrossRef]
- Bhat, A.; Raveesha, K.A. Antifungal activity of Pimenta dioica (L.) merril an aromatic medicinal tree. Int. J. Pharm. Pharm. Sci. 2016, 8, 92. [Google Scholar] [CrossRef][Green Version]
- Jan, R.; Khan, M.; Asaf, S.; Lubna; Asif, S.; Kim, K.-M. Bioactivity and Therapeutic Potential of Kaempferol and Quercetin: New Insights for Plant and Human Health. Plants 2022, 11, 2623. [Google Scholar] [CrossRef]
- Shaheen, H.A.; Issa, M.Y. In Vitro and In Vivo Activity of Peganum harmala L. Alkaloids against Phytopathogenic Bacteria. Sci. Hortic. 2020, 264, 108940. [Google Scholar] [CrossRef]
- Vardhan, S.; Sahoo, S.K. In Silico ADMET and Molecular Docking Study on Searching Potential Inhibitors from Limonoids and Triterpenoids for COVID-19. Comput. Biol. Med. 2020, 124, 103936. [Google Scholar] [CrossRef]
- Irfan, A.; Imran, M.; Khalid, N.; Hussain, R.; Basra, M.A.R.; Khaliq, T.; Shahzad, M.; Hussien, M.; Shah, A.T.; Qayyum, M.A.; et al. Isolation of Phytochemicals from Malva Neglecta Wallr and Their Quantum Chemical, Molecular Docking Exploration as Active Drugs against COVID-19. J. Saudi Chem. Soc. 2021, 25, 101358. [Google Scholar] [CrossRef]
- Singh, P.; Chauhan, S.S.; Pandit, S.; Sinha, M.; Gupta, S.; Gupta, A.; Parthasarathi, R. The Dual Role of Phytochemicals on SARS-CoV-2 Inhibition by Targeting Host and Viral Proteins. J. Tradit. Complement. Med. 2022, 12, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Neupane, N.P.; Karn, A.K.; Mukeri, I.H.; Pathak, P.; Kumar, P.; Singh, S.; Qureshi, I.A.; Jha, T.; Verma, A. Molecular Dynamics Analysis of Phytochemicals from Ageratina Adenophora against COVID-19 Main Protease (Mpro) and Human Angiotensin-Converting Enzyme 2 (ACE2). Biocatal. Agric. Biotechnol. 2021, 32, 101924. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Zia, K.; Ashraf, S.; Khan, A.; Ul-Haq, Z. Theoretical Investigation of Selective Ligand Binding Mode of Galanin Receptors. J. Biomol. Struct. Dyn. 2022, 40, 12964–12974. [Google Scholar] [CrossRef] [PubMed]
- Noureddine, O.; Issaoui, N.; Al-Dossary, O. DFT and Molecular Docking Study of Chloroquine Derivatives as Antiviral to Coronavirus COVID-19. J. King Saud Univ.-Sci. 2021, 33, 101248. [Google Scholar] [CrossRef] [PubMed]
- Sherif, Y.E.; Gabr, S.A.; Hosny, N.M.; Alghadir, A.H.; Alansari, R. Phytochemicals of Rhus Spp. as Potential Inhibitors of the SARS-CoV-2 Main Protease: Molecular Docking and Drug-Likeness Study. Evid.-Based Complement. Altern. Med. 2021, 2021, 8814890. [Google Scholar] [CrossRef]
- Belhassan, A.; Zaki, H.; Chtita, S.; Alaqarbeh, M.; Alsakhen, N.; Benlyas, M.; Lakhlifi, T.; Bouachrine, M. Camphor, Artemisinin and Sumac Phytochemicals as Inhibitors against COVID-19: Computational Approach. Comput. Biol. Med. 2021, 136, 104758. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhang, L.; Wang, L. Kaempferol, a Potential Neuroprotective Agent in Neurodegenerative Diseases: From Chemistry to Medicine. Biomed. Pharmacother. 2023, 165, 115215. [Google Scholar] [CrossRef]
- Arts, I.C.W.; Sesink, A.L.A.; Faassen-Peters, M.; Hollman, P.C.H. The Type of Sugar Moiety Is a Major Determinant of the Small Intestinal Uptake and Subsequent Biliary Excretion of Dietary Quercetin Glycosides. Br. J. Nutr. 2004, 91, 841–847. [Google Scholar] [CrossRef]
- Oladele, J.O.; Oyeleke, O.M.; Oladele, O.T.; Oladiji, A.T. COVID-19 Treatment: Investigation on the Phytochemical Constituents of Vernonia Amygdalina as Potential Coronavirus-2 Inhibitors. Comput. Toxicol. 2021, 18, 100161. [Google Scholar] [CrossRef]
