Influence of Spider Silk Protein Structure on Mechanical and Biological Properties for Energetic Material Detection
Abstract
1. Introduction
2. Relationship between Structural and Mechanical Properties of Spider Silk Proteins and Their Structural Benefits in Biosensor Applications
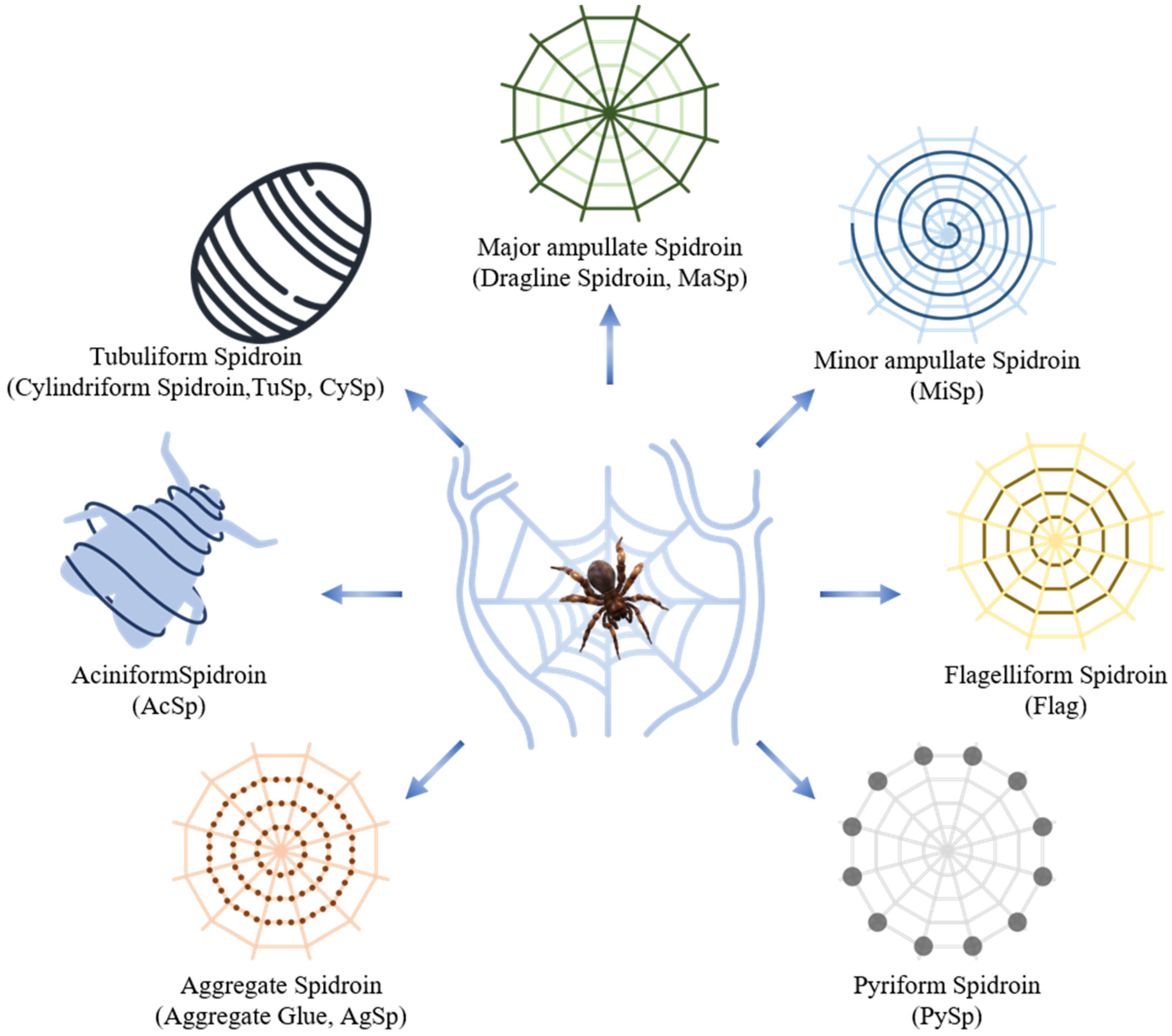
2.1. Structure of Spider Silk Proteins
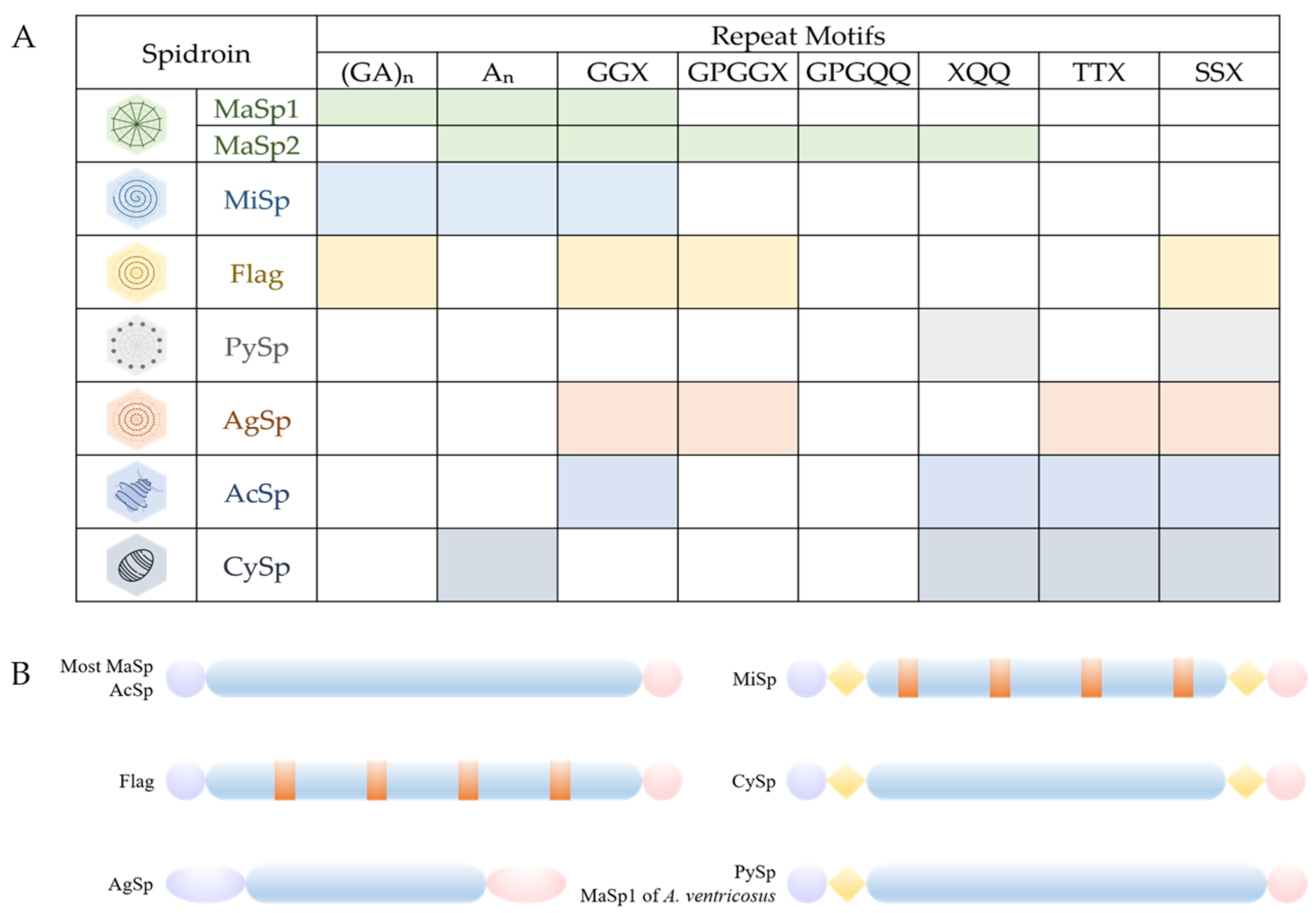
| Spider Silk | Molecular Weight | Main Amino Acids | Main Motifs of Repeat Domain | References |
|---|---|---|---|---|
| MaSp | >250 | Alanine Glycine | polyA or An polyGA or (GA)n GGX GPGXX | [37,38] |
| MiSp | >250 | Alanine Glycine | polyA polyGA GGX Spacer N-linker | [37,39,40] |
| Flag | >250 | Alanine Glycine | polyGA GGX GPGXX Spacer | [37] |
| PySp | 200–400 | Proline Glutamine | polyPX or (PX)n QQ-containing regions N-linker | [8,37,41] |
| AgSp | 450–1400 | Glutamine | Distinct repeat motifs Q-rich regions | [37] |
| AcSp | 300–500 | / | Long and complex motifs | [37] |
| CySp | 180 | Alanine Glutamine Serine | polyA QQ-containing regions S-rich motifs Linkers | [37,42] |
2.2. Relationship between Different Functional Domains of Spider Silk Proteins and Their Mechanical Properties
2.2.1. Repetitive Domains Mostly Influence Mechanical Properties of Spidroins
2.2.2. Non-Repetitive Terminal Domain Controls Spidroin Assembly
2.2.3. Linker Regions Influence Silk Fiber Properties and Assembly
2.2.4. Spider Silk Glues and Viscidity of Silk Fiber
2.3. Benefits of Spider Silk Structure When Silk Is Utilized in Biosensors
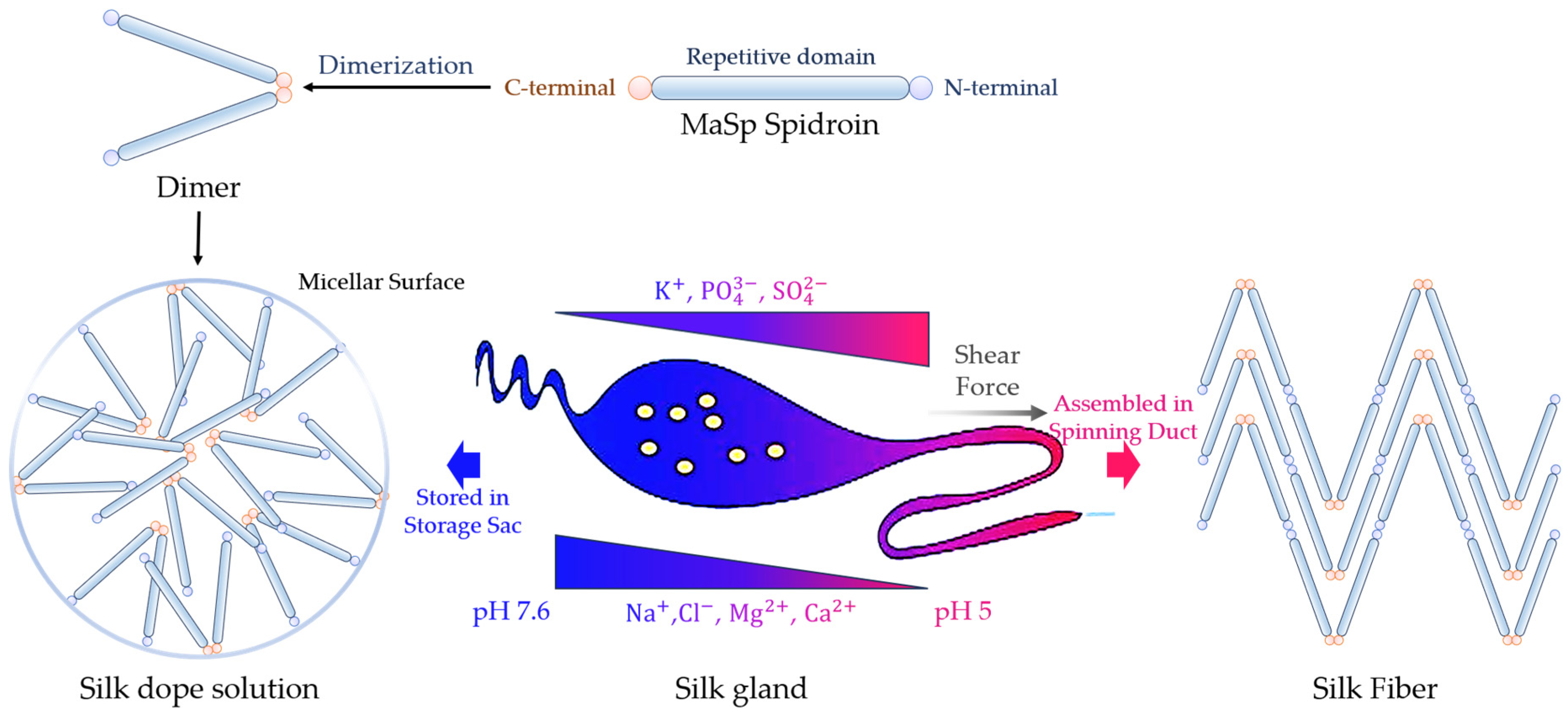
3. Influence of Production and Spinning of Spider Silk Proteins on Their Properties and Their Influences in Spidroin Biosensing Research
3.1. Properties of Recombinant Spider Silk Do Not Compare with Those of Native Silk Fibers
3.2. Spinning Process Has Important Influence on Silk Properties
3.2.1. Influences of Natural Spinning Process of Native Spider Silk Proteins on Silk Properties
3.2.2. Artificial Spinning Technology of Recombinant Spider Silk Proteins and Their Properties
3.3. Influence of Production and Spinning on Spidrion Biosensor Fabrication
4. Applications of Spider Silk Proteins
5. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vendrely, C.; Scheibel, T. Biotechnological production of spider-silk proteins enables new applications. Macromol. Biosci. 2007, 7, 401–409. [Google Scholar] [CrossRef]
- Lamont, S.M.; Vink, C.J.; Seldon, D.S.; Holwell, G.I. Spider diversity and community composition in native broadleaf-podocarp forest fragments of northern Hawke’s Bay, New Zealand. N. Z. J. Zool. 2017, 44, 129–143. [Google Scholar] [CrossRef]
- Xu, L.; Lefevre, T.; Orrell, K.E.; Meng, Q.; Auger, M.; Liu, X.-Q.; Rainey, J.K. Structural and Mechanical Roles for the C-Terminal Nonrepetitive Domain Become Apparent in Recombinant Spider Aciniform Silk. Biomacromolecules 2017, 18, 3678–3686. [Google Scholar] [CrossRef]
- Li, X.; Qi, X.M.; Cai, Y.M.; Sun, Y.; Wen, R.; Zhang, R.; Johansson, J.; Meng, Q.; Chen, G.F. Customized Flagelliform Spidroins Form Spider Silk-like Fibers at pH 8.0 with Outstanding Tensile Strength. ACS Biomater. Sci. Eng. 2022, 8, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Amarpuri, G.; Dhopatkar, N.; Blackledge, T.A.; Dhinojwala, A. Molecular Changes in Spider Viscid Glue As a Function of Relative Humidity Revealed Using Infrared Spectroscopy. ACS Biomater. Sci. Eng. 2022, 8, 3354–3360. [Google Scholar] [CrossRef]
- Shanafelt, M.; Larracas, C.; Dyrness, S.; Hekman, R.; La Mattina-Hawkins, C.; Rabara, T.; Wu, W.; Vierra, C.A. Egg Case Protein 3: A Constituent of Black Widow Spider Tubuliform Silk. Molecules 2021, 26, 10. [Google Scholar] [CrossRef]
- Wen, R.; Wang, K.K.; Yang, D.; Yu, T.T.; Zan, X.J.; Meng, Q. The novel aciniform silk protein (AcSp2-v2) reveals the unique repetitive domain with high acid and thermal stability and self-assembly capability. Int. J. Biol. Macromol. 2022, 202, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.N.; Rising, A.; Johansson, J.; Zhang, X.H.; Lin, Y.; Zhang, L.; Yi, T.; Mi, J.P.; Meng, Q. Tensile properties of synthetic pyriform spider silk fibers depend on the number of repetitive units as well as the presence of N- and C-terminal domains. Int. J. Biol. Macromol. 2020, 154, 765–772. [Google Scholar] [CrossRef]
- Bittencourt, D.M.D.; Oliveira, P.; Michalczechen-Lacerda, V.A.; Rosinha, G.M.S.; Jones, J.A.; Rech, E.L. Bioengineering of spider silks for the production of biomedical materials. Front. Bioeng. Biotechnol. 2022, 10, 16. [Google Scholar] [CrossRef]
- Kozlowska, A.K.; Florczak, A.; Smialek, M.; Dondajewska, E.; Mackiewicz, A.; Kortylewski, M.; Dams-Kozlowska, H. Functionalized bioengineered spider silk spheres improve nuclease resistance and activity of oligonucleotide therapeutics providing a strategy for cancer treatment. Acta Biomater. 2017, 59, 221–233. [Google Scholar] [CrossRef]
- Kuhbier, J.W.; Coger, V.; Mueller, J.; Liebsch, C.; Schlottmann, F.; Bucan, V.; Vogt, P.M.; Strauss, S. Influence of direct or indirect contact for the cytotoxicity and blood compatibility of spider silk. J. Mater. Sci.-Mater. Med. 2017, 28, 127. [Google Scholar] [CrossRef] [PubMed]
- De Tommasi, D.; Puglisi, G.; Saccomandi, G. Damage, Self-Healing, and Hysteresis in Spider Silks. Biophys. J. 2010, 98, 1941–1948. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Kumar, A.; Patel, A.; Vijay, S.; Saurabh, S.; Kumar, N. Biomechanical characterization of spider webs. J. Mech. Behav. Biomed. Mater. 2017, 67, 101–109. [Google Scholar] [CrossRef]
