Phytochemicals, Health-Promoting Effects, and Enzyme Inhibition Traits of Phlomis stewartii Extracts
Abstract
1. Introduction
2. Results and Discussion
2.1. Methanolic Extracts
2.2. Model Fitting
2.3. Total Phenolic Contents (TPC)
2.4. Total Flavonoid Contents (TFC) of Methanolic Extracts
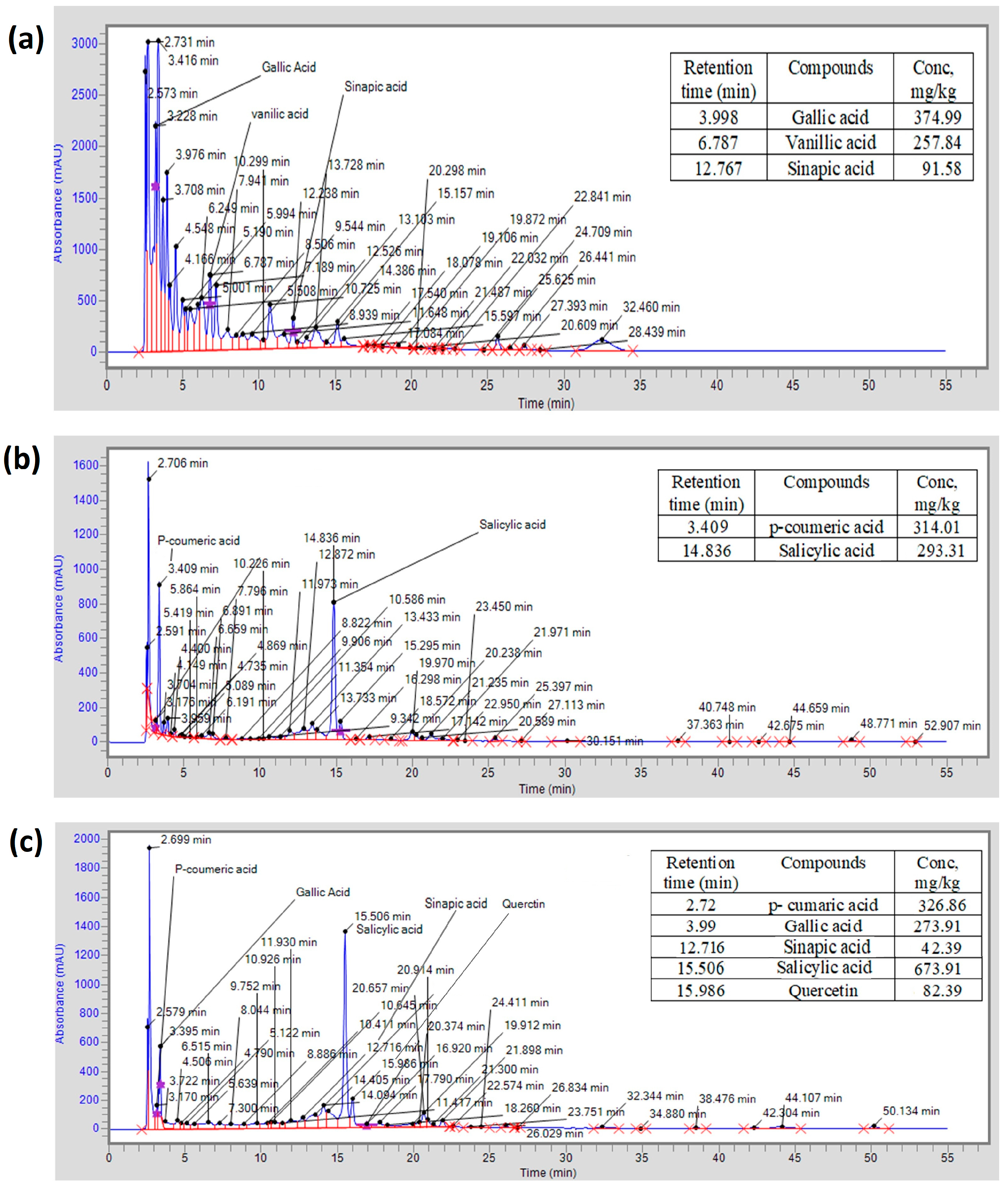
2.5. HPLC Analysis of Methanolic Extracts
2.6. Enzyme Inhibition
2.6.1. α-Amylase Enzyme Inhibition Activity
2.6.2. α-Glucosidase Inhibition Activity
3. Materials and Methods
3.1. Collection, Identification, and Preparation of Plant Parts
3.2. Preparation of Plant Extract
3.3. Total Phenolic Contents (TPC)
3.4. Total Flavonoid Content (TFC)
3.5. HPLC Analysis of Phenolic Acids
3.6. α-Amylase Inhibition Assessment
3.7. α-Glucosidase Inhibition Assay
3.8. Optimization Design
3.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Olufunmilayo, E.O.; Gerke-Duncan, M.B.; Holsinger, R.D. Oxidative Stress and Antioxidants in Neurodegenerative disorders. Antioxidants 2013, 12, 517. [Google Scholar] [CrossRef]
- Lourenço, S.C.; Moldão-Martins, M.; Alves, V.D. Antioxidants of natural plant origins: From sources to food industry applications. Molecules 2019, 24, 4132. [Google Scholar] [CrossRef]
- Abbas, A.; Naqvi, S.A.R.; Rasool, M.H.; Noureen, A.; Mubarik, M.S.; Tareen, R.B. Phytochemical Analysis, Antioxidant and Antimicrobial Screening of Seriphidium Oliverianum Plant Extracts. Dose Response 2021, 19, 15593258211004739. [Google Scholar] [CrossRef] [PubMed]
- Bitwell, C.; Sen, I.S.; Luke, C.; Kakoma, M.K. A review of modern and conventional extraction techniques and their applications for extracting phytochemicals from plants. Sci. Afr. 2023, 19, e01585. [Google Scholar] [CrossRef]
- Kovács, Z.; Molnár, C.; Štangar, U.L.; Cristea, V.M.; Pap, Z.; Hernadi, K.; Baia, L. Optimization Method of the Solvothermal Parameters Using Box–Behnken Experimental Design—The Case Study of ZnO Structural and Catalytic Tailoring. Nanomaterials 2022, 11, 1334. [Google Scholar] [CrossRef]
- Ullah, M.; Ullah, A.; Aman, A.; Afridi, H.H.; Naeem, M.A. Ethnomedicinal Importance of Family Lamiaceae and Family Rosaceae Among Local Communities of Dir Kohistan, Khyber Pakhtunkhwa, Pakistan. J. Bioresour. Manag. 2023, 10, 12. [Google Scholar]
- Farooq, A.; Ali, S.; Ullah, H.; Khan, A.; Jahan, N.; Agha, I.; Tareen, R.B. 30. Evaluation of antioxidant, antimicrobial activity and GC-MS analysis of Phlomis stewartii. Pure Appl. Biol. (PAB) 2019, 8, 2420–2433. [Google Scholar]
- Firuzi, O.; Javidnia, K.; Gholami, M.; Soltani, M.; Miri, R. Antioxidant activity and total phenolic content of 24 Lamiaceae species growing in Iran. Nat. Prod. Commun. 2010, 5, 193–199. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.Z. Phenolic composition and antioxidant activities of two Phlomis species: A correlation study. Comptes Rendus Biol. 2009, 332, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Morteza-Semnani, K.; Saeedi, M.; Mahdavi, M.R.; Rahimi, F. Antimicrobial studies on extracts of three species of Phlomis. Pharm. Biol. 2010, 44, 426–429. [Google Scholar] [CrossRef]
