Salvianolic Acid B Significantly Suppresses the Migration of Melanoma Cells via Direct Interaction with β-Actin
Abstract
1. Introduction
2. Results
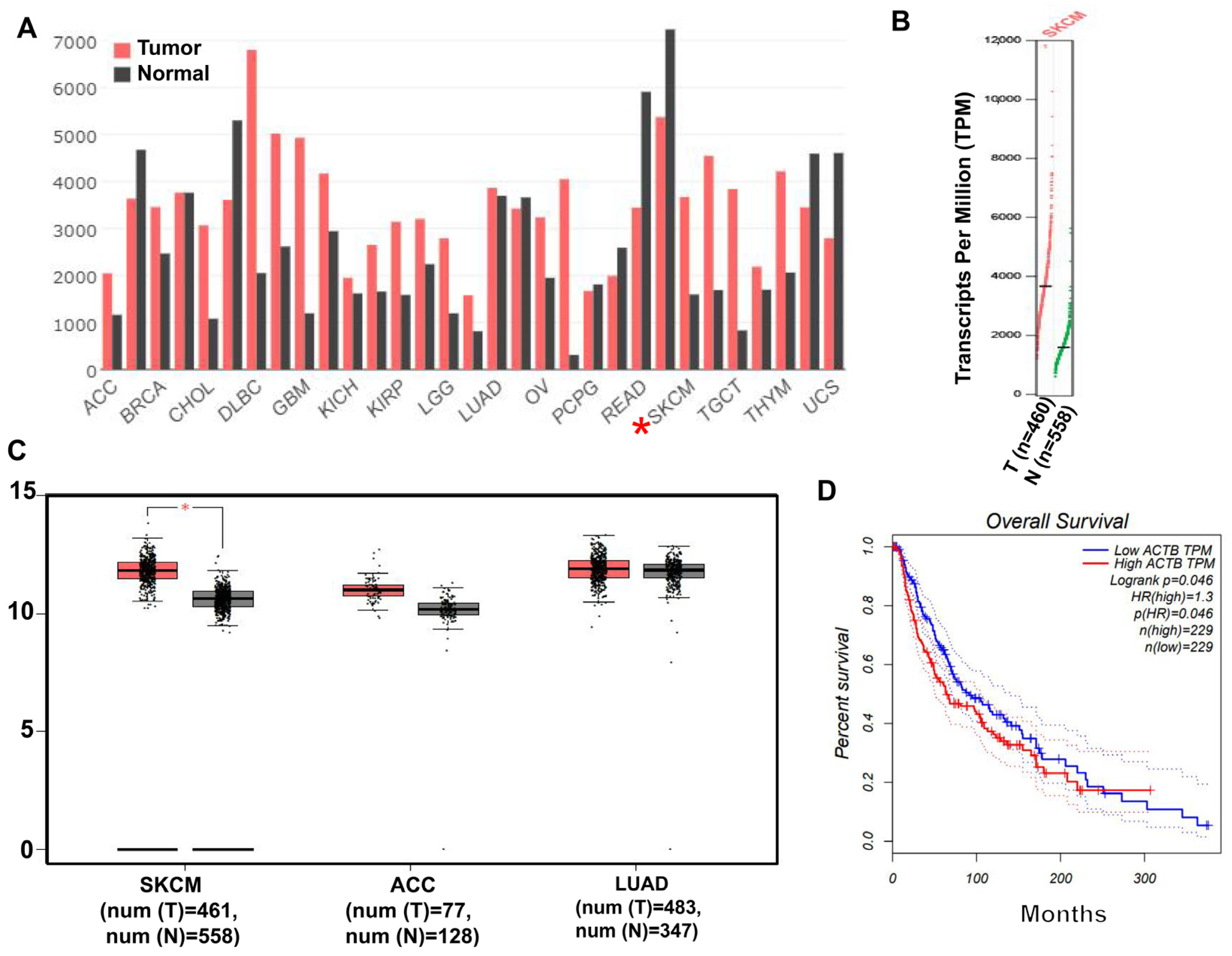
2.1. β-Actin Protein Is Highly Expressed in Skin Cancer
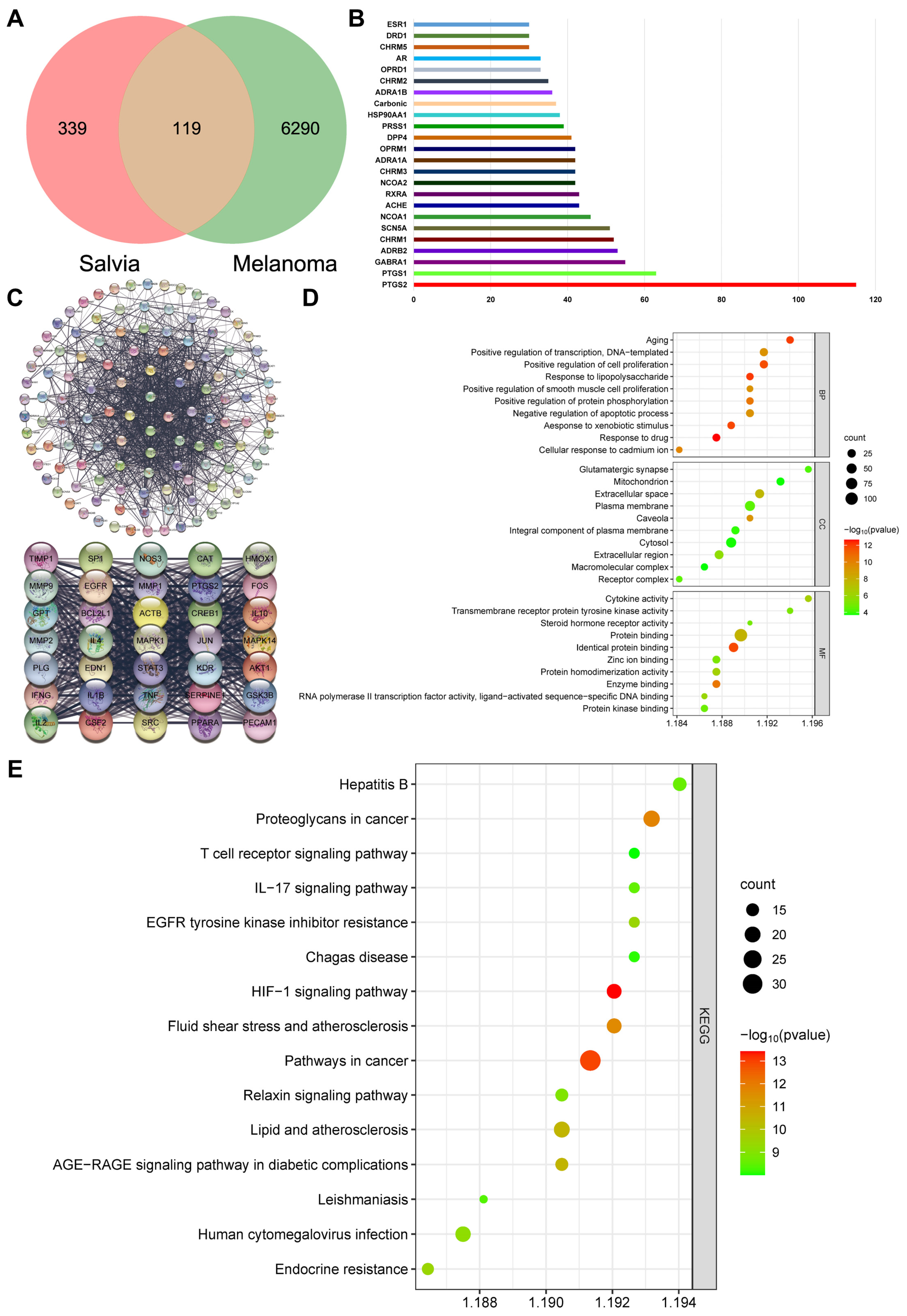
2.2. β-Actin Is the Target of Salvia miltiorrhiza for the Treatment of Melanoma through Network Pharmacology Analysis
2.3. SAB Significantly Inhibited the Proliferation of HaCaT, A375, and B16 Cells
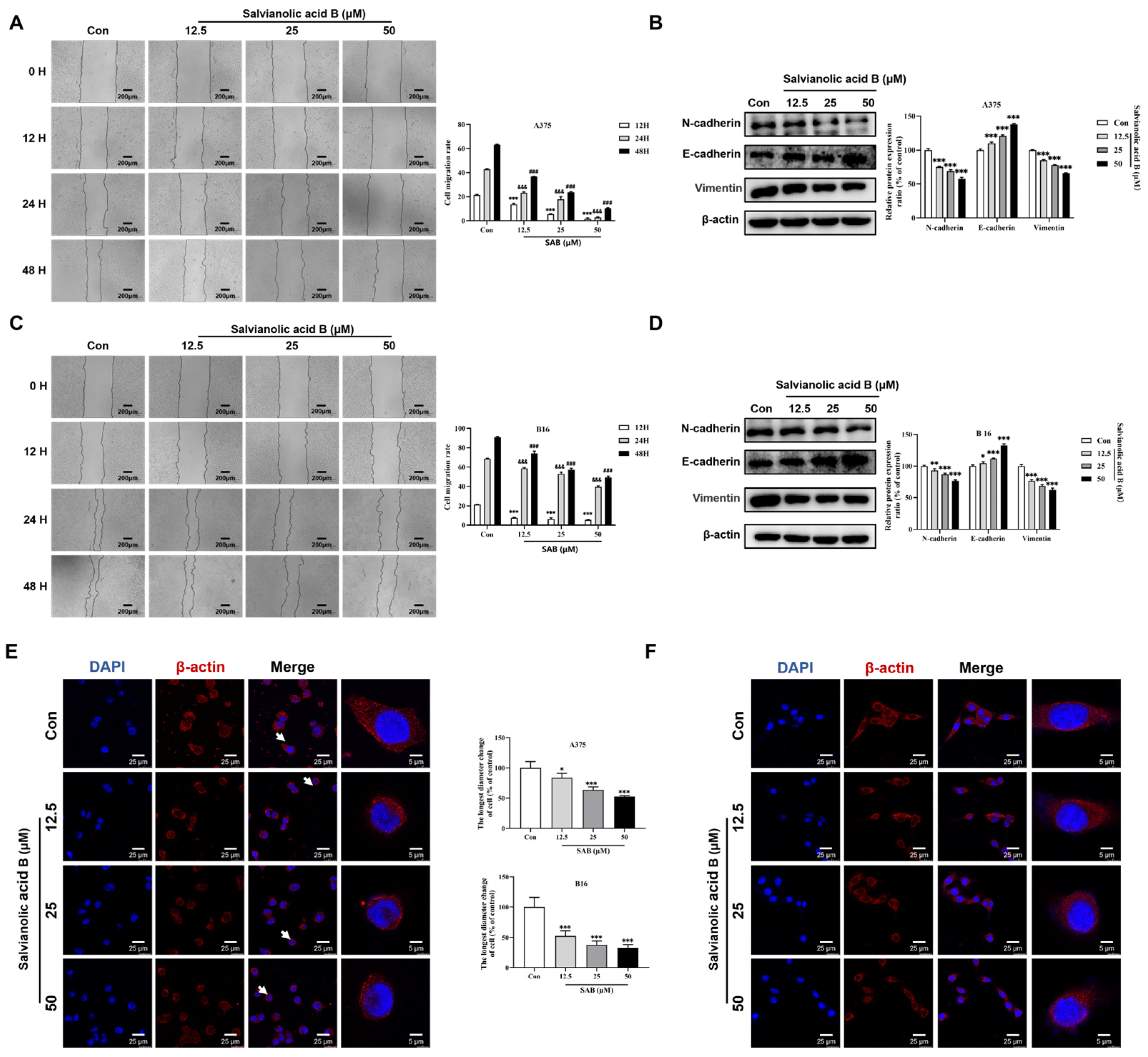
2.4. SAB Significantly Suppressed the Migration of A375 and B16 Cells
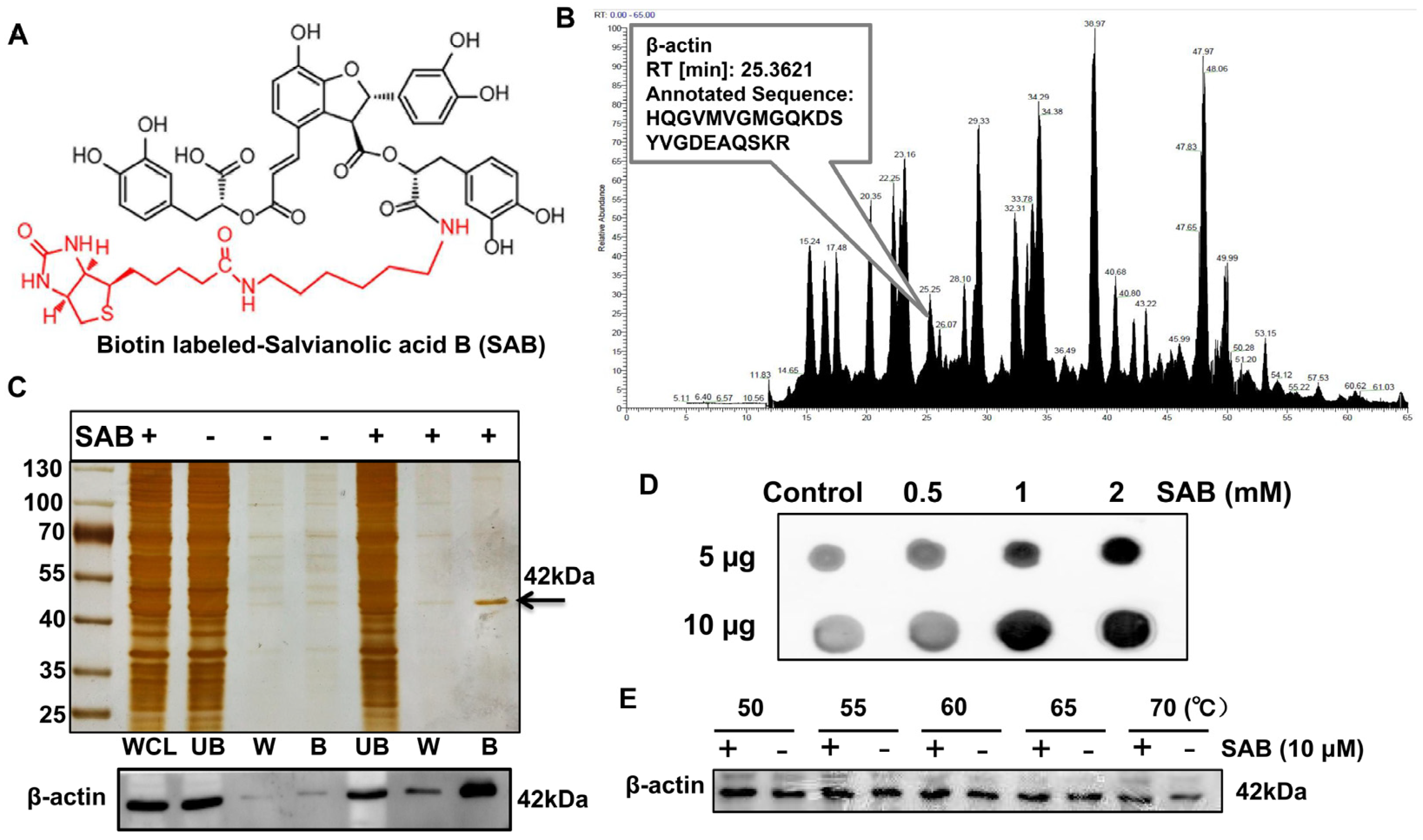
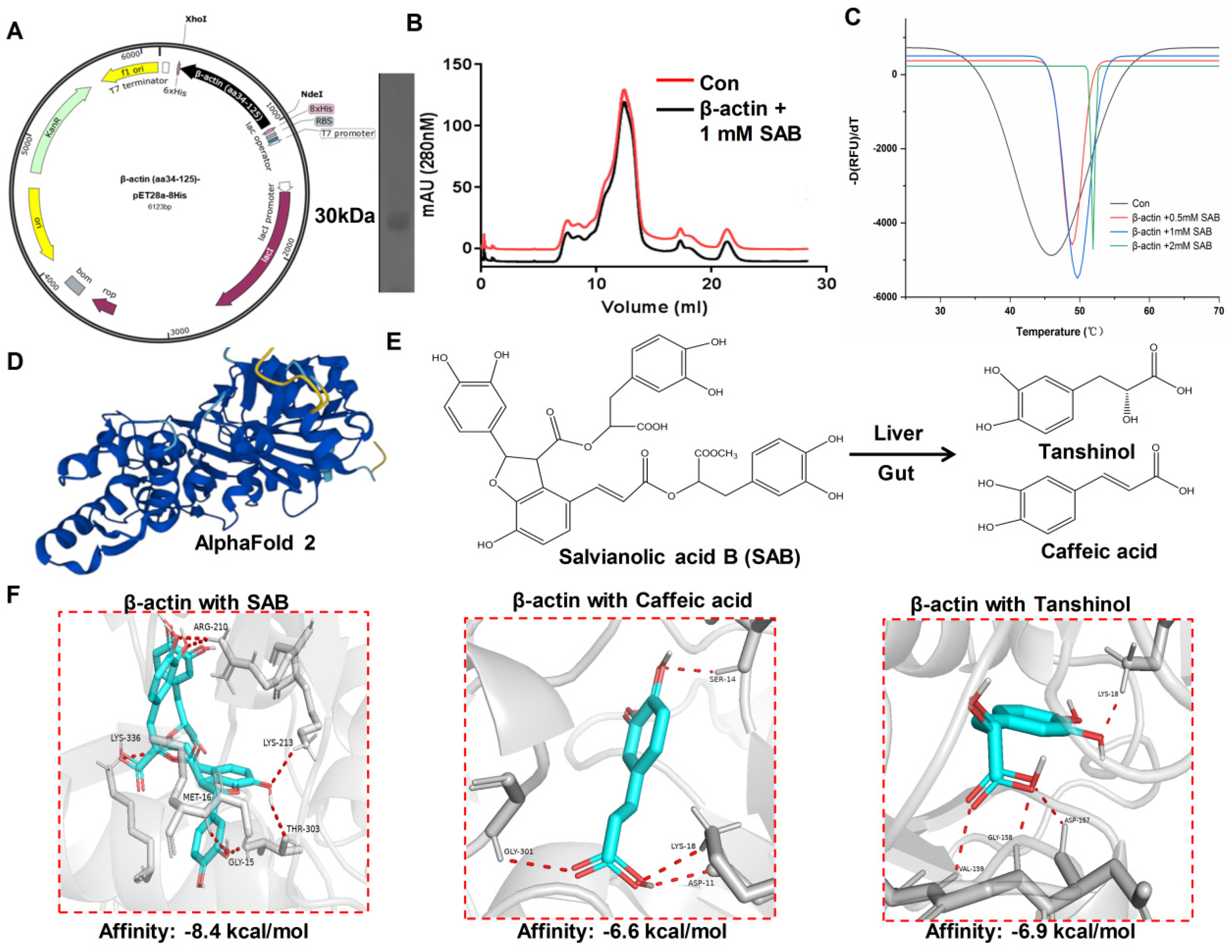
2.5. SAB Directly Interacted with β-actin
2.6. The Binding Mode of SAB and β-actin Protein Revealed by Molecular Docking
3. Discussion
4. Material and Methods
4.1. Cell Lines
4.2. Nature Product
4.3. MTT Assay
4.4. EdU Assay
4.5. Wound Healing Assay
4.6. Western Blot Analysis
4.7. Immunofluorescence
4.8. Biotin Labeled SAB and Molecular Fishing
4.9. Clone of β-actin into the pET-28a-psp Expression Vector
4.10. Expression and Purification of Recombinant Human β-actin
4.11. Protein Thermal Shift Assay
4.12. Dot Blot Assay
4.13. Molecular Docking
4.14. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Namikawa, K.; Yamazaki, N. Targeted Therapy and Immunotherapy for Melanoma in Japan. Curr. Treat. Options Oncol. 2019, 20, 7. [Google Scholar] [CrossRef]
- Long, G.V.; Swetter, S.M.; Menzies, A.M.; Gershenwald, J.E.; Scolyer, R.A. Cutaneous melanoma. Lancet 2023, 402, 485–502. [Google Scholar] [CrossRef]
- Cabrera, R.; Recule, F. Unusual Clinical Presentations of Malignant Melanoma: A Review of Clinical and Histologic Features with Special Emphasis on Dermatoscopic Findings. Am. J. Clin. Dermatol. 2018, 19 (Suppl. S1), 15–23. [Google Scholar] [CrossRef] [PubMed]