- Lin, S.; Zhu, J.; Li, X.; Guo, Y.; Fang, Y.; Cheng, H.; Zhu, H. Water-driven actuation of Ornithoctonus huwena spider silk fibers. Appl. Phys. Lett. 2017, 110, 053103. [Google Scholar] [CrossRef]
- Scheibel, T. Spider silks: Recombinant synthesis, assembly, spinning, and engineering of synthetic proteins. Microb. Cell Factories 2004, 3, 14. [Google Scholar] [CrossRef]
- Yuan, X. The development and application of spider silk. Beijing Text. J. 2005, 26, 30–32. [Google Scholar]
- Prince, J.T.; McGrath, K.P.; Digirolamo, C.M.; Kaplan, D.L. Construction, cloning, and expression of synthetic genes encoding spider dragline silk. Biochemistry 1995, 34, 10879–10885. [Google Scholar] [CrossRef]
- Xia, X.-X.; Qian, Z.-G.; Ki, C.S.; Park, Y.H.; Kaplan, D.L.; Lee, S.Y. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc. Natl. Acad. Sci. USA 2010, 107, 14059–14063. [Google Scholar] [CrossRef]
- Fahnestock, S.R.; Irwin, S.L. Synthetic spider dragline silk proteins and their production in Escherichia coli. Appl. Microbiol. Biotechnol. 1997, 47, 23–32. [Google Scholar] [CrossRef]
- Fahnestock, S.R.; Bedzyk, L.A. Production of synthetic spider dragline silk protein in Pichia pastoris. Appl. Microbiol. Biotechnol. 1997, 47, 33–39. [Google Scholar] [CrossRef]
- Miao, Y.; Zhang, Y.; Nakagaki, K.; Zhao, T.; Zhao, A.; Meng, Y.; Nakagaki, M.; Park, E.Y.; Maenaka, K. Expression of spider flagelliform silk protein in Bombyx mori cell line by a novel Bac-to-Bac/BmNPV baculovirus expression system. Appl. Microbiol. Biotechnol. 2006, 71, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Zhao, T.F.; Zhao, A.C.; Nakagaki, M. Stably express spider flagelliform silk protein in Bombyx mori cell line by piggyBac transposon-derived vector. In Advanced Textile Materials, Pts 1–3; Qian, X.M., Liu, H.W., Eds.; Trans Tech Publications Ltd.: Tianjin, China, 2011; Volume 332–334, pp. 779–782. [Google Scholar]
- Piruzian, E.S.; Bogush, V.G.; Sidoruk, K.V.; Goldenkova, I.V.; Mysiychuk, K.A.; Debabov, V.G. Construction of synthetic genes for analogs of spider silk spidroin 1 and their expression in tobacco plants. Mol. Biol. 2003, 37, 554–560. [Google Scholar] [CrossRef]
- Barr, L.A.; Fahnestock, S.R.; Yang, J.J. Production and purification of recombinant DP1B silk-like protein in plants. Mol. Breed. 2004, 13, 345–356. [Google Scholar] [CrossRef]
- Peng, C.A.; Russo, J.; Gravgaard, C.; McCartney, H.; Gaines, W.; Marcotte, W.R. Spider silk-like proteins derived from transgenic Nicotiana tabacum. Transgenic Res. 2016, 25, 517–526. [Google Scholar] [CrossRef]
- Casem, M.L.; Turner, D.; Houchin, K. Protein and amino acid composition of silks from the cob weaver, Latrodectus hesperus (black widow). Int. J. Biol. Macromol. 1999, 24, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Lazaris, A.; Arcidiacono, S.; Huang, Y.; Zhou, J.F.; Duguay, F.; Chretien, N.; Welsh, E.A.; Soares, J.W.; Karatzas, C.N. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science 2002, 295, 472–476. [Google Scholar] [CrossRef]
- Rathore, O.; Sogah, D.Y. Self-assembly of beta-sheets into nanostructures by poly(alanine) segments incorporated in multiblock copolymers inspired by spider silk. J. Am. Chem. Soc. 2001, 123, 5231–5239. [Google Scholar] [CrossRef]
- Kluge, J.A.; Rabotyagova, U.; Leisk, G.G.; Kaplan, D.L. Spider silks and their applications. Trends Biotechnol. 2008, 26, 244–251. [Google Scholar] [CrossRef]
- Thamm, C.; Scheibel, T. Recombinant Production, Characterization, and Fiber Spinning of an Engineered Short Major Ampullate Spidroin (MaSp1s). Biomacromolecules 2017, 18, 1365–1372. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, N.A.; Garb, J.E.; Tinghitella, R.M.; Collin, M.A.; Hayashi, C.Y. Blueprint for a High-Performance Biomaterial: Full-Length Spider Dragline Silk Genes. PLoS ONE 2007, 2, e514. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, T.A.; Hayashi, C.Y. Silken toolkits: Biomechanics of silk fibers spun by the orb web spider Argiope argentata (Fabricius 1775). J. Exp. Biol. 2006, 209, 2452–2461. [Google Scholar] [CrossRef]
- Aparecido dos Santos-Pinto, J.R.; Arcuri, H.A.; Priewalder, H.; Salles, H.C.; Palma, M.S.; Lubec, G. Structural Model for the Spider Silk Protein Spidroin-1. J. Proteome Res. 2015, 14, 3859–3870. [Google Scholar] [CrossRef]
- Simmons, J.R.; Gasmi-Seabrook, G.; Rainey, J.K. Structural features, intrinsic disorder, and modularity of a pyriform spidroin 1 core repetitive domain. Biochem. Cell Biol. 2023, 13, 271–283. [Google Scholar] [CrossRef]
- Sintya, E.; Alam, P. Self-assembled semi-crystallinity at parallel beta-sheet nanocrystal interfaces in clustered MaSp1 (spider silk) proteins. Mater. Sci. Eng. C-Mater. Biol. Appl. 2016, 58, 366–371. [Google Scholar] [CrossRef]
- Kono, N.; Nakamura, H.; Ohtoshi, R.; Moran, D.; Shinohara, A.; Yoshida, Y.; Fujiwara, M.; Mori, M.; Tomita, M.; Arakawa, K. Orb-weaving spider Araneus ventricosus genome elucidates the spidroin gene catalogue. Sci. Rep. 2019, 9, 13. [Google Scholar] [CrossRef]
- Ramezaniaghdam, M.; Nahdi, N.D.; Reski, R. Recombinant Spider Silk: Promises and Bottlenecks. Front. Bioeng. Biotechnol. 2022, 10, 18. [Google Scholar] [CrossRef]
- Gatesy, J.; Hayashi, C.; Motriuk, D.; Woods, J.; Lewis, R. Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science 2001, 291, 2603–2605. [Google Scholar] [CrossRef]
- Colgin, M.A.; Lewis, R.V. Spider minor ampullate silk proteins contain new repetitive sequences and highly conserved non-silk-like “spacer regions”. Protein Sci. 1998, 7, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, T.; Pezolet, M. Unexpected beta-sheets and molecular orientation in flagelliform spider silk as revealed by Raman spectromicroscopy. Soft Matter 2012, 8, 6350–6357. [Google Scholar] [CrossRef]