- Elshafie, H.S.; Camele, I.; Mohamed, A.A. A Comprehensive review on the biological, agricultural and pharmaceutical properties of secondary metabolites based-plant origin. Int. J. Mol. Sci. 2023, 24, 3266. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, I.; Mirghani, M.E.; Yusof, F. Effects of time, temperature, and solvent ratio on the extraction of non-extractable polyphenols with anticancer activity of barhi date palm kernels extracts using response surface methodology. Preprints 2019, 2019070055. [Google Scholar]
- Thouri, A.; Chahdoura, H.; El Arem, A.; Omri Hichri, A.; Ben Hassin, R.; Achour, L. Effect of solvents extraction on phytochemical components and biological activities of Tunisian date seeds (var. Korkobbi and Arechti). BMC Complement. Altern. Med. 2017, 17, 248. [Google Scholar] [CrossRef] [PubMed]
- Aydar, A.Y. Utilization of response surface methodology in optimization of extraction of plant materials. In Statistical Approaches with Emphasis on Design of Experiments Applied to Chemical Processes; InTech: London, UK, 2017; pp. 157–169. [Google Scholar]
- Amin, M.; Bhatti, H.N.; Sadaf, S.; Bilal, M. Optimization of lipase production by response surface methodology and its application for efficient biodegradation of polyester vylon-200. Catal. Lett. 2017, 151, 3603–3616. [Google Scholar] [CrossRef]
- Mallieswaran, K.; Padmanabhan, R.; Balasubramanian, V. Friction stir welding parameters optimization for tailored welded blank sheets of AA1100 with AA6061 dissimilar alloy using response surface methodology. Adv. Mater. Process. Technol. 2018, 4, 142–157. [Google Scholar] [CrossRef]
- Merouane, A.; Saadi, A.; Noui, A.; Bader, A. Evaluation of phenolic contents and antioxidant properties of the leaves and flowers of Phlomis biloba Desf. Int. Food Res. J. 2019, 26, 167–173. [Google Scholar]
- Eruygur, N.; Kirci, D.; Ayaz, F.; Dogu, S.; Bagci, Y. Biological activities of three Phlomis species. J. Res. Pharm. 2022, 26, 255–262. [Google Scholar] [CrossRef]
- Tarhan, L.; Urek, R.O.; Oner, A.; Nakiboglu, M. Evaluation of phenolic profiles, antioxidant activities, and cytotoxic and apoptotic potentials of Phlomis angustissima and Phlomis fruticosa, medicinal plants from Turkey. Eur. J. Integr. Med. 2022, 55. [Google Scholar] [CrossRef]
- Rahman, M.M.; Rahaman, M.S.; Islam, M.R.; Rahman, F.; Mithi, F.M.; Alqahtani, T.; Uddin, M.S. Role of phenolic compounds in human disease: Current knowledge and future prospects. Molecules 2021, 27, 233. [Google Scholar] [CrossRef]