- Melendez-Moreno, A.; Yeşiltaş, Y.S.; Wrenn, J.; Singh, A.D. Iris melanoma: Prognostication for metastasis. Surv. Ophthalmol. 2023, 68, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Gao, W.D.; Lin, J.Y.; Xu, S.; Zhang, L.J.; Lu, X.C.; Luan, X.; Peng, J.Q.; Chen, Y. Nanotechnology-based photo-immunotherapy: A new hope for inhibition of melanoma growth and metastasis. J. Drug Target. 2023, 31, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Shattuck-Brandt, R.L.; Chen, S.C.; Murray, E.; Johnson, C.A.; Crandall, H.; O’Neal, J.F.; Al-Rohil, R.N.; Nebhan, C.A.; Bharti, V.; Dahlman, K.B.; et al. Metastatic Melanoma Patient-Derived Xenografts Respond to MDM2 Inhibition as a Single Agent or in Combination with BRAF/MEK Inhibition. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 3803–3818. [Google Scholar] [CrossRef]
- Timis, T.; Bergthorsson, J.T.; Greiff, V.; Cenariu, M.; Cenariu, D. Pathology and Molecular Biology of Melanoma. Curr. Issues Mol. Biol. 2023, 45, 5575–5597. [Google Scholar] [CrossRef] [PubMed]
- Bunnell, T.M.; Burbach, B.J.; Shimizu, Y.; Ervasti, J.M. β-Actin specifically controls cell growth, migration, and the G-actin pool. Mol. Biol. Cell 2011, 22, 4047–4058. [Google Scholar] [CrossRef] [PubMed]
- Fritz-Laylin, L.K.; Titus, M.A. The evolution and diversity of actin-dependent cell migration. Mol. Biol. Cell 2023, 34, pe6. [Google Scholar] [CrossRef]
- Yang, Z.; Zhou, L.; Si, T.; Chen, S.; Liu, C.; Ng, K.K.; Wang, Z.; Chen, Z.; Qiu, C.; Liu, G.; et al. Lysyl hydroxylase LH1 promotes confined migration and metastasis of cancer cells by stabilizing Septin2 to enhance actin network. Mol. Cancer 2023, 22, 21. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, L.; Liu, S. Determination of reference genes for ovine pulmonary adenocarcinoma infected lung tissues using RNA-seq transcriptome profiling. J. Virol. Methods 2020, 284, 113923. [Google Scholar] [CrossRef]