- Babb, P.L.; Lahens, N.F.; Correa-Garhwal, S.M.; Nicholson, D.N.; Kim, E.J.; Hogenesch, J.B.; Kuntner, M.; Higgins, L.; Hayashi, C.Y.; Agnarsson, I.; et al. The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat. Genet. 2017, 49, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Jorge, I.; Ruiz, V.; Lavado-Garcia, J.; Vazquez, J.; Hayashi, C.; Rojo, F.J.; Atienza, J.M.; Elices, M.; Guinea, G.V.; Perez-Rigueiro, J. Expression of spidroin proteins in the silk glands of golden orb-weaver spiders. J. Exp. Zool. Part B 2022, 338, 241–253. [Google Scholar] [CrossRef]
- Bergmann, F.; Stadlmayr, S.; Millesi, F.; Zeitlinger, M.; Naghilou, A.; Radtke, C. The properties of native Trichonephila dragline silk and its biomedical applications. Biomater. Adv. 2022, 140, 213089. [Google Scholar] [CrossRef] [PubMed]
- Htut, K.Z.; Alicea-Serrano, A.M.; Singla, S.; Agnarsson, I.; Garb, J.E.; Kuntner, M.; Gregoric, M.; Haney, R.A.; Marhabaie, M.; Blackledge, T.A.; et al. Correlation between protein secondary structure and mechanical performance for the ultra-tough dragline silk of Darwin’s bark spider. J. R. Soc. Interface 2021, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Omenetto, F.G.; KapLan, D.L. A new route for silk. Nat. Photonics 2008, 2, 641–643. [Google Scholar] [CrossRef]
- Xing, C.; Munro, T.; White, B.; Ban, H.; Copeland, C.G.; Lewis, R.V. Thermophysical properties of the dragline silk of Nephila clavipes spider. Polymer 2014, 55, 4226–4231. [Google Scholar] [CrossRef]
- Krishnaji, S.T.; Bratzel, G.; Kinahan, M.E.; Kluge, J.A.; Staii, C.; Wong, J.Y.; Buehler, M.J.; Kaplan, D.L. Sequence-Structure-Property Relationships of Recombinant Spider Silk Proteins: Integration of Biopolymer Design, Processing, and Modeling. Adv. Funct. Mater. 2013, 23, 241–253. [Google Scholar] [CrossRef]
- Tokareva, O.S.; Lin, S.; Jacobsen, M.M.; Huang, W.; Rizzo, D.; Li, D.; Simon, M.; Staii, C.; Cebe, P.; Wong, J.Y.; et al. Effect of sequence features on assembly of spider silk block copolymers. J. Struct. Biol. 2014, 186, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Anton, A.M.; Heidebrecht, A.; Mahmood, N.; Beiner, M.; Scheibel, T.; Kremer, F. Foundation of the Outstanding Toughness in Biomimetic and Natural Spider Silk. Biomacromolecules 2017, 18, 3954–3962. [Google Scholar] [CrossRef] [PubMed]
- Simmons, A.; Ray, E.; Jelinski, L.W. Solid-state 13C nmr of nephila-clavipes dragline silk establishes structure and identity of crystalline regions. Macromolecules 1994, 27, 5235–5237. [Google Scholar] [CrossRef]
- Kummerlen, J.; van Beek, J.D.; Vollrath, F.; Meier, B.H. Local structure in spider dragline silk investigated by two-dimensional spin-diffusion nuclear magnetic resonance. Macromolecules 1996, 29, 2920–2928. [Google Scholar] [CrossRef]
- Guerette, P.A.; Ginzinger, D.G.; Weber, B.H.F.; Gosline, J.M. Silk properties determined by gland-specific expression of a spider fibroin gene family. Science 1996, 272, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N. The underlying mechanisms behind the hydration-induced and mechanical response of spider silk. J. Mech. Phys. Solids 2023, 172, 13. [Google Scholar] [CrossRef]
- Bratzel, G.; Buehler, M.J. Sequence-structure correlations in silk: Poly-Ala repeat of N. clavipes MaSp1 is naturally optimized at a critical length scale. J. Mech. Behav. Biomed. Mater. 2012, 7, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Keten, S.; Xu, Z.; Ihle, B.; Buehler, M.J. Nanoconfinement controls stiffness, strength and mechanical toughness of beta-sheet crystals in silk. Nat. Mater. 2010, 9, 359–367. [Google Scholar] [CrossRef]
- Wen, R.; Wang, S.Y.; Wang, K.K.; Yang, D.; Zan, X.J.; Meng, Q. Complete gene sequence and mechanical property of the fourth type of major ampullate silk protein. Acta Biomater. 2023, 155, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, D.; Lee, S.-M.; Choi, J.-u.; You, M.; So, H.-M.; Han, J.; Nah, J.; Seol, J.H. Effects of beta-sheet crystals and a glycine-rich matrix on the thermal conductivity of spider dragline silk. Int. J. Biol. Macromol. 2017, 96, 384–391. [Google Scholar] [CrossRef]
- Sintya, E.; Alam, P. Localised semicrystalline phases of MaSp1 proteins show high sensitivity to overshearing in beta-sheet nanocrystals. Int. J. Biol. Macromol. 2016, 92, 1006–1011. [Google Scholar] [CrossRef]
- Heim, M.; Roemer, L.; Scheibel, T. Hierarchical structures made of proteins. The complex architecture of spider webs and their constituent silk proteins. Chem. Soc. Rev. 2010, 39, 156–164. [Google Scholar] [CrossRef]
- Bittencourt, D.M.D.; Oliveira, P.F.; Souto, B.M.; de Freitas, S.M.; Silva, L.P.; Murad, A.M.; Michalczechen-Lacerda, V.A.; Lewis, R.V.; Rech, E.L. Molecular Dynamics of Synthetic Flagelliform Silk Fiber Assembly (vol 306, 2000530, 2021). Macromol. Mater. Eng. 2022, 307, 2200368. [Google Scholar] [CrossRef]
- Onofrei, D.; Stengel, D.; Jia, D.; Johnson, H.R.; Trescott, S.; Soni, A.; Addison, B.; Muthukumar, M.; Holland, G.P. Investigating the Atomic and Mesoscale Interactions that Facilitate Spider Silk Protein Pre-Assembly. Biomacromolecules 2021, 22, 3377–3385. [Google Scholar] [CrossRef]
- Keten, S.; Buehler, M.J. Nanostructure and molecular mechanics of spider dragline silk protein assemblies. J. R. Soc. Interface 2010, 7, 1709–1721. [Google Scholar] [CrossRef]
- Li, X.; Shi, C.-H.; Tang, C.-L.; Cai, Y.-M.; Meng, Q. The correlation between the length of repetitive domain and mechanical properties of the recombinant flagelliform spidroin. Biol. Open 2017, 6, 333–339. [Google Scholar] [CrossRef]