- Chen, C. Sinapic acid and its derivatives as medicine in oxidative stress-induced diseases and aging. Oxidative Med. Cell. Longev. 2016, 7, 12–20. [Google Scholar] [CrossRef]
- Kahkeshani, N.; Farzaei, F.; Fotouhi, M.; Alavi, S.S.; Bahramsoltani, R.; Naseri, R.; Bishayee, A. Pharmacological effects of gallic acid in health and diseases: A mechanistic review. Iran. J. Basic Med. Sci. 2019, 22, 225–232. [Google Scholar]
- Ingole, A.; Kadam, M.P.; Dalu, A.P.; Kute, S.M.; Mange, P.R.; Theng, V.D.; Patil, P.A. A review of the pharmacological characteristics of vanillic acid. J. Drug Deliv. Ther. 2021, 11, 200–204. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, X.; Zhu, G.; Liu, H.; Chen, J.; Wang, Y.; He, X. Quercetin inhibits TNF-α induced HUVECs apoptosis and inflammation via downregulating NF-kB and AP-1 signaling pathway in vitro. Medicine 2020, 99, e22241. [Google Scholar] [CrossRef]
- Telagari, M.; Hullatti, K. In-vitro α-amylase and α-glucosidase inhibitory activity of Adiantum caudatum Linn. and Celosia argentea Linn. extracts and fractions. Indian J. Pharmacol. 2015, 47, 425. [Google Scholar]
- Savini, I.; Catani, M.V.; Evangelista, D.; Gasperi, V.; Avigliano, L. Obesity-associated oxidative stress: Strategies finalized to improve redox state. Int. J. Mol. Sci. 2013, 14, 10497–10538. [Google Scholar] [CrossRef]
- Zatterale, F.; Longo, M.; Naderi, J.; Raciti, G.A.; Desiderio, A.; Miele, C.; Beguinot, F. Chronic adipose tissue inflammation linking obesity to insulin resistance and type 2 diabetes. Front. Physiol. 2020, 10, 1607. [Google Scholar] [CrossRef] [PubMed]
- Jabeen, B.; Riaz, N.; Saleem, M.; Naveed, M.A.; Ashraf, M.; Alam, U.; Jabbar, A. Isolation of natural compounds from Phlomis stewartii showing α-glucosidase inhibitory activity. Phytochemistry 2013, 96, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Zhang, W.; Feng, F.; Zhang, Y.; Kang, W. α-Glucosidase inhibitors isolated from medicinal plants. Food Sci. Hum. Wellness 2014, 3, 136–174. [Google Scholar] [CrossRef]
- Naqvi, S.A.R.; Waseem, R.; Mahmood, N.; Hussain, Z.; Khan, Z.A.; Shahzad, S.A.; Hussain, A.I. Phenolic acid content, antioxidant properties, and antibacterial potential of flowers and fruits from selected Pakistani indigenous medicinal plants. Sci. Asia 2013, 39, 340–345. [Google Scholar] [CrossRef]
- Ying, X.; Wang, R.; Xu, J.; Zhang, W.; Li, H.; Zhang, C.; Li, F. HPLC determination of eight polyphenols in the leaves of Crataegus pinnatifida Bge. var. major. J. Chromatogr. Sci. 2009, 47, 201–205. [Google Scholar] [CrossRef]
- Bhutkar, M.A.; Bhise, S.B. In vitro assay of alpha-amylase inhibitory activity of some indigenous plants. Int. J. Chem. Sci. 2012, 10, 457–462. [Google Scholar]
- Montgomery, D.C. Design and Analysis of Experiments; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
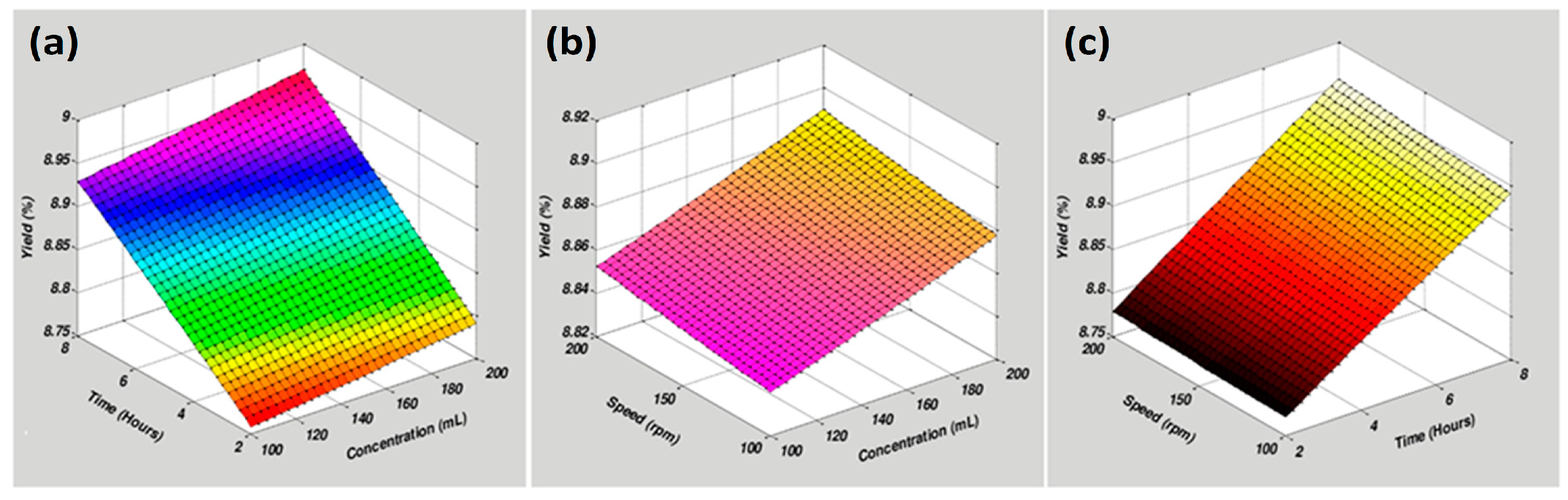
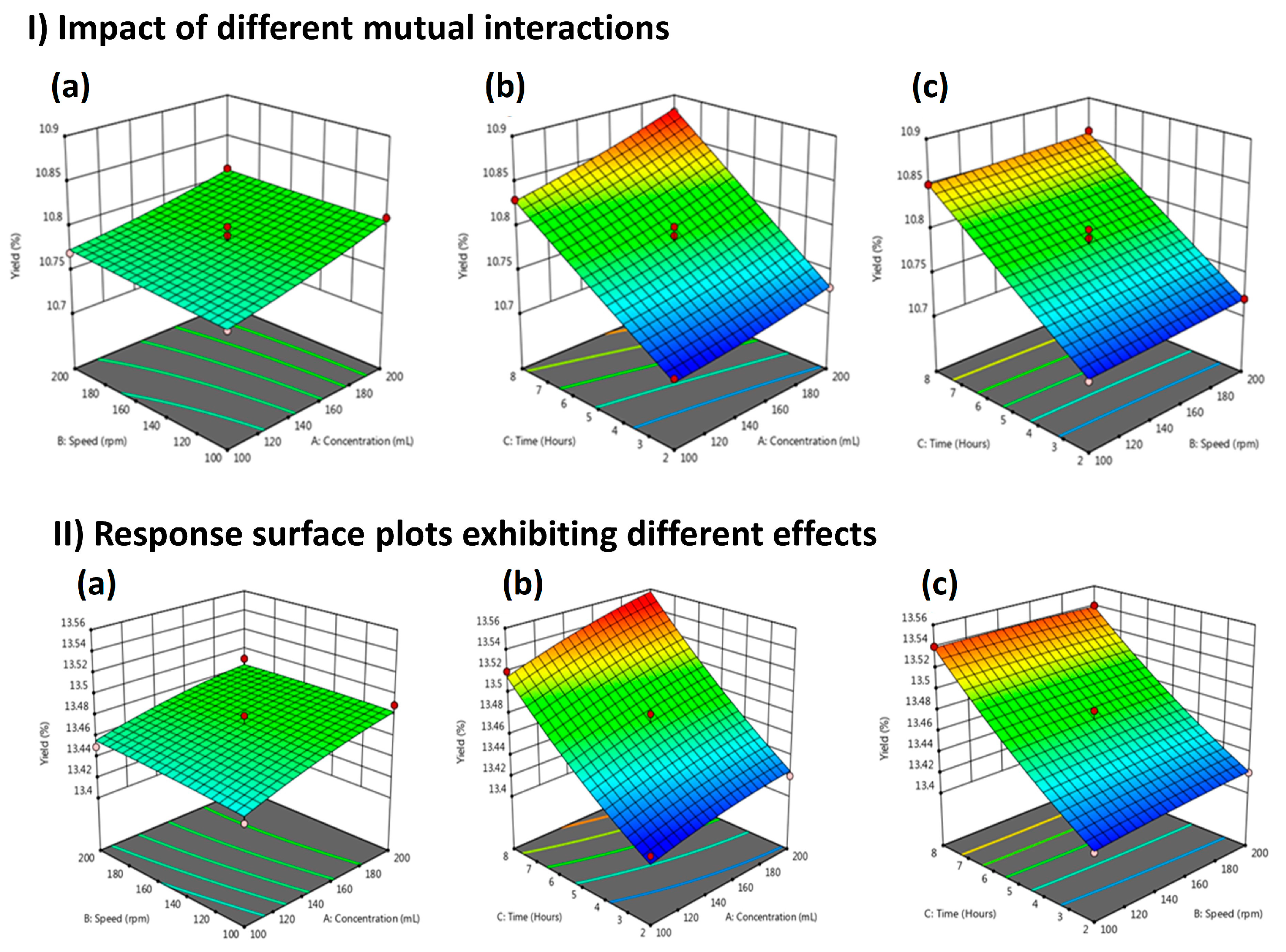