- Herath, S.; Dai, H.; Erlich, J.; Au, A.Y.; Taylor, K.; Succar, L.; Endre, Z.H. Selection and validation of reference genes for normalisation of gene expression in ischaemic and toxicological studies in kidney disease. PLoS ONE 2020, 15, e0233109. [Google Scholar] [CrossRef]
- Ben-Ze’ev, A. The cytoskeleton in cancer cells. Biochim. Biophys. Acta 1985, 780, 197–212. [Google Scholar] [CrossRef]
- Fäßler, F.; Javoor, M.G.; Schur, F.K. Deciphering the molecular mechanisms of actin cytoskeleton regulation in cell migration using cryo-EM. Biochem. Soc. Trans. 2023, 51, 87–99. [Google Scholar] [CrossRef]
- Nowak, D.; Skwarek-Maruszewska, A.; Zemanek-Zboch, M.; Malicka-Błaszkiewicz, M. Beta-actin in human colon adenocarcinoma cell lines with different metastatic potential. Acta Biochim. Pol. 2005, 52, 461–468. [Google Scholar] [CrossRef]
- Choudhary, B.S.; Chaudhary, N.; Shah, M.; Dwivedi, N.; PK, S.; Das, M.; Dalal, S.N. Lipocalin 2 inhibits actin glutathionylation to promote invasion and migration. FEBS Lett. 2023, 597, 1086–1097. [Google Scholar] [CrossRef]
- Mukherjee, D.; Previs, R.A.; Haines, C.; Al Abo, M.; Juras, P.K.; Strickland, K.C.; Chakraborty, B.; Artham, S.; Whitaker, R.S.; Hebert, K.; et al. Targeting CaMKK2 Inhibits Actin Cytoskeletal Assembly to Suppress Cancer Metastasis. Cancer Res. 2023, 83, 2889–2907. [Google Scholar] [CrossRef]
- Shestakova, E.A.; Wyckoff, J.; Jones, J.; Singer, R.H.; Condeelis, J. Correlation of beta-actin messenger RNA localization with metastatic potential in rat adenocarcinoma cell lines. Cancer Res. 1999, 59, 1202–1205. [Google Scholar]