- Adrianos, S.L.; Teule, F.; Hinman, M.B.; Jones, J.A.; Weber, W.S.; Yarger, J.L.; Lewis, R.V. Nephila clavipes Flagelliform Silk-Like GGX Motifs Contribute to Extensibility and Spacer Motifs Contribute to Strength in Synthetic Spider Silk Fibers. Biomacromolecules 2013, 14, 1751–1760. [Google Scholar] [CrossRef]
- Du, N.; Yang, Z.; Liu, X.Y.; Li, Y.; Xu, H.Y. Structural Origin of the Strain-Hardening of Spider Silk. Adv. Funct. Mater. 2011, 21, 772–778. [Google Scholar] [CrossRef]
- Aparecido dos Santos-Pinto, J.R.; Lamprecht, G.; Chen, W.-Q.; Heo, S.; Hardy, J.G.; Priewalder, H.; Scheibel, T.R.; Palma, M.S.; Lubec, G. Structure and post-translational modifications of the web silk protein spidroin-1 from Nephila spiders. J. Proteom. 2014, 105, 174–185. [Google Scholar] [CrossRef] [PubMed]
- van Beek, J.D.; Hess, S.; Vollrath, F.; Meier, B.H. The molecular structure of spider dragline silk: Folding and orientation of the protein backbone. Proc. Natl. Acad. Sci. USA 2002, 99, 10266–10271. [Google Scholar] [CrossRef]
- Grubb, D.T.; Ji, G.D. Molecular chain orientation in supercontracted and re-extended spider silk. Int. J. Biol. Macromol. 1999, 24, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Korley, L.T.J.; Pate, B.D.; Thomas, E.L.; Hammond, P.T. Effect of the degree of soft and hard segment ordering on the morphology and mechanical behavior of semicrystalline segmented polyurethanes. Polymer 2006, 47, 3073–3082. [Google Scholar] [CrossRef]
- Ittah, S.; Barak, N.; Gat, U. A Proposed Model for Drag line Spider Silk Self-Assembly: Insights from the Effect of the Repetitive Domain Size on Fiber Progenies. Biopolymers 2010, 93, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Krishnaji, S.; Tokareva, O.R.; Kaplan, D.; Cebe, P. Tunable crystallization, degradation, and self-assembly of recombinant protein block copolymers. Polymer 2017, 117, 107–116. [Google Scholar] [CrossRef]
- Humenik, M.; Magdeburg, M.; Scheibel, T. Influence of repeat numbers on self-assembly rates of repetitive recombinant spider silk proteins. J. Struct. Biol. 2014, 186, 431–437. [Google Scholar] [CrossRef]
- Malay, A.D.; Craig, H.C.; Chen, J.M.; Oktaviani, N.A.; Numata, K. Complexity of Spider Dragline Silk. Biomacromolecules 2022, 23, 1827–1840. [Google Scholar] [CrossRef]
- Bauer, J.; Scheibel, T. Dimerization of the Conserved N-Terminal Domain of a Spider Silk Protein Controls the Self-Assembly of the Repetitive Core Domain. Biomacromolecules 2017, 18, 2521–2528. [Google Scholar] [CrossRef]
- Otikovs, M.; Chen, G.; Nordling, K.; Landreh, M.; Meng, Q.; Joernvall, H.; Kronqvist, N.; Rising, A.; Johansson, J.; Jaudzems, K. Diversified Structural Basis of a Conserved Molecular Mechanism for pH-Dependent Dimerization in Spider Silk N-Terminal Domains. ChemBioChem 2015, 16, 1720–1724. [Google Scholar] [CrossRef]
- Askarieh, G.; Hedhammar, M.; Nordling, K.; Saenz, A.; Casals, C.; Rising, A.; Johansson, J.; Knight, S.D. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature 2010, 465, 236-U125. [Google Scholar] [CrossRef]
- Sede, M.; Fridmanis, J.; Otikovs, M.; Johansson, J.; Rising, A.; Kronqvist, N.; Jaudzems, K. Solution Structure of Tubuliform Spidroin N-Terminal Domain and Implications for pH Dependent Dimerization. Front. Mol. Biosci. 2022, 9, 13. [Google Scholar] [CrossRef]
- Chakraborty, R.; Fan, J.S.; Lai, C.C.; Raghuvamsi, P.V.; Chee, P.X.; Anand, G.S.; Yang, D.W. Structural Basis of Oligomerization of N-Terminal Domain of Spider Aciniform Silk Protein. Int. J. Mol. Sci. 2020, 21, 15. [Google Scholar] [CrossRef]
- Jin, H.J.; Kaplan, D.L. Mechanism of silk processing in insects and spiders. Nature 2003, 424, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Finnigan, W.; Roberts, A.D.; Ligorio, C.; Scrutton, N.S.; Breitling, R.; Blaker, J.J.; Takano, E. The effect of terminal globular domains on the response of recombinant mini-spidroins to fiber spinning triggers. Sci. Rep. 2020, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Heidebrecht, A.; Eisoldt, L.; Diehl, J.; Schmidt, A.; Geffers, M.; Lang, G.; Scheibel, T. Biomimetic Fibers Made of Recombinant Spidroins with the Same Toughness as Natural Spider Silk. Adv. Mater. 2015, 27, 2189–2194. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Scheibel, T. Conformational Stability and Interplay of Helical N- and C-Terminal Domains with Implications on Major Ampullate Spidroin Assembly. Biomacromolecules 2017, 18, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Hagn, F.; Eisoldt, L.; Hardy, J.G.; Vendrely, C.; Coles, M.; Scheibel, T.; Kessler, H. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature 2010, 465, 239–242. [Google Scholar] [CrossRef]
- Zhou, M.; Qian, Z.; Song, W.; Xia, X. Smart hydrogels assembled from the genetically engineered C-terminal domain of spider silk. J. Control. Release 2017, 259, E95–E96. [Google Scholar] [CrossRef]
- Strickland, M.; Tudorica, V.; Rezac, M.; Thomas, N.R.; Goodacre, S.L. Conservation of a pH-sensitive structure in the C-terminal region of spider silk extends across the entire silk gene family. Heredity 2018, 120, 574–580. [Google Scholar] [CrossRef]
- Hagn, F.; Thamm, C.; Scheibel, T.; Kessler, H. pH-Dependent Dimerization and Salt-Dependent Stabilization of the N-terminal Domain of Spider Dragline Silk-Implications for Fiber Formation. Angew. Chem.-Int. Ed. 2011, 50, 310–313. [Google Scholar] [CrossRef]
- Jaudzems, K.; Askarieh, G.; Landreh, M.; Nordling, K.; Hedhammar, M.; Jornvall, H.; Rising, A.; Knight, S.D.; Johansson, J. pH-Dependent Dimerization of Spider Silk N-Terminal Domain Requires Relocation of a Wedged Tryptophan Side Chain. J. Mol. Biol. 2012, 422, 477–487. [Google Scholar] [CrossRef]