- Dennington, R.; Keith, T.; Millam, J. GaussView, version 6.0.16; Semichem Inc.: Sahawnee Mission, KS, USA, 2019. [Google Scholar]
- Frisch, M.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G. Gaussian 09, Rev. D. 01; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Adnyana, I.M.D.M.; Sudaryati, N.L.G. Phytochemical Analysis of the Antioxidant Compounds of Baper Tea and Its Potential as an Immunomodulatory Agent and Candidate for Standardized Herbal Medicine. Trends Sci. 2022, 20, 6391. [Google Scholar] [CrossRef]
- Jin, Y.-H.; Cai, L.; Cheng, Z.-S.; Cheng, H.; Deng, T.; Fan, Y.-P.; Fang, C.; Huang, D.; Huang, L.-Q.; Huang, Q.; et al. A Rapid Advice Guideline for the Diagnosis and Treatment of 2019 Novel Coronavirus (2019-NCoV) Infected Pneumonia (Standard Version). Mil. Med. Res. 2020, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Rutwick Surya, U.; Praveen, N. A Molecular Docking Study of SARS-CoV-2 Main Protease against Phytochemicals of Boerhavia diffusa Linn. for Novel COVID-19 Drug Discovery. VirusDisease 2021, 32, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Elmaaty, A.A.; Darwish, K.M.; Khattab, M.; Elhady, S.S.; Salah, M.; Hamed, M.I.A.; Al-Karmalawy, A.A.; Saleh, M.M. In a Search for Potential Drug Candidates for Combating COVID-19: Computational Study Revealed Salvianolic Acid B as a Potential Therapeutic Targeting 3CLpro and Spike Proteins. J. Biomol. Struct. Dyn. 2022, 40, 8866–8893. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Aloufi, B.H.; Snoussi, M.; Sulieman, A.M.E. Antiviral Efficacy of Selected Natural Phytochemicals against SARS-CoV-2 Spike Glycoprotein Using Structure-Based Drug Designing. Molecules 2022, 27, 2401. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, H.; Tian, L.; Li, Q.; Luo, J.; Zhang, Y. Analysis of the Physicochemical Properties of Acaricides Based on Lipinski’s Rule of Five. J. Comput. Biol. 2020, 27, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Adel, A.; Elnaggar, M.S.; Albohy, A.; Elrashedy, A.A.; Mostafa, A.; Kutkat, O.; Abdelmohsen, U.R.; Al-Sayed, E.; Rabeh, M.A. Evaluation of Antiviral Activity of Carica Papaya Leaves against SARS-CoV-2 Assisted by Metabolomic Profiling. RSC Adv. 2022, 12, 32844–32852. [Google Scholar] [CrossRef]
- Zhu, K.; Borrelli, K.W.; Greenwood, J.R.; Day, T.; Abel, R.; Farid, R.S.; Harder, E. Docking Covalent Inhibitors: A Parameter Free Approach to Pose Prediction and Scoring. J. Chem. Inf. Model. 2014, 54, 1932–1940. [Google Scholar] [CrossRef]
| Phytochemicals Codes | (MF) | EHUMO | ELUMO | H-L Egap | η | µ | ω | N |
|---|---|---|---|---|---|---|---|---|
| P-1 | C12H16O7 | −5.63 | −0.02 | 5.60 | −2.80 | −2.83 | 2.02 | −5.60 |
| P-2 | C10H14O | −5.74 | −0.29 | 5.45 | −2.73 | −3.01 | 2.21 | −5.45 |
| P-3 | C15H14O6 | −6.52 | −1.91 | 4.61 | −2.31 | −4.21 | 3.65 | −4.61 |
| P-4 | C9H6O2 | −5.50 | −0.05 | 5.45 | −2.73 | −2.78 | 2.04 | −5.45 |
| P-5 | C12H12N2O | −5.68 | −0.79 | 4.89 | −2.45 | −3.24 | 2.65 | −4.89 |
| P-6 | C13H14N2O | −5.10 | −0.82 | 4.27 | −2.14 | −2.96 | 2.77 | −4.27 |
| P-7 | C15H10O6 | −5.54 | −1.92 | 3.62 | −1.81 | −3.73 | 4.12 | −3.62 |
| P-8 | C15H10O7 | −5.43 | −1.95 | 3.48 | −1.74 | −3.69 | 4.24 | −3.48 |
| P-9 | C11H12N2O | −5.27 | −0.50 | 4.77 | −2.39 | −2.89 | 2.42 | −4.77 |
| P-10 | C11H10N2O2 | −6.26 | −1.11 | 5.15 | −2.58 | −3.68 | 2.86 | −5.15 |