- Lei, F.; Xu, X.; Huang, J.; Su, D.; Wan, P. Drosophila RhoGAP18B regulates actin cytoskeleton during border cell migration. PLoS ONE 2023, 18, e0280652. [Google Scholar] [CrossRef]
- Du, W.W.; Qadir, J.; Du, K.Y.; Chen, Y.; Wu, N.; Yang, B.B. Nuclear Actin Polymerization Regulates Cell Epithelial-Mesenchymal Transition. Adv. Sci. 2023, 10, e2300425. [Google Scholar] [CrossRef]
- Shin, A.E.; Giancotti, F.G.; Rustgi, A.K. Metastatic colorectal cancer: Mechanisms and emerging therapeutics. Trends Pharmacol. Sci. 2023, 44, 222–236. [Google Scholar] [CrossRef]
- Rout-Pitt, N.; Farrow, N.; Parsons, D.; Donnelley, M. Epithelial mesenchymal transition (EMT): A universal process in lung diseases with implications for cystic fibrosis pathophysiology. Respir. Res. 2018, 19, 136. [Google Scholar] [CrossRef]
- Kaplan, H.J.; Wang, W.; Piri, N.; Dean, D.C. Metabolic rescue of cone photoreceptors in retinitis pigmentosa. Taiwan. J. Ophthalmol. 2021, 11, 331–335. [Google Scholar] [CrossRef]
- Zhang, X.; Qiu, H.; Li, C.; Cai, P.; Qi, F. The positive role of traditional Chinese medicine as an adjunctive therapy for cancer. Biosci. Trends 2021, 15, 283–298. [Google Scholar] [CrossRef]
- Wang, K.; Chen, Q.; Shao, Y.; Yin, S.; Liu, C.; Liu, Y.; Wang, R.; Wang, T.; Qiu, Y.; Yu, H. Anticancer activities of TCM and their active components against tumor metastasis. Biomed. Pharmacother. 2021, 133, 111044. [Google Scholar] [CrossRef]
- Huang, J.; Li, J.X.; Ma, L.R.; Xu, D.H.; Wang, P.; Li, L.Q.; Yu, L.L.; Li, Y.; Li, R.Z.; Zhang, H.; et al. Traditional Herbal Medicine: A Potential Therapeutic Approach for Adjuvant Treatment of Non-small Cell Lung Cancer in the Future. Integr. Cancer Ther. 2022, 21, 15347354221144312. [Google Scholar] [CrossRef]