- Oktaviani, N.A.; Malay, A.D.; Matsugami, A.; Hayashi, F.; Numata, K. Unusual pKa Values Mediate the Self-Assembly of Spider Dragline Silk Proteins. Biomacromolecules 2023, 24, 1604–1616. [Google Scholar] [CrossRef]
- Kronqvist, N.; Otikovs, M.; Chmyrov, V.; Chen, G.; Andersson, M.; Nordling, K.; Landreh, M.; Sarr, M.; Jornvall, H.; Wennmalm, S.; et al. Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation. Nat. Commun. 2014, 5, 3254. [Google Scholar] [CrossRef]
- Andersson, M.; Chen, G.; Otikovs, M.; Landreh, M.; Nordling, K.; Kronqvist, N.; Westermark, P.; Jornvall, H.; Knight, S.; Ridderstrale, Y.; et al. Carbonic Anhydrase Generates CO2 and H+ That Drive Spider Silk Formation Via Opposite Effects on the Terminal Domains. PLoS Biol. 2014, 12, e1001921. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Schaal, D.; Eisoldt, L.; Schweimer, K.; Schwarzinger, S.; Scheibel, T. Acidic Residues Control the Dimerization of the N-terminal Domain of Black Widow Spiders’ Major Ampullate Spidroin 1. Sci. Rep. 2016, 6, 34442. [Google Scholar] [CrossRef] [PubMed]
- Ortlepp, C.S.; Gosline, J.M. Consequences of forced silking. Biomacromolecules 2004, 5, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Chaw, R.C.; Saski, C.A.; Hayashi, C.Y. Complete gene sequence of spider attachment silk protein (PySp1) reveals novel linker regions and extreme repeat homogenization. Insect Biochem. Mol. Biol. 2017, 81, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Lin, Z.; Huang, W.; Lai, C.C.; Fan, J.-S.; Yang, D. Structural Characterization of Minor Ampullate Spidroin Domains and Their Distinct Roles in Fibroin Solubility and Fiber Formation. PLoS ONE 2013, 8, e56142. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.K.; Wen, R.; Wang, S.Z.; Tian, L.Y.; Xiao, J.H.; Meng, Q. The molecular structure of novel pyriform spidroin (PySp2) reveals extremely complex central repetitive region. Int. J. Biol. Macromol. 2020, 145, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.K.; Wen, R.; Meng, Q. Properties of two spliceoforms of major ampullate spidroin 1 reveal unique functions of N-linker region. Int. J. Biol. Macromol. 2020, 157, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.K.; Wen, R.; Jia, Q.P.; Liu, X.Q.; Xiao, J.H.; Meng, Q. Analysis of the Full-Length Pyriform Spidroin Gene Sequence. Genes 2019, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Wolff, J.O.; van der Meijden, A.; Herberstein, M.E. Distinct spinning patterns gain differentiated loading tolerance of silk thread anchorages in spiders with different ecology. Proc. Biol. Sci. 2017, 284, 20171124. [Google Scholar] [CrossRef]
- Vollrath, F.; Tillinghast, E.K. Glycoprotein glue beneath a spider webs aqueous coat. Naturwissenschaften 1991, 78, 557–559. [Google Scholar] [CrossRef]
- Denny, M. Physical-properties of spiders silk and their role in design of orb-webs. J. Exp. Biol. 1976, 65, 483–506. [Google Scholar] [CrossRef]
- Sun, Y.; Lee, S.M.; Ku, B.J.; Moon, M.J. Fine structural aspects on the web glue production in the golden orb-web spider Trichonephila clavata. Anim. Cells Syst. 2023, 27, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Stellwagen, S.D.; Renberg, R.L. Toward Spider Glue: Long Read Scaffolding for Extreme Length and Repetitious Silk Family Genes AgSp1 and AgSp2 with Insights into Functional Adaptation. G3-Genes Genomes Genet. 2019, 9, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, N.A.; Friend, K.; Clarke, T.; Baker, R.; Correa-Garhwal, S.M.; Crean, A.; Dendev, E.; Foster, D.; Hoff, L.; Kelly, S.D.; et al. Protein Composition and Associated Material Properties of Cobweb Spiders’ Gumfoot Glue Droplets. Integr. Comp. Biol. 2021, 61, 1459–1480. [Google Scholar] [CrossRef]
- Liu, X.; Shi, L.X.; Wan, X.Z.; Dai, B.; Chen, Y.P.; Wang, S.T. Recent Progress of Spider-Silk-Inspired Adhesive Materials. ACS Mater. Lett. 2021, 3, 1453–1467. [Google Scholar] [CrossRef]
- Vasanthavada, K.; Hu, X.; Tuton-Blasingame, T.; Hsia, Y.; Sampath, S.; Pacheco, R.; Freeark, J.; Falick, A.M.; Tang, S.; Fong, J.; et al. Spider Glue Proteins Have Distinct Architectures Compared with Traditional Spidroin Family Members. J. Biol. Chem. 2012, 287, 35986–35999. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, F.; Fairbrother, W.J.; Williams, R.J.P.; Tillinghast, E.K.; Bernstein, D.T.; Gallagher, K.S.; Townley, M.A. Compounds in the droplets of the orb spiders viscid spiral. Nature 1990, 345, 526–528. [Google Scholar] [CrossRef]
- Kelly, S.D.; Opell, B.D.; Correa-Garwhal, S.M. Correlated evolution between orb weaver glue droplets and supporting fibres maintains their distinct biomechanical roles in adhesion. J. Evol. Biol. 2022, 35, 879–890. [Google Scholar] [CrossRef]
- Sahni, V.; Blackledge, T.A.; Dhinojwala, A. Viscoelastic solids explain spider web stickiness. Nat. Commun. 2010, 1, 19. [Google Scholar] [CrossRef]
- Amarpuri, G.; Zhang, C.; Diaz, C.; Opell, B.D.; Blackledge, T.A.; Dhinojwala, A. Spiders Tune Glue Viscosity to Maximize Adhesion. ACS Nano 2015, 9, 11472–11478. [Google Scholar] [CrossRef]
- Opell, B.D.; Karinshak, S.E.; Sigler, M.A. Environmental response and adaptation of glycoprotein glue within the droplets of viscous prey capture threads from araneoid spider orb-webs. J. Exp. Biol. 2013, 216, 3023–3034. [Google Scholar] [CrossRef]
- Opell, B.D.; Karinshak, S.E.; Sigler, M.A. Humidity affects the extensibility of an orb-weaving spider’s viscous thread droplets. J. Exp. Biol. 2011, 214, 2988–2993. [Google Scholar] [CrossRef]
- Qiao, C.D.; Ma, X.G.; Zhang, J.L.; Yao, J.S. Effect of hydration on water state, glass transition dynamics and crystalline structure in chitosan films. Carbohydr. Polym. 2019, 206, 602–608. [Google Scholar] [CrossRef]