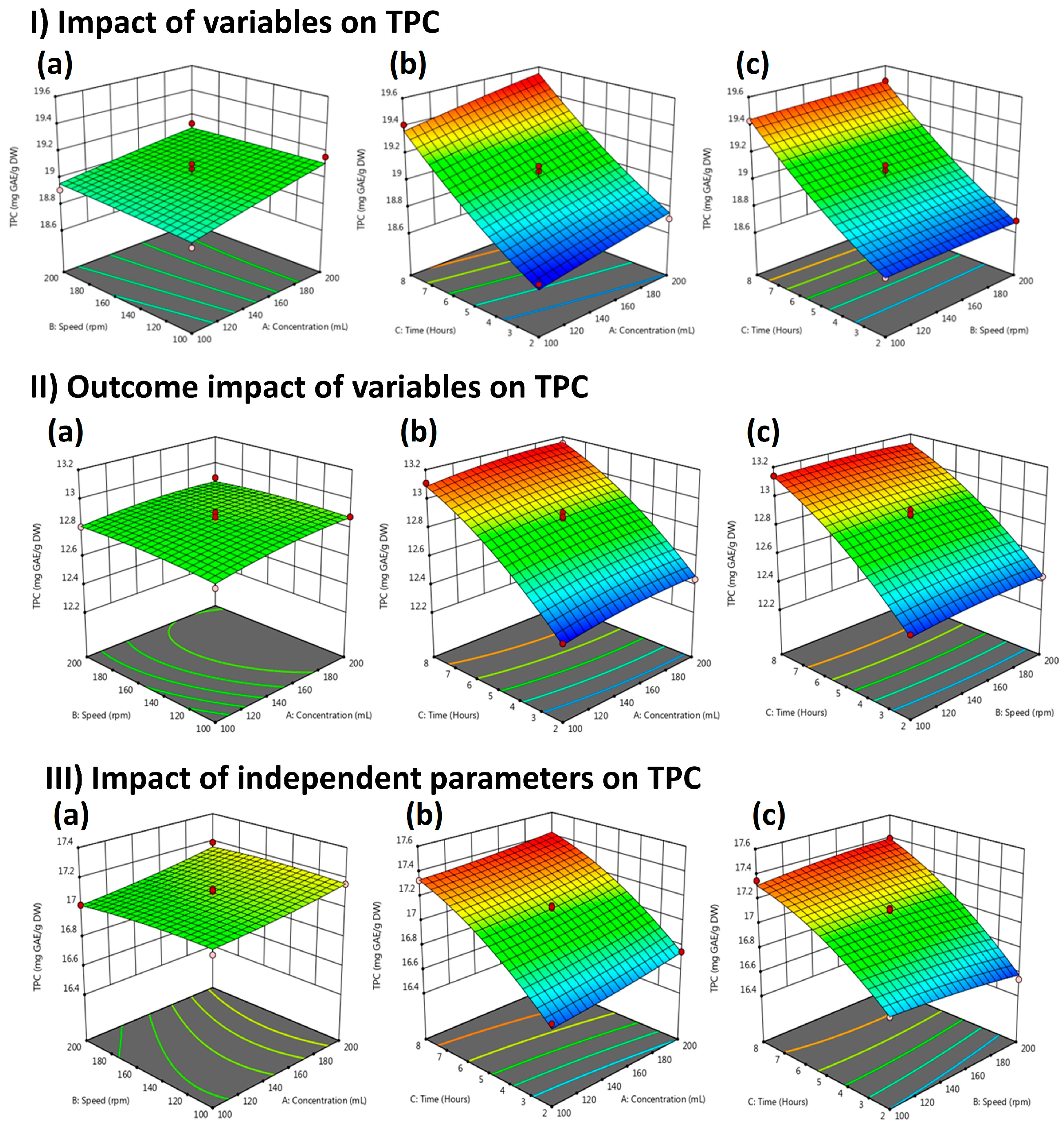
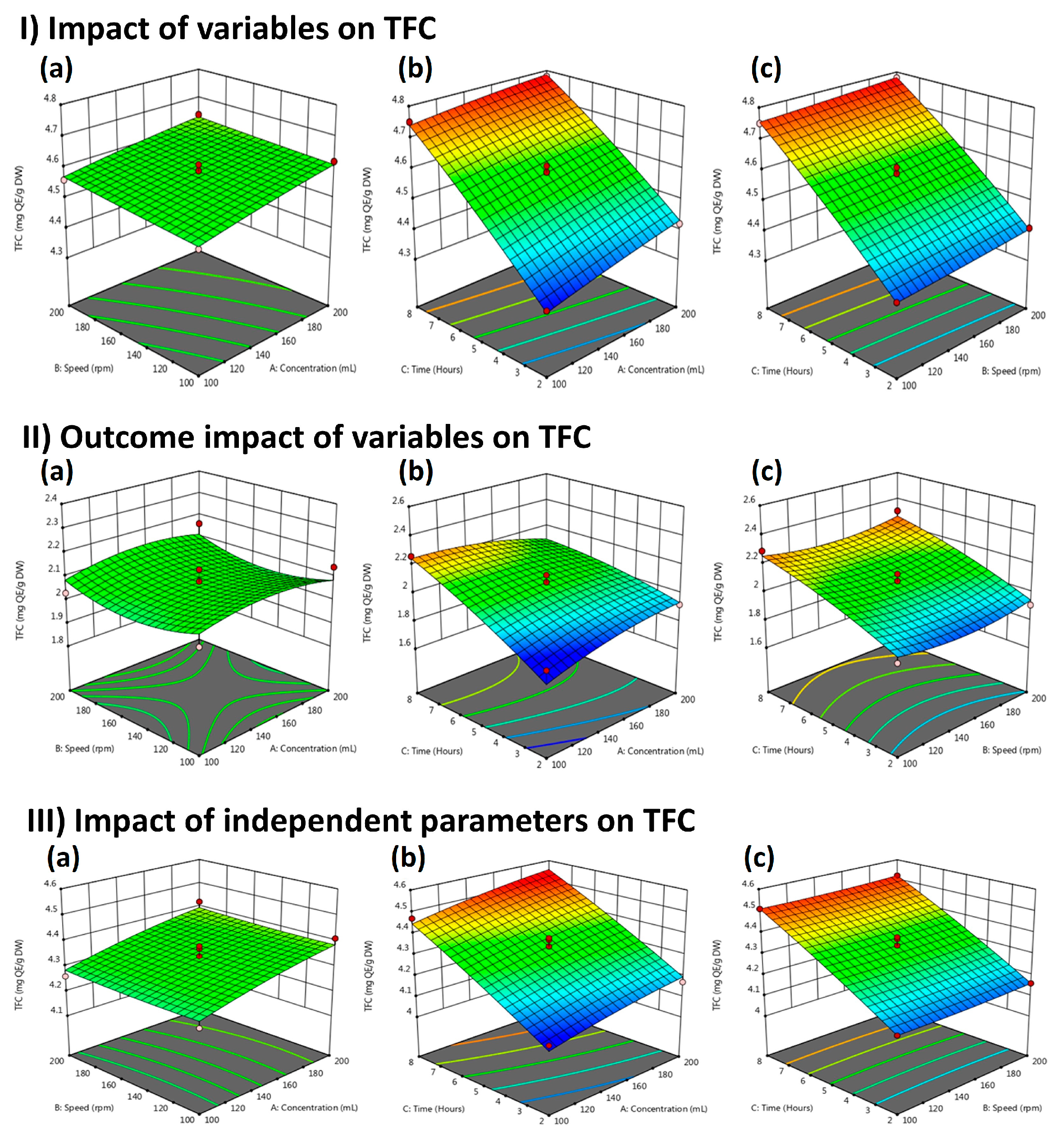
| Run | Solvent (mL) | Extraction Time (h) | Speed (rpm) | Yield (%) | ||
|---|---|---|---|---|---|---|
| LME | FME | WPME | ||||
| 1 | 200 (+1) | 8 (+1) | 150 (0) | 8.97 ± 0.38 ab | 10.88 ± 0.28 d | 13.55 ± 0.11 fg |
| 2 (C1) | 150 (0) | 5 (0) | 150 (0) | 8.87 ± 0.25 d | 10.8 ± 0.21 e | 13.48± 0.18 c |
| 3 (C2) | 150 (0) | 5 (0) | 150 (0) | 8.86 ± 0.30 d | 10.79 ± 0.29 f | 13.48 ± 0.27 d |
| 4 | 150 (0) | 8 (+1) | 200 (+1) | 8.96 ± 0.34 ab | 10.86 ± 0.19 ab | 13.54 ± 0.28 hi |
| 5 | 200 (+1) | 5 (0) | 200 (+1) | 8.89 ± 0.34 c | 10.81 ± 0.22 fg | 13.49 ± 0.36 f |
| 6 (C3) | 150 (0) | 5 (0) | 150 (0) | 8.86 ± 0.24 df | 10.78 ± 0.31 c | 13.47 ± 0.41 m |
| 7 (C4) | 150 (0) | 5 (0) | 150 (0) | 8.87 ± 0.27 d | 10.78 ± 0.33 h | 13.47 ± 0.18 n |
| 8 (C5) | 150 (0) | 5 (0) | 150 (0) | 8.86 ± 0.31 d | 10.77 ± 0.23 i | 13.46 ± 0.30 e |
| 9 | 200 (+1) | 2 (−1) | 150 (0) | 8.79 ± 0.19 fg | 10.73 ± 0.25 ba | 13.42 ± 0.34 a |
| 10 | 150 (0) | 8 (+1) | 100 (−1) | 8.94 ± 0.28 b | 10.85 ± 0.18 a | 13.54 ± 0.16 hf |
| 11 | 150 (0) | 2 (−1) | 100 (−1) | 8.77 ± 0.22 f | 10.71 ± 0.25 d | 13.41 ± 0.21 d |
| 12 | 100 (−1) | 5 (0) | 100 (−1) | 8.84 ± 0.30 f | 10.76 ± 0.38 h | 13.44 ± 0.26 l |
| 13 | 100 (−1) | 5 (0) | 200 (+1) | 8.85 ± 0.22 f | 10.77 ± 0.14 mn | 13.45 ± 0.31 q |
| 14 | 100 (−1) | 8 (+1) | 150 (0) | 8.93 ± 0.33 bc | 10.83 ± 0.39 a | 13.52 ± 0.25 s |
| 15 | 100 (−1) | 2 (−1) | 150 (0) | 8.76 ± 0.23 f | 10.71 ± 0.40 f | 13.41 ± 0.16 h |
| 16 | 200 (+1) | 5 (0) | 100 (−1) | 8.88 ± 0.29 dk | 10.81 ± 0.29 k | 13.49 ± 0.32 d |
| 17 | 150 (0) | 2 (−1) | 200 (+1) | 8.78 ± 0.12 fg | 10.72 ± 0.19 f | 13.42 ± 0.22 m |
| Source of Variation | Response Parameters | |||||||
|---|---|---|---|---|---|---|---|---|
| DF | Yield of LME | Yield of FME | Yield of WPME | |||||
| MS | p-Value | MS | p-Value | MS | p-Value | |||
| Model | 9 | 0.0072 | 0.0001 | 0.0046 | 0.0001 | 0.0036 | 0.0001 | |
| Linear Effects | A-Concentration | 1 | 0.0028 | 0.0001 | 0.0005 | 0.0021 | 0.0017 | 0.0017 |
| B-Speed | 1 | 0.0003 | 0.0060 | 0.2877 | 0.0001 | 0.4717 | 0.4717 | |
| C-Time | 1 | 0.0613 | 0.0001 | 0.0001 | 0.0300 | 0.0001 | 0.0001 | |
| Interaction Effects | AB | 1 | 0.0000 | 1.0000 | 0.6044 | 0.0000 | 0.6074 | 0.6074 |
| AC | 1 | 0.0000 | 0.3083 | 0.1478 | 0.0001 | 0.3178 | 0.3178 | |
| BC | 1 | 0.0000 | 0.3083 | 1.0000 | 0.0000 | 0.6074 | 0.6074 | |
| Quadratic Effects | A2 | 1 | 1.053 × 10−6 | 0.8281 | 0.5257 | 0.0001 | 0.4651 | 0.4651 |
| B2 | 1 | 1.053 × 10−6 | 0.8281 | 0.9145 | 4.211 × 10−6 | 0.8316 | 0.8316 | |
| C2 | 1 | 0.0000 | 0.3972 | 0.9145 | 0.0002 | 0.1945 | 0.1945 | |
| Residual | 7 | 0.0000 | - | 0.0001 | - | 0.0001 | ||
| Lack of Fit | 3 | 8.333 × 10−6 | 0.8395 | 0.0000 | 0.8966 | 0.0001 | 0.3329 | |
| Pure Error | 4 | 0.0000 | - | 0.0001 | 0.0001 | |||