| Functions | P-1 | P-2 | P-3 | P-4 | P-5 | P-6 | P-7 | P-8 | P-9 | P-10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Surface area (Grid) (Å2) | 4.73 × 102 | 7.61 × 102 | 1.10 × 103 | 7.73 × 102 | 8.78 × 102 | 9.36 × 102 | 2.73 × 101 | 1.00 × 103 | 4.48 × 101 | 9.22 × 102 |
| Volume (Å3) | 7.83 × 102 | 1.45 × 103 | 2.14 × 103 | 1.28 × 103 | 1.77 × 103 | 1.84 × 103 | 2.02 × 103 | 2.02 × 103 | 2.15 × 101 | 1.75 × 103 |
| Hydration energy (kcal/mole) | −2.23 × 101 | −3.26 × 105 | −3.71 × 106 | −3.71 × 106 | −3.34 × 106 | −1.04 × 106 | −6.37 × 100 | −1.68 × 105 | 1.338 × 101 | −7.94 × 104 |
| Log P | −9.55 × 10−1 | −2.95 × 101 | −4.80 × 101 | −2.64 × 101 | −3.83 × 101 | −4.20 × 101 | −4.54 × 101 | −4.59 × 101 | −6.50 × 10−1 | −3.51 × 101 |
| Refractivity (Å3) | 6.34 × 101 | 8.74 × 101 | 1.36 × 102 | 7.23 × 101 | 1.09 × 102 | 1.19 × 102 | 1.29 × 102 | 1.33 × 102 | 1.60 × 100 | 1.03 × 102 |
| Polarizability (Å3) | 2.57 × 101 | 3.83 × 101 | 5.84 × 101 | 3.10 × 101 | 4.63 × 101 | 5.04 × 101 | 5.53 × 101 | 5.68 × 101 | 7.70 × 10−1 | 4.35 × 101 |
| Complex | Binding Affinity (kcal/mol) | ||
|---|---|---|---|
| 1st Most Stable Orientation | 2nd Most Stable Orientation | 3rd Most Stable Orientation | |
| P-1@6LU7 | −6.8 | −6.6 | −6.5 |
| P-2@6LU7 | −4.7 | −4.6 | −4.6 |
| P-3@6LU7 | −7.2 | −7.1 | −7.1 |
| P-4@6LU7 | −5.0 | −5.0 | −5.0 |
| P-5@6LU7 | −6.3 | −6.0 | −5.9 |
| P-6@6LU7 | −6.3 | −6.0 | −5.7 |
| P-7@6LU7 | −7.7 | −7.3 | −7.1 |
| P-8@6LU7 | −7.5 | −7.4 | −7.4 |
| P-9@6LU7 | −5.7 | −5.3 | −5.3 |
| P-10@6LU7 | −5.9 | −5.8 | −5.7 |
| Phytochemical’s Codes | MW | L | HBD | HBA | RB | SAscore | ESOL Log S | ESOL Solubility (mol/L) | GI | BBB |
|---|---|---|---|---|---|---|---|---|---|---|
| P-1 | 272.25 | 0 | 5 | 7 | 3 | 4.18 | −0.71 | 1.94 × 10−1 | High | No |
| P-2 | 150.22 | 0 | 1 | 1 | 1 | 1 | −3.31 | 4.92 × 10−4 | High | Yes |
| P-3 | 290.27 | 0 | 0 | 2 | 0 | 2.74 | −2.29 | 5.08 × 10−3 | High | Yes |
| P-4 | 146.14 | 0 | 5 | 6 | 1 | 3.50 | −2.22 | 5.08 × 10−3 | High | No |
| P-5 | 200.24 | 0 | 2 | 2 | 0 | 2.51 | −2.63 | 2.36 × 10−3 | High | Yes |
| P-6 | 214.26 | 0 | 1 | 2 | 1 | 2.58 | −2.82 | 1.50 × 10−3 | High | Yes |
| P-7 | 286.24 | 0 | 6 | 4 | 1 | 3.14 | −3.31 | 4.90 × 10−4 | High | No |
| P-8 | 302.24 | 0 | 5 | 7 | 1 | 3.23 | −3.16 | 6.98 × 10−4 | High | No |
| P-9 | 188.23 | 0 | 1 | 2 | 0 | 3.36 | −1.60 | 2.50 × 10−2 | High | No |
| P-10 | 202.21 | 0 | 1 | 3 | 0 | 2.75 | −1.91 | 1.22 × 10−2 | High | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salamat, A.; Kosar, N.; Mohyuddin, A.; Imran, M.; Zahid, M.N.; Mahmood, T. SAR, Molecular Docking and Molecular Dynamic Simulation of Natural Inhibitors against SARS-CoV-2 Mpro Spike Protein. Molecules 2024, 29, 1144. https://doi.org/10.3390/molecules29051144
Salamat A, Kosar N, Mohyuddin A, Imran M, Zahid MN, Mahmood T. SAR, Molecular Docking and Molecular Dynamic Simulation of Natural Inhibitors against SARS-CoV-2 Mpro Spike Protein. Molecules. 2024; 29(5):1144. https://doi.org/10.3390/molecules29051144
Chicago/Turabian StyleSalamat, Aqsa, Naveen Kosar, Ayesha Mohyuddin, Muhammad Imran, Muhammad Nauman Zahid, and Tariq Mahmood. 2024. "SAR, Molecular Docking and Molecular Dynamic Simulation of Natural Inhibitors against SARS-CoV-2 Mpro Spike Protein" Molecules 29, no. 5: 1144. https://doi.org/10.3390/molecules29051144
APA StyleSalamat, A., Kosar, N., Mohyuddin, A., Imran, M., Zahid, M. N., & Mahmood, T. (2024). SAR, Molecular Docking and Molecular Dynamic Simulation of Natural Inhibitors against SARS-CoV-2 Mpro Spike Protein. Molecules, 29(5), 1144. https://doi.org/10.3390/molecules29051144