- Chen, W.; Chen, G. Danshen (Salvia miltiorrhiza Bunge): A Prospective Healing Sage for Cardiovascular Diseases. Curr. Pharm. Des. 2017, 23, 5125–5135. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Y.; Wang, S.; Zhao, X.; Zhao, L.; Wang, Y. Salvianolic acid B and ferulic acid synergistically promote angiogenesis in HUVECs and zebrafish via regulating VEGF signaling. J. Ethnopharmacol. 2022, 283, 114667. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Fu, L.; Wang, X.; Yini, X.; Ling, T.; Shen, X. Salvianolic acid B ameliorates myocardial fibrosis in diabetic cardiomyopathy by deubiquitinating Smad7. Chin. Med. 2023, 18, 161. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ma, Y.; Guo, M.; Yang, H.; Guan, X. Salvianolic acid B suppresses EMT and apoptosis to lessen drug resistance through AKT/mTOR in gastric cancer cells. Cytotechnology 2021, 73, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Begum, G.; Singh, N.D.; Leishangthem, G.D.; Banga, H.S. Amelioration of bleomycin induced pulmonary fibrosis by administration of Salvianolic acid B in mice. Vet. Ital. 2022, 58, 87–101. [Google Scholar] [PubMed]
- Tang, Q.; Zhong, H.; Xie, F.; Xie, J.; Chen, H.; Yao, G. Expression of miR-106b-25 induced by salvianolic acid B inhibits epithelial-to-mesenchymal transition in HK-2 cells. Eur. J. Pharmacol. 2014, 741, 97–103. [Google Scholar] [CrossRef]
- Chen, J.; Yuan, S.; Zhou, J.; Huang, X.; Wu, W.; Cao, Y.; Liu, H.; Hu, Q.; Li, X.; Guan, X.; et al. Danshen injection induces autophagy in podocytes to alleviate nephrotic syndrome via the PI3K/AKT/mTOR pathway. Phytomed. Int. J. Phytother. Phytopharm. 2022, 107, 154477. [Google Scholar] [CrossRef]
- Gong, L.; Di, C.; Xia, X.; Wang, J.; Chen, G.; Shi, J.; Chen, P.; Xu, H.; Zhang, W. AKT/mTOR signaling pathway is involved in salvianolic acid B-induced autophagy and apoptosis in hepatocellular carcinoma cells. Int. J. Oncol. 2016, 49, 2538–2548. [Google Scholar] [CrossRef]