| Cor. Total | 16 | - | - | - | - | |||
| Response Parameter | Regression Form | Regression Equation |
|---|---|---|
| Yield of LME | Coded | R1 = +8.86 + 0.0188A + 0.0063B + 0.0875C + 0.0000AB + 0.0025AC + 0.0025BC + 0.0005A2 + 0.0005B2 − 0.0020C2 |
| Actual | R1 = +8.67161 + 0.000232 Con − 0.000018 Speed +0.026389Time − 3.35785 × 10−19 Con * Speed + 0.000017 Con * Time + 0.000017 Speed * Time + 2.00000 × 10−7 Con2 + 2.00000 × 10−7 Speed2 − 0.000222 Time2 | |
| Yield of FME | Coded | R2 = +10.78 + 0.0200A + 0.0037B + 0.0687C − 0.0025AB + 0.0075AC + 0.0000BC + 0.0030A2 + 0.0005B2 + 0.0005C2 |
| Actual | R2 = +10.64606 − 0.000060Con + 0.000165Speed + 0.014861Time − 1.00000 × 10−6 Con * Speed + 0.000050 Con * Time + 2.78315 × 10−20 Speed * Time + 1.20000 × 10−6 Con2 + 2.00000 × 10−7 Speed2 + 0.000056 Time2 | |
| Yield of WPME | Coded | R3 = +13.47 + 0.0163A + 0.0025B + 0.0612C − 0.0025AB + 0.0050AC − 0.0025BC − 0.0035A2 − 0.0010B2 + 0.0065C2 |
| Actual | R3 = +13.28122 + 0.000728Con + 0.000403Speed + 0.010694Time − 1.00000 × 10−6 Con * Speed + 0.000033 Con * Time − 0.000017 Speed * Time − 1.40000 × 10−6 Con2 − 4.00000 × 10−7 Speed2 + 0.000722 Time2 |
| Run | Solvent (mL) | Extraction Time (h) | Speed (rpm) | TPC (mg GAE/g DW) | ||
|---|---|---|---|---|---|---|
| LME | FME | WPME | ||||
| 1 | 200 (+1) | 8 (+1) | 150 (0) | 19.51 ± 0.34 i | 13.15 ± 0.27 d | 17.39 ± 0.30 dk |
| 2 (C1) | 150 (0) | 5 (0) | 150 (0) | 19.11 ± 0.19 j | 12.88 ± 0.39 a | 17.12 ± 0.37 s |
| 3 (C2) | 150 (0) | 5 (0) | 150 (0) | 19.07 ± 0.32 kl | 12.87 ± 0.29 i | 17.13 ± 0.25 f |
| 4 | 150 (0) | 8 (+1) | 200 (+1) | 19.48 ± 0.36 d | 13.11 ± 0.28 dj | 17.396 ± 0.38 i |
| 5 | 200 (+1) | 5 (0) | 200 (+1) | 19.13 ± 0.29 ab | 12.89 ± 0.11 g | 17.19 ± 0.33 hi |
| 6 (C3) | 150 (0) | 5 (0) | 150 (0) | 19.01 ± 0.24 cj | 12.86 ± 0.37 a | 17.12 ± 0.13 i |
| 7 (C4) | 150 (0) | 5 (0) | 150 (0) | 18.99 ± 0.17 f | 12.91 ± 0.24 lj | 17.11 ± 0.27 gk |
| 8 (C5) | 150 (0) | 5 (0) | 150 (0) | 18.95 ± 0.39 f | 12.83 ± 0.18 c | 17.04 ± 0.20 w |
| 9 | 200 (+1) | 2 (−1) | 150 (0) | 18.71 ± 0.21 h | 12.44 ± 0.11 e | 16.75 ± 0.21 k |
| 10 | 150 (0) | 8 (+1) | 100 (−1) | 19.43 ± 0.30 hi | 13.14 ± 0.32 ij | 17.35 ± 0.17 ab |
| 11 | 150 (0) | 2 (−1) | 100 (−1) | 18.68 ± 0.26 e | 12.43 ± 0.24 d | 16.71 ± 0.22 hi |
| 12 | 100 (−1) | 5 (0) | 100 (−1) | 18.87 ± 0.19 df | 12.74 ± 0.22 g | 17.03 ± 0.31 ab |
| 13 | 100 (−1) | 5 (0) | 200 (+1) | 18.91 ± 0.22 i | 12.81 ± 0.13 f | 17.02 ± 0.24 l |
| 14 | 100 (−1) | 8 (+1) | 150 (0) | 19.41 ± 0.38 ij | 13.11 ± 0.33 hi | 17.33 ± 0.33 f |
| 15 | 100 (−1) | 2 (−1) | 150 (0) | 18.64 ± 0.40 j | 12.39 ± 0.38 b | 16.64 ± 0.22 g |
| 16 | 200 (+1) | 5 (0) | 100 (−1) | 19.16 ± 0.19 d | 12.88 ± 0.29 f | 17.16 ± 0.22 ik |
| 17 | 150 (0) | 2 (−1) | 200 (+1) | 18.69 ± 0.24 m | 12.44 ± 0.25 d | 16.54 ± 0.24 y |
| Source of Variation | Response Parameter | |||||||