- Jain, D.; Amarpuri, G.; Fitch, J.; Blackledge, T.A.; Dhinojwala, A. Role of Hygroscopic Low Molecular Mass Compounds in Humidity Responsive Adhesion of Spider’s Capture Silk. Biomacromolecules 2018, 19, 3048–3057. [Google Scholar] [CrossRef]
- Spizzo, F.; Greco, G.; Del Bianco, L.; Coïsson, M.; Pugno, N.M. Magnetostrictive and Electroconductive Stress-Sensitive Functional Spider Silk. Adv. Funct. Mater. 2022, 32, 14. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Z.H.; Zhang, Y.; Zhang, Y.X.; Zhang, J.Z.; Yang, X.H.; Yuan, L.B. Spider Silk as a Flexible Light Waveguide for Temperature Sensing. J. Light. Technol. 2023, 41, 1884–1889. [Google Scholar] [CrossRef]
- Monks, J.N.; Yan, B.; Hawkins, N.; Vollrath, F.; Wang, Z.B. Spider Silk: Mother Nature’s Bio-Superlens. Nano Lett. 2016, 16, 5842–5845. [Google Scholar] [CrossRef] [PubMed]
- Huby, N.; Vié, V.; Renault, A.; Beaufils, S.; Lefèvre, T.; Paquet-Mercier, F.; Pézolet, M.; Bêche, B. Native spider silk as a biological optical fiber. Appl. Phys. Lett. 2013, 102, 3. [Google Scholar] [CrossRef]
- Lin, L.Q.; Zhong, Y.; Lin, H.Y.; Wang, C.L.; Yang, Z.F.; Wu, Q.; Zhang, D.; Zhu, W.G.; Zhong, Y.C.; Pan, Y.W.; et al. Spider Silk-Improved Quartz-Enhanced Conductance Spectroscopy for Medical Mask Humidity Sensing. Molecules 2022, 27, 11. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, P.; Zemke, F.; Wagermaier, W.; Linder, M.B. Interfacial Crystallization and Supramolecular Self-Assembly of Spider Silk Inspired Protein at the Water-Air Interface. Materials 2021, 14, 13. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, H.G.; Zhang, M.; Ning, Y.G.; Liu, Z.H.; Zhang, Y.X.; Ji, X.H.; Zhang, J.Z.; Yuan, L.B. Spider dragline silk for PH sensing. Opt. Commun. 2022, 506, 4. [Google Scholar] [CrossRef]
- Zhang, G.M.; Xu, T.; Du, H.Z.; Qiao, Y.Y.; Guo, X.H.; Shi, L.H.; Zhang, Y.; Shuang, S.M.; Dong, C.; Ma, H.M. A reversible fluorescent pH-sensing system based on the one-pot synthesis of natural silk fibroin-capped copper nanoclusters. J. Mater. Chem. C 2016, 4, 3540–3545. [Google Scholar] [CrossRef]
- Wang, J.L.; Zhang, H.; Tang, Y.J.; Wen, M.C.; Yao, B.J.; Yuan, S.; Zhang, W.N.; Lei, H.X. Metal-Nanostructure-Decorated Spider Silk for Highly Sensitive Refractive Index Sensing. ACS Biomater. Sci. Eng. 2022, 8, 1060–1066. [Google Scholar] [CrossRef] [PubMed]
- Tow, K.H.; Chow, D.M.; Vollrath, F.; Dicaire, I.; Gheysens, T.; Thévenaz, L. Exploring the Use of Native Spider Silk as an Optical Fiber for Chemical Sensing. J. Light. Technol. 2018, 36, 1138–1144. [Google Scholar] [CrossRef]
- Yang, Y.-X.; Qian, Z.-G.; Zhong, J.-J.; Xia, X.-X. Hyper-production of large proteins of spider dragline silk MaSp2 by Escherichia coli via synthetic biology approach. Process Biochem. 2016, 51, 484–490. [Google Scholar] [CrossRef]
- Chung, H.; Kim, T.Y.; Lee, S.Y. Recent advances in production of recombinant spider silk proteins. Curr. Opin. Biotechnol. 2012, 23, 957–964. [Google Scholar] [CrossRef]
- Edlund, A.M.; Jones, J.; Lewis, R.; Quinn, J.C. Economic feasibility and environmental impact of synthetic spider silk production from Escherichia coli. New Biotechnol. 2018, 42, 12–18. [Google Scholar] [CrossRef]
- Jastrzebska, K.; Felcyn, E.; Kozak, M.; Szybowicz, M.; Buchwald, T.; Pietralik, Z.; Jesionowski, T.; Mackiewicz, A.; Dams-Kozlowska, H. The method of purifying bioengineered spider silk determines the silk sphere properties. Sci. Rep. 2016, 6, 28106. [Google Scholar] [CrossRef]
- Dams-Kozlowska, H.; Majer, A.; Tomasiewicz, P.; Lozinska, J.; Kaplan, D.L.; Mackiewicz, A. Purification and cytotoxicity of tag-free bioengineered spider silk proteins. J. Biomed. Mater. Res. Part A 2013, 101, 456–464. [Google Scholar] [CrossRef]
- Peng, X.Y.; Cui, Y.T.; Chen, J.H.; Gao, C.A.; Yang, Y.; Yu, W.F.; Rai, K.; Zhang, M.; Nian, R.; Bao, Z.X.; et al. High-Strength Collagen-Based Composite Films Regulated by Water-Soluble Recombinant Spider Silk Proteins and Water Annealing. ACS Biomater. Sci. Eng. 2022, 8, 3341–3353. [Google Scholar] [CrossRef]
- Tucker, C.L.; Jones, J.A.; Brinshurst, H.N.; Copeland, C.G.; Addison, J.B.; Weber, W.S.; Mou, Q.S.; Yarger, J.L.; Lewis, R.V. Mechanical and Physical Properties of Recombinant Spider Silk Films Using Organic and Aqueous Solvents. Biomacromolecules 2014, 15, 3158–3170. [Google Scholar] [CrossRef]
- Lewis, R.V.; Hinman, M.; Kothakota, S.; Fournier, M.J. Expression and purification of a spider silk protein: A new strategy for producing repetitive proteins. Protein Expr. Purif. 1996, 7, 400–406. [Google Scholar] [CrossRef]
- Simmons, J.R.; Xu, L.; Rainey, J.K. Recombinant Pyriform Silk Fiber Mechanics Are Modulated by Wet Spinning Conditions. ACS Biomater. Sci. Eng. 2019, 5, 4985–4993. [Google Scholar] [CrossRef]
- Jansson, R.; Lau, C.H.; Ishida, T.; Ramstrom, M.; Sandgren, M.; Hedhammar, M. Functionalized silk assembled from a recombinant spider silk fusion protein (Z-4RepCT) produced in the methylotrophic yeast Pichia pastoris. Biotechnol. J. 2016, 11, 687–699. [Google Scholar] [CrossRef]
- Lefevre, T.; Auger, M. Spider silk as a blueprint for greener materials: A review. Int. Mater. Rev. 2016, 61, 127–153. [Google Scholar] [CrossRef]
- Giesa, T.; Perry, C.C.; Buehler, M.J. Secondary Structure Transition and Critical Stress for a Model of Spider Silk Assembly. Biomacromolecules 2016, 17, 427–436. [Google Scholar] [CrossRef]
- Lefevre, T.; Boudreault, S.; Cloutier, C.; Pezolet, M. Diversity of Molecular Transformations Involved in the Formation of Spider Silks. J. Mol. Biol. 2011, 405, 238–253. [Google Scholar] [CrossRef]