- Katary, M.A.; Abdelsayed, R.; Alhashim, A.; Abdelhasib, M.; Elmarakby, A.A. Salvianolic Acid B Slows the Progression of Breast Cancer Cell Growth via Enhancement of Apoptosis and Reduction of Oxidative Stress, Inflammation, and Angiogenesis. Int. J. Mol. Sci. 2019, 20, 5653. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Wang, Y.; Liu, T.; Gao, J.; Duan, F.; Chen, M.; Yang, Y.; Wu, C. Salvianolic acid B acts against non-small cell lung cancer A549 cells via inactivation of the MAPK and Smad2/3 signaling pathways. Mol. Med. Rep. 2022, 25, 184. [Google Scholar] [CrossRef]
- Liu, Q.; Chu, H.; Ma, Y.; Wu, T.; Qian, F.; Ren, X.; Tu, W.; Zhou, X.; Jin, L.; Wu, W.; et al. Salvianolic Acid B Attenuates Experimental Pulmonary Fibrosis through Inhibition of the TGF-β Signaling Pathway. Sci. Rep. 2016, 6, 27610. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, J.; Cao, Y.; Wang, T. Salvianolic Acid B Suppresses Non-Small-Cell Lung Cancer Metastasis through PKM2-Independent Metabolic Reprogramming. Evid.-Based Complement. Altern. Med. eCAM 2022, 2022, 9302403. [Google Scholar] [CrossRef]
- Bachari, A.; Nassar, N.; Schanknecht, E.; Telukutla, S.; Piva, T.J.; Mantri, N. Rationalizing a prospective coupling effect of cannabinoids with the current pharmacotherapy for melanoma treatment. WIREs Mech. Dis. 2024, 16, e1633. [Google Scholar] [CrossRef]
- Tian, Y.; Li, M.W.; Liu, Q.K.; Kang, H. Clinical features and prognosis of cutaneous melanoma. Zhonghua Zhong Liu Za Zhi 2022, 44, 1146–1154. [Google Scholar]
- Zhong, Z.; Liu, Z.Z. Quality control index for standardized diagnosis and treatment of melanoma in China (2022 edition). Zhonghua Zhong Liu Za Zhi 2022, 44, 908–912. [Google Scholar]