|---|---|---|---|---|---|---|---|---|
| DF | TPC of LME | TPC of FME | TPC of WPME | |||||
| MS | p-Value | MS | p-Value | MS | p-Value | |||
| Model | 9 | 0.1421 | 0.0001 | 0.1143 | 0.0001 | 0.1195 | 0.0001 | |
| Linear Effects | A-Concentration | 0.0578 | 0.0087 | 0.0120 | 0.0107 | 0.0276 | 0.0068 | 0.0068 |
| B-Speed | 0.0006 | 0.7218 | 0.0004 | 0.5260 | 0.0014 | 0.4296 | 0.4296 | |
| C-Time | 1.21 | 0.0001 | 0.9870 | 0.0001 | 0.9983 | 0.0001 | 0.0001 | |
| Interaction Effects | AB | 0.0012 | 0.6163 | 0.0009 | 0.3768 | 0.0004 | 0.6622 | 0.6622 |
| AC | 0.0002 | 0.8286 | 0.0000 | 0.8795 | 0.0006 | 0.5865 | 0.5865 | |
| BC | 0.0004 | 0.7732 | 0.0004 | 0.5493 | 0.0117 | 0.0433 | 0.0433 | |
| Quadratic Effects | A2 | 0.0001 | 0.8705 | 0.0024 | 0.1692 | 0.0006 | 0.5845 | 0.5845 |
| B2 | 0.0000 | 0.9291 | 0.0011 | 0.3291 | 0.0011 | 0.4720 | 0.4720 | |
| C2 | 0.0093 | 0.1918 | 0.0229 | 0.0021 | 0.0332 | 0.0043 | 0.0043 | |
| Residual | 7 | 0.0045 | - | 0.0010 | - | 0.0019 | - | |
| Lack of Fit | 3 | 0.0050 | 0.4123 | 0.0012 | 0.3558 | 0.0027 | 0.2506 | |
| Pure Error | 4 | 0.0041 | - | 0.0008 | - | 0.0013 | - | |
| Cor. Total | 16 | - | - | - | - | - | - | |
| Response Parameter | Regression Form | Regression Equation |
|---|---|---|
| TPC of LME | Coded | R4 = +19.03 + 0.0850A + 0.0088B + 0.3888C0.0175AB + 0.0075AC + 0.0100BC − 0.0055A2 − 0.0030B2 + 0.0470B2 |
| Actual | R4 = +18.08089 + 0.003160Con + 0.001252Speed + 0.059861Time − 7.00000 × 10−6 Con * Speed + 0.000050 Con * Time + 0.000067 Speed * Time − 2.20000 × 10−6 Con2 − 1.20000 × 10−6 Speed2 + 0.005222Time2 | |
| TPC of FME | Coded | R5 = +12.87 + 0.0387A + 0.0075B + 0.3513C − 0.0150AB − 0.0025AC − 0.0100BC − 0.0237A2 − 0.0162B2 − 0.0738C2 |
| Actual | R5 = +11.38347 + 0.004608Con + 0.003333Speed + 0.211528Time − 6.00000 × 10−6 Con * Speed − 0.000017 Con * Time − 0.000067 Speed * Time − 9.50000 × 10−6 Con2 − 6.50000 × 10−6 Speed2 − 0.008194Tim2 | |
| TPC of WPME | Coded | R6 = +17.10 + 0.0588A − 0.0130B + 0.3533C + 0.0100AB0.0125AC + 0.0540BC + 0.0123A2 − 0.0162B2 − 0.0888C2 |
| Actual | R6 = +16.39297 − 0.000478Con − 0.000710Speed + 0.174861Time + 4.00000 × 10−6 Con * Speed − 0.000083 Con * Time + 0.000360 Speed * Time + 4.90000 × 10−6 Con2 − 6.50000 × 10−6 Speed2 − 0.009861Tim2 |
| Run | Solvent (mL) | Extraction Time (h) | Speed (rpm) | TFC (mg QE/g DW) | ||
|---|---|---|---|---|---|---|
| LME | FME | WPME | ||||
| 1 | 200 (+1) | 8 (+1) | 150 (0) | 4.78 ± 0.34 a | 2.01 ± 0.33 p | 4.53 ± 0.12 g |
| 2 (C1) | 150 (0) | 5 (0) | 150 (0) | 4.61 ± 0.37 gh | 2.13 ± 0.24 f | 4.38 ± 0.23 ik |
| 3 (C2) | 150 (0) | 5 (0) | 150 (0) | 4.59 ± 0.24 b | 2.08 ± 0.10 l | 4.37 ± 0.20 de |
| 4 | 150 (0) | 8 (+1) | 200 (+1) | 4.78 ± 0.28 c | 2.31 ± 0.40 t | 4.52 ± 0.25 df |
| 5 | 200 (+1) | 5 (0) | 200 (+1) | 4.64 ± 0.14 ik | 2.16 ± 0.33 e | 4.42 ± 0.32 d |