- Jahn, T.R.; Makin, O.S.; Morris, K.L.; Marshall, K.E.; Tian, P.; Sikorski, P.; Serpell, L.C. The Common Architecture of Cross-beta Amyloid. J. Mol. Biol. 2010, 395, 717–727. [Google Scholar] [CrossRef]
- Knight, D.P.; Vollrath, F. Changes in element composition along the spinning duct in a Nephila spider. Naturwissenschaften 2001, 88, 179–182. [Google Scholar] [CrossRef]
- Vollrath, F.; Knight, D.P. Liquid crystalline spinning of spider silk. Nature 2001, 410, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, T.; Boudreault, S.; Cloutier, C.; Pezolet, M. Conformational and orientational transformation of silk proteins in the major ampullate gland of Nephila clavipes spiders. Biomacromolecules 2008, 9, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Guinea, G.V.; Elices, M.; Perez-Rigueiro, J.; Plaza, G.R. Stretching of supercontracted fibers: A link between spinning and the variability of spider silk. J. Exp. Biol. 2005, 208, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shao, Z.Z.; Vollrath, F. Relationships between supercontraction and mechanical properties of spider silk. Nat. Mater. 2005, 4, 901–905. [Google Scholar] [CrossRef]
- Ling, S.; Qin, Z.; Li, C.; Huang, W.; Kaplan, D.L.; Buehler, M.J. Polymorphic regenerated silk fibers assembled through bioinspired spinning. Nat. Commun. 2017, 8, 1387. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhang, L.; Peng, Q.; Sun, M.; Zhang, Y.; Shao, H.; Hu, X. Tough silk fibers prepared in air using a biomimetic microfluidic chip. Int. J. Biol. Macromol. 2014, 66, 319–324. [Google Scholar] [CrossRef]
- Greiner, A.; Wendorff, J.H. Electrospinning: A fascinating method for the preparation of ultrathin fibres. Angew. Chem.-Int. Ed. 2007, 46, 5670–5703. [Google Scholar] [CrossRef]
- Madurga, R.; Guinea, G.V.; Elices, M.; Perez-Rigueiro, J.; Ganan-Calvo, A.M. Straining flow spinning: Simplified model of a bioinspired process to mass produce regenerated silk fibers controllably. Eur. Polym. J. 2017, 97, 26–39. [Google Scholar] [CrossRef]
- Shao, Z.Z.; Vollrath, F. Materials: Surprising strength of silkworm silk. Nature 2002, 418, 741. [Google Scholar] [CrossRef] [PubMed]
- Madsen, B.; Shao, Z.Z.; Vollrath, F. Variability in the mechanical properties of spider silks on three levels: Interspecific, intraspecific and intraindividual. Int. J. Biol. Macromol. 1999, 24, 301–306. [Google Scholar] [CrossRef]
- Holland, C.; O’Neil, K.; Vollrath, F.; Dicko, C. Distinct structural and optical regimes in natural silk spinning. Biopolymers 2012, 97, 368–373. [Google Scholar] [CrossRef]
- Xu, M.; Lewis, R.V. Structure of a protein superfiber-spider dragline silk. Proc. Natl. Acad. Sci. USA 1990, 87, 7120–7124. [Google Scholar] [CrossRef]
- Garrido, M.A.; Elices, M.; Viney, C.; Perez-Rigueiro, J. Active control of spider silk strength: Comparison of drag line spun on vertical and horizontal surfaces. Polymer 2002, 43, 1537–1540. [Google Scholar] [CrossRef]
- Jia, Z.R.; Gong, J.L.; Zeng, Y.; Ran, J.H.; Liu, J.; Wang, K.F.; Xie, C.M.; Lu, X.; Wang, J. Bioinspired Conductive Silk Microfiber Integrated Bioelectronic for Diagnosis and Wound Healing in Diabetes. Adv. Funct. Mater. 2021, 31, 13. [Google Scholar] [CrossRef]
- Florczak, A.; Mackiewicz, A.; Dams-Kozlowska, H. Functionalized Spider Silk Spheres As Drug Carriers for Targeted Cancer Therapy. Biomacromolecules 2014, 15, 2971–2981. [Google Scholar] [CrossRef]
- Paquet-Mercier, F.; Lefèvre, T.; Auger, M.; Pézolet, M. Evidence by infrared spectroscopy of the presence of two types of β-sheets in major ampullate spider silk and silkworm silk. Soft Matter 2013, 9, 208–215. [Google Scholar] [CrossRef]
- Guo, C.; Zhang, J.; Wang, X.; Nguyen, A.T.; Liu, X.Y.; Kaplan, D.L. Comparative Study of Strain-Dependent Structural Changes of Silkworm Silks: Insight into the Structural Origin of Strain-Stiffening. Small 2017, 13, 1702266. [Google Scholar] [CrossRef]
- Jeon, J.W.; Cho, S.Y.; Jeong, Y.J.; Shin, D.S.; Kim, N.R.; Yun, Y.S.; Kim, H.-T.; Choi, S.B.; Hong, W.G.; Kim, H.J.; et al. Pyroprotein-Based Electronic Textiles with High Stability. Adv. Mater. 2017, 29, 1605479. [Google Scholar] [CrossRef]
- Bai, J.; Ma, T.; Chu, W.; Wang, R.; Silva, L.; Michal, C.; Chiao, J.-C.; Chiao, M. Regenerated spider silk as a new biomaterial for MEMS. Biomed. Microdevices 2006, 8, 317–323. [Google Scholar] [CrossRef]
- Sammi, A.; Divya; Mahapatra, S.; Kumar, R.; Chandra, P. Nano-bio-engineered silk matrix based devices for molecular bioanalysis. Biotechnol. Bioeng. 2022, 119, 784–806. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Purohit, B.; Maurya, P.K.; Pandey, L.M.; Chandra, P. Engineered Nanomaterial Assisted Signal-amplification Strategies for Enhancing Analytical Performance of Electrochemical Biosensors. Electroanalysis 2019, 31, 1615–1629. [Google Scholar] [CrossRef]
- Yu, C.P.; Gu, B.N.; Bao, M.H.; Chen, J.H.; Shi, W.; Ye, J.H.; Zhang, W.C. In Situ Electrochemical Construction of CuN3@CuCl Hybrids for Controllable Energy Release and Self-Passivation Ability. Inorg. Chem. 2024, 63, 1642–1651. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.; Boczkaj, G.; Lassi, U. An Overview of Treatment Approaches for Octahydro-1, 3, 5, 7-tetranitro-1, 3, 5, 7-tetrazocine (HMX) Explosive in Soil, Groundwater, and Wastewater. Int. J. Environ. Res. Public Health 2022, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.H.; Ma, S.X.; Zhao, C.J.; Dong, D.M. Application of Molecular Emissions in Laser-Induced Breakdown Spectroscopy: A Review. Front. Phys. 2022, 10, 7. [Google Scholar] [CrossRef]
- Lee, W.; Lee, K. Recent Research Trends in Explosive Detection through Electrochemical Methods. Appl. Chem. Eng. 2019, 30, 399–407. [Google Scholar]