- Wu, Z.; Yi, H.; Li, C.; Cui, Q.P.; Liu, H.Y.; Guo, F.Q.; Xiang, D.H.; Liu, X.Q.; Sun, X.L. Advantage analysis of flow-through cell method in quality evaluation of Chinese patent medicine: A case study of Danshen Tablets. Zhongguo Zhong Yao Za Zhi 2023, 48, 5548–5557. [Google Scholar] [PubMed]
- Xin, G.J.; Fu, J.H.; Han, X.; Li, L.; Guo, H.; Meng, H.X.; Zhao, Y.W.; Jia, F.F.; Liu, J.X. Salvianolic acid B regulates mitochondrial autophagy mediated by NIX to protect H9c2 cardiomyocytes from hypoxia/reoxygenation injury. Zhongguo Zhong Yao Za Zhi 2020, 45, 2960–2965. [Google Scholar] [PubMed]
- Khanfar, M.A.; Alqtaishat, S. Discovery of Potent Natural-Product-Derived SIRT2 Inhibitors Using Structure- Based Exploration of SIRT2 Pharmacophoric Space Coupled with QSAR Analyses. Anticancer Agents Med. Chem. 2021, 21, 2278–2286. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Wang, J.; Gao, W.; Liu, X.; Wu, S.; Wan, B.; Xu, L.; Li, Y. Salvianolic acid B reverses multidrug resistance in nude mice bearing human colon cancer stem cells. Mol. Med. Rep. 2018, 18, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Z.; Ardah, M.; Haikal, C.; Svanbergsson, A.; Diepenbroek, M.; Vaikath, N.N.; Li, W.; Wang, Z.Y.; Outeiro, T.F.; El-Agnaf, O.M.; et al. Dihydromyricetin and Salvianolic acid B inhibit alpha-synuclein aggregation and enhance chaperone-mediated autophagy. Transl. Neurodegener. 2019, 8, 18. [Google Scholar] [CrossRef]
- Tian, Y.P.; Cui, Y.S.; Zheng, X.; Liu, B.L.; Zhang, Y.P.; Wei, K.P.; Zhang, Z.; Hu, W.N.; Zhang, X.M.; Sun, G.G. Dihydromyricetin mediates epithelial mesenchymal transformation and regulates the proliferation and apoptosis of esophageal squamous cell carcinoma cells. Zhonghua Zhong Liu Za Zhi 2022, 44, 326–333. [Google Scholar]