| 6 (C3) | 150 (0) | 5 (0) | 150 (0) | 4.59 ± 0.18 e | 2.07 ± 0.38 m | 4.34 ± 0.17 c |
| 7 (C4) | 150 (0) | 5 (0) | 150 (0) | 4.58 ± 0.19 h | 2.07 ± 0.22 t | 4.32 ± 0.16 ba |
| 8 (C5) | 150 (0) | 5 (0) | 150 (0) | 4.57 ± 0.19 c | 2.03 ± 0.32 h | 4.29 ± 0.16 l |
| 9 | 200 (+1) | 2 (−1) | 150 (0) | 4.42 ± 0.36 bc | 1.92 ± 0.21 f | 4.17 ± 0.20 s |
| 10 | 150 (0) | 8 (+1) | 100 (−1) | 4.75 ± 0.28 c | 2.29 ± 0.38 i | 4.51 ± 0.31 kj |
| 11 | 150 (0) | 2 (−1) | 100 (−1) | 4.37 ± 0.29 dc | 1.89 ± 0.15 a | 4.15 ± 0.38 b |
| 12 | 100 (−1) | 5 (0) | 100 (−1) | 4.52 ± 0.17 di | 2.03 ± 0.21 k | 4.25 ± 0.32 d |
| 13 | 100 (−1) | 5 (0) | 200 (+1) | 4.56 ± 0.18 i | 2.03 ± 0.23 hi | 4.26 ± 0.09 id |
| 14 | 100 (−1) | 8 (+1) | 150 (0) | 4.75 ± 0.22 f | 2.26 ± 0.34 d | 4.47 ± 0.27 l |
| 15 | 100 (−1) | 2 (−1) | 150 (0) | 4.34 ± 0.23 i | 1.85 ± 0.17 e | 4.11 ± 0.11 k |
| 16 | 200 (+1) | 5 (0) | 100 (−1) | 4.62 ± 0.30 f | 2.14 ± 0.19 n | 4.41 ± 0.37 cd |
| 17 | 150 (0) | 2 (−1) | 200 (+1) | 4.41 ± 0.10 i | 1.91 ± 0.24 ij | 4.16 ± 0.23 i |
| Source of Variation | Response Parameter | |||||||
|---|---|---|---|---|---|---|---|---|
| DF | TFC of LME | TFC of FME | TFC of WPME | |||||
| MS | p-Value | MS | p-Value | MS | p-Value | |||
| Model | 9 | 0.0336 | 0.0001 | 0.0286 | 0.0203 | 0.0316 | 0.0003 | |
| Linear Effects | A-Concentration | 1 | 0.0105 | 0.0005 | 0.7830 | 0.0242 | 0.0050 | 0.0050 |
| B-Speed | 1 | 0.0021 | 0.0005 | 0.7830 | 0.0002 | 0.7245 | 0.7245 | |
| C-Time | 1 | 0.2888 | 0.2112 | 0.0004 | 0.2592 | 0.0001 | 0.0001 | |
| Interaction Effects | AB | 1 | 0.0001 | 0.0001 | 0.8965 | 0.0000 | 1.0000 | 1.0000 |
| AC | 1 | 0.0006 | 0.0256 | 0.0678 | 0.0000 | 1.0000 | 1.0000 | |
| BC | 1 | 0.0001 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 1.0000 | |
| Quadratic Effects | A2 | 1 | 0.0001 | 0.0061 | 0.3278 | 0.0004 | 0.6110 | 0.6110 |
| B2 | 1 | 0.004 | 0.0114 | 0.1932 | 0.0001 | 0.7978 | 0.7978 | |
| C2 | 1 | 0.0006 | 0.0033 | 0.4637 | 0.0004 | 0.6110 | 0.6110 | |
| Residual | 7 | 0.0002 | - | 0.0055 | - | 0.0015 | - | |
| Lack of Fit | 3 | 0.0002 | 0.4712 | 0.0111 | 0.0317 | 0.0017 | 0.4070 | |
| Pure Error | 4 | 0.0002 | - | 0..0013 | - | 0.0014 | - | |
| Cor. Total | 16 | - | - | - | - | - | - | |
| Response Parameter | Regression Form | Regression Equation |
|---|---|---|
| TFC of LME | Coded | R7 = +4.59 + 0.0363A + 0.0163B + 0.1900C − 0.0050AB − 0.0125AC − 0.0025BC − 0.0040A2 + 0.0010B2 − 0.0115C2 |
| Actual | R7 = +3.93489 + 0.001922Con + 0.000588Speed + 0.091111Time − 2.00000 × 10−6 Con * Speed − 0.000083 Con * Time − 0.000017 Speed * Time − 1.60000 × 10−6 Con2 + 4.00000 × 10−7 Speed2 − 0.001278Time2 | |
| TFC of FME | Coded | R8 = +2.08 + 0.0075A + 0.0075B + 0.1625C + 0.0050AB − 0.0800AC + 0.0000BC − 0.0380A2 + 0.0520B2 − 0.0280C2 |