- Wang, J.M.; Yu, R.H.; Tao, F.R.; Cui, Y.Z.; Li, T.D. Determination of Nitroaromatics Using a Double-Layer of Gelatin Nanofibers and a Pyrene-Doped Polystyrene Membrane. Anal. Lett. 2018, 51, 2878–2894. [Google Scholar] [CrossRef]
- Guidetti, G.; d’Amone, L.; Kim, T.; Matzeu, G.; Mogas-Soldevila, L.; Napier, B.; Ostrovsky-Snider, N.; Roshko, J.; Ruggeri, E.; Omenetto, F.G. Silk materials at the convergence of science, sustainability, healthcare, and technology. Appl. Phys. Rev. 2022, 9, 50. [Google Scholar] [CrossRef]
- Singh, A.; Hede, S.; Sastry, M. Spider silk as an active scaffold in the assembly of gold nanoparticles and application of the gold-silk bioconjugate in vapor sensing. Small 2007, 3, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, J.D.; Zhang, M.; Li, S.; Liu, Z.H.; Zhang, Y.X.; Zhang, J.Z.; Yuan, L.B. Long-period fiber grating humidity sensor based on spider silks. Sens. Actuator A-Phys. 2022, 342, 7. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Z.H.; Zhang, Y.; Zhang, Y.X.; Yang, X.H.; Zhang, J.Z.; Yang, J.; Yuan, L.B. Spider dragline silk-based humidity alarm sensor with ultra-high sensitivity. Opt. Commun. 2022, 519, 6. [Google Scholar] [CrossRef]
- Gong, Z.Y.; Wu, T.L.; Chen, X.X.; Guo, J.H.; Zhang, Y.; Li, Y.C. Upconversion Nanoparticle Decorated Spider Silks as Single-Cell Thermometers. Nano Lett. 2021, 21, 1469–1476. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, W.F.; Sun, D.T.; Huang, J.H.; Qiu, X.Q.; Li, Z.X.; Wu, X.X. Very Strong, Super-Tough, Antibacterial, and Biodegradable Polymeric Materials with Excellent UV-Blocking Performance. ChemSusChem 2020, 13, 4974–4984. [Google Scholar] [CrossRef]
- Kucharczyk, K.; Rybka, J.D.; Hilgendorff, M.; Krupinski, M.; Slachcinski, M.; Mackiewicz, A.; Giersig, M.; Dams-Kozlowska, H. Composite spheres made of bioengineered spider silk and iron oxide nanoparticles for theranostics applications. PLoS ONE 2019, 14, 20. [Google Scholar] [CrossRef]
- Chu, M.Q.; Sun, Y. Self-assembly method for the preparation of near-infrared fluorescent spider silk coated with CdTe nanocrystals. Smart Mater. Struct. 2007, 16, 2453–2456. [Google Scholar] [CrossRef]
- Chang, S.Q.; Kang, B.; Dai, Y.D.; Chen, D. A novel route to synthesize CdS quantum dots on the surface of silk fibers via gamma-radiation. Mater. Lett. 2008, 62, 3447–3449. [Google Scholar] [CrossRef]
- Pan, H.; Zhang, Y.P.; Shao, H.L.; Hu, X.C.; Li, X.H.; Tian, F.; Wang, J. Nanoconfined crystallites toughen artificial silk. J. Mat. Chem. B 2014, 2, 1408–1414. [Google Scholar] [CrossRef] [PubMed]
- Chen, W. Index to Data of Agricultural. Agric. Archaeol. 1991, 1, 373–377. [Google Scholar]
- Butulija, S.; Spasojevic, V.; Brankovic, G.; Dordevic, M.P.; Radulovic, T.; Zarubica, A.; Matovic, B. Synthesis, characterization and magnetic properties of spider silk coated with maghemite (gamma-Fe2O3) nanoparticles. Mater. Lett. 2022, 314, 4. [Google Scholar] [CrossRef]
- Trigona, C.; Cunsolo, C.; Cardillo, G.D.; Rizza, M.; Baglio, S. Exploitation of a Spider Silk based Sensing Element. In Proceedings of the IEEE International Instrumentation and Measurement Technology Conference (I2MTC), Ottawa, ON, Canada, 16–19 May 2022; IEEE: Ottawa, ON, Canada, 2022. [Google Scholar]
- Chakravarty, S.; Bhardwaj, N.; Mandal, B.B.; Sen Sarma, N. Silk fibroin-carbon nanoparticle composite scaffolds: A cost effective supramolecular ‘turn off’ chemiresistor for nitroaromatic explosive vapours. J. Mater. Chem. C 2016, 4, 8920–8929. [Google Scholar] [CrossRef]


| Protein | Protein Weight (kDa) | Production | Reference |
|---|---|---|---|
| MaSp2 | 45 | 10 mg/g | [131] |
| MaSp1 | 284.9 | 2.7 g/L | [18] |
| MaSp2 | 201 | 3.6 g/L | [124] |
| PySp1 | 47.9 | 30–60 mg/L | [132] |
| Protein | Host | Production | Reference |
|---|---|---|---|
| DP-1B (MaSp) | Pichia pastoris | 1 g/L | [20] |
| Z-4RepCT (fusion protein) | Pichia pastoris | / | [133] |
| Flagelliform silk | Bombyx mori | 0.08 mg/flask (2 × 106 cells, 6 mL) | [21] |
| MaSp 1 | Nicotiana tabacum | 0.5% TSP | [23] |
| DP1B (MaSp) | Arabidopsis | 0.34% TSP (64 kDa) 0.03% TSP (127 kDa) | [24] |
| MaSp1 | Nicotiana tabacum | 0.7% TSP | [25] |
| MaSp2 | Nicotiana tabacum | 1.9% TSP | [25] |
| ADF-3 (MaSp) | Goat mammary epithelial cell | 2.8–8.3% | [26,27] |
| ADF-3 (MaSp) | Bovine mammary epithelial alveolar cells | 10–28% | [26,27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, X.; Liu, Z.; Gao, J.; Zhang, Y.; Wang, H.; Li, C.; Lv, X.; Gao, Y.; Deng, H.; Zhao, B.; et al. Influence of Spider Silk Protein Structure on Mechanical and Biological Properties for Energetic Material Detection. Molecules 2024, 29, 1025. https://doi.org/10.3390/molecules29051025
Peng X, Liu Z, Gao J, Zhang Y, Wang H, Li C, Lv X, Gao Y, Deng H, Zhao B, et al. Influence of Spider Silk Protein Structure on Mechanical and Biological Properties for Energetic Material Detection. Molecules. 2024; 29(5):1025. https://doi.org/10.3390/molecules29051025
Chicago/Turabian StylePeng, Xinying, Zhiyong Liu, Junhong Gao, Yuhao Zhang, Hong Wang, Cunzhi Li, Xiaoqiang Lv, Yongchao Gao, Hui Deng, Bin Zhao, and et al. 2024. "Influence of Spider Silk Protein Structure on Mechanical and Biological Properties for Energetic Material Detection" Molecules 29, no. 5: 1025. https://doi.org/10.3390/molecules29051025
APA StylePeng, X., Liu, Z., Gao, J., Zhang, Y., Wang, H., Li, C., Lv, X., Gao, Y., Deng, H., Zhao, B., Gao, T., & Li, H. (2024). Influence of Spider Silk Protein Structure on Mechanical and Biological Properties for Energetic Material Detection. Molecules, 29(5), 1025. https://doi.org/10.3390/molecules29051025