- Xiong, B.H.; Li, S.S.; Ren, Z.Y.; Zhang, Z.; Liu, Y.Z.; Sun, Y.; Chi, J.L.; Luo, H.Y. Inhibition of GAS5 promoted invasion, migration and epithelial-mesenchymal transition of colorectal cancer cells via miR-21/PTEN/Akt axis. Zhonghua Zhong Liu Za Zhi 2022, 44, 1168–1174. [Google Scholar]
- Wu, C.; Chen, W.; Ding, H.; Li, D.; Wen, G.; Zhang, C.; Lu, W.; Chen, M.; Yang, Y. Salvianolic acid B exerts anti-liver fibrosis effects via inhibition of MAPK-mediated phospho-Smad2/3 at linker regions in vivo and in vitro. Life Sci. 2019, 239, 116881. [Google Scholar] [CrossRef]
- Dhapare, S.; Sakagami, M. Salvianolic acid B as an anti-emphysema agent I: In vitro stimulation of lung cell proliferation and migration, and protection against lung cell death, and in vivo lung STAT3 activation and VEGF elevation. Pulm. Pharmacol. Ther. 2018, 53, 107–115. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhai, W.; Fan, M.; Wu, J.; Wang, C. Salvianolic Acid B Significantly Suppresses the Migration of Melanoma Cells via Direct Interaction with β-Actin. Molecules 2024, 29, 906. https://doi.org/10.3390/molecules29040906
Zhang Y, Zhai W, Fan M, Wu J, Wang C. Salvianolic Acid B Significantly Suppresses the Migration of Melanoma Cells via Direct Interaction with β-Actin. Molecules. 2024; 29(4):906. https://doi.org/10.3390/molecules29040906
Chicago/Turabian StyleZhang, Ying, Wenjuan Zhai, Minqi Fan, Jinjun Wu, and Caiyan Wang. 2024. "Salvianolic Acid B Significantly Suppresses the Migration of Melanoma Cells via Direct Interaction with β-Actin" Molecules 29, no. 4: 906. https://doi.org/10.3390/molecules29040906
APA StyleZhang, Y., Zhai, W., Fan, M., Wu, J., & Wang, C. (2024). Salvianolic Acid B Significantly Suppresses the Migration of Melanoma Cells via Direct Interaction with β-Actin. Molecules, 29(4), 906. https://doi.org/10.3390/molecules29040906