| Actual | R8 = +1.45339Con − 0.006390Speed + 0.165278Time + 2.00000 × 10−6 Con * Speed − 0.000533Con * Time − 3.54202 × 10−20 Speed * Time − 0.000015Con2 + 0.000021Speed2 − 0.003111Time2 | |
| TFC of WPME | Coded | R9 = +4.34 + 0.0550A + 0.0050B + 0.1800C + 0.0000AB + 0.0000AC + 0.0000BC − 0.0100A2 + 0.0050B2 − 0.0100C2 |
| Actual | R9 = +3.78722 + 0.002300Con − 0.000500Speed + 0.071111time − 5.85380 × 10−19 Conc * Speed + 1.32185 × 10−18 Con * Time + 2.10381 × 10−20 Speed * Time − 4.00000 × 10−6 Con2 + 2.00000 × 10−6 Speed2 − 0.001111Time2 |
| Sample | Concentration (µg/mL) | % of Inhibition α-Amylase | % of Inhibition α-Glucosidase | α-Amylase IC50 Value (µg/mL) | α-Glucosidase IC50 Value (µg/mL) |
|---|---|---|---|---|---|
| Acarbose | 25 | 46.66 ± 0.30 d | 45.71 ± 0.29 d | 33.29 ± 0.34 | 37.29 ± 0.28 |
| 50 | 55.01 ± 0.39 c | 54.22 ± 0.34 c | |||
| 100 | 66.22 ± 0.44 b | 65.12 ± 0.44 b | |||
| 200 | 88.11 ± 0.54 a | 87.18 ± 0.55 a | |||
| LME | 25 | 44.27 ± 0.31 d | 43.08 ± 0.3 d | 46.86 ± 0.21 | 46.81 ± 0.17 |
| 50 | 52.27 ± 0.34 c | 52.67 ± 0.35 c | |||
| 100 | 63.17 ± 0.45 b | 63.32 ± 0.47 b | |||
| 200 | 83.43 ± 0.58 a | 82.49 ± 0.57 a | |||
| WPME | 25 | 41.72 ± 0.34 d | 41.91 ± 0.34 d | 53.88 ± 0.11 | 51.19 ± 0.30 |
| 50 | 51.85 ± 0.34 c | 51.87 ± 0.34 c | |||
| 100 | 63.09 ± 0.34 b | 62.55 ± 0.34 b | |||
| 200 | 81.15 ± 0.54 a | 81.89 ± 0.54 a | |||
| FME | 25 | 40.11 ± 0.34 d | 40.68 ± 0.34 d | 58.88 ± 0.12 | 56.68 ± 0.16 |
| 50 | 50.77 ± 0.34 c | 50.77 ± 0.34 c | |||
| 100 | 62.08 ± 0.34 b | 61.28 ± 0.34 b | |||
| 200 | 81.88 ± 0.50 a | 80.22 ± 0.52 a |
| Independent Variable | Unit | Coded Level | ||
|---|---|---|---|---|
| −1 | 0 | +1 | ||
| Extraction time | Second | 2 | 5 | 8 |
| Speed | (m/s) | 100 | 150 | 200 |
| Solvent concentration | mL | 100 | 150 | 200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rasheed, M.U.; Naqvi, S.A.R.; Al-Asmari, F.; Rahim, M.A.; Ramadan, M.F. Phytochemicals, Health-Promoting Effects, and Enzyme Inhibition Traits of Phlomis stewartii Extracts. Molecules 2024, 29, 1049. https://doi.org/10.3390/molecules29051049
Rasheed MU, Naqvi SAR, Al-Asmari F, Rahim MA, Ramadan MF. Phytochemicals, Health-Promoting Effects, and Enzyme Inhibition Traits of Phlomis stewartii Extracts. Molecules. 2024; 29(5):1049. https://doi.org/10.3390/molecules29051049
Chicago/Turabian StyleRasheed, Mamoon Ur, Syed Ali Raza Naqvi, Fahad Al-Asmari, Muhammad Abdul Rahim, and Mohamed Fawzy Ramadan. 2024. "Phytochemicals, Health-Promoting Effects, and Enzyme Inhibition Traits of Phlomis stewartii Extracts" Molecules 29, no. 5: 1049. https://doi.org/10.3390/molecules29051049
APA StyleRasheed, M. U., Naqvi, S. A. R., Al-Asmari, F., Rahim, M. A., & Ramadan, M. F. (2024). Phytochemicals, Health-Promoting Effects, and Enzyme Inhibition Traits of Phlomis stewartii Extracts. Molecules, 29(5), 1049. https://doi.org/10.3390/molecules29051049










