Fungi Tryptophan Synthases: What Is the Role of the Linker Connecting the α and β Structural Domains in Hemileia vastatrix TRPS? A Molecular Dynamics Investigation
Abstract
1. Introduction
2. Results
2.1. TRPS Sequence Extraction
2.2. TRPS Apo 3D Structure: Homology Modeling Step
2.3. Target 3D Structure: MD Simulations
2.4. Domain Movements
2.5. Tunnel Detection and Evolution during the MD
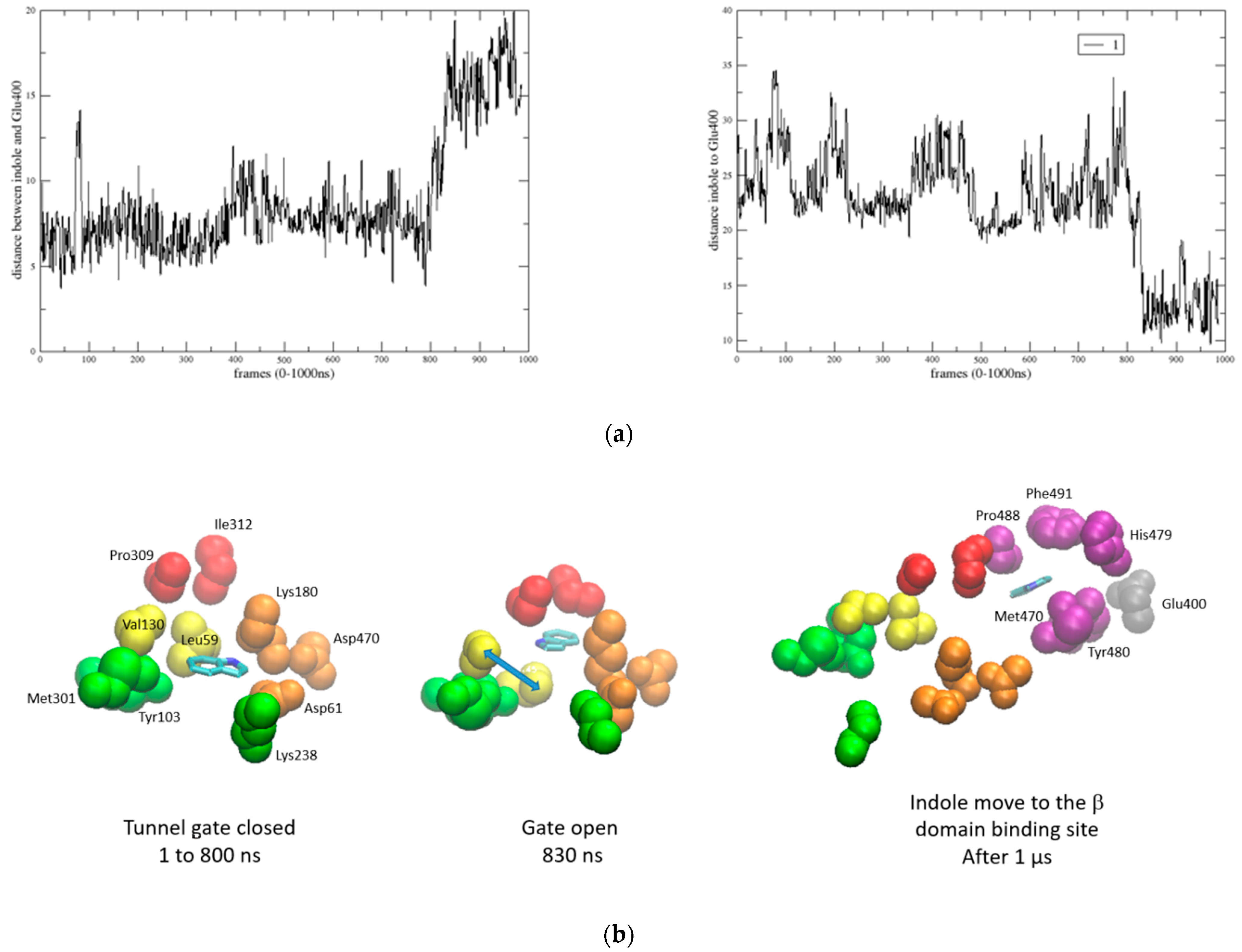
2.6. Channeling Process through the Tunnel
2.7. No-Linker Simulation
3. Discussion
4. Materials and Methods
4.1. Tryptophan Synthase Gene Detection in the Hemileia vastatrix Transcript
4.2. TRPS Target 3D Model Refinement: Molecular Dynamics Simulations (MD) Step
4.3. Domain Movements
4.4. Tunnel Detection and Indole Transfer Simulation
4.5. Removing the Linker
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miles, E.W. The Tryptophan Synthase α2β2 Complex: A Model for Substrate Channeling, Allosteric Communication, and Pyridoxal Phosphate Catalysis. J. Biol. Chem. 2013, 288, 10084–10091. [Google Scholar] [CrossRef]
- Barends, T.R.; Dunn, M.F.; Schlichting, I. Tryptophan synthase, an allosteric molecular factory. Curr. Opin. Chem. Biol. 2008, 12, 593–600. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, R.N.; Boehr, D.D. Allostery, engineering and inhibition of tryptophan synthase. Curr. Opin. Struct. Biol. 2023, 82, 102657. [Google Scholar] [CrossRef]
- Raboni, S.; Bettati, S.; Mozzarelli, A. Tryptophan synthase: A mine for enzymologists. Cell. Mol. Life Sci. 2009, 66, 2391–2403. [Google Scholar] [CrossRef] [PubMed]
- Lott, J.S. The tryptophan biosynthetic pathway is essential for Mycobacterium tuberculosis to cause disease. Biochem. Soc. Trans. 2020, 48, 2029–2037. [Google Scholar] [CrossRef]
- Bosken, Y.K.; Ai, R.; Hilario, E.; Ghosh, R.K.; Dunn, M.F.; Kan, S.; Niks, D.; Zhou, H.; Ma, W.; Mueller, L.J.; et al. Discovery of antimicrobial agent targeting tryptophan synthase. Protein Sci. 2022, 31, 432–442. [Google Scholar] [CrossRef]
- Yin, R.; Frey, M.; Gierl, A.; Glawischnig, E. Plants contain two distinct classes of functional tryptophan synthase beta proteins. Phytochemistry 2010, 71, 1667–1672. [Google Scholar] [CrossRef]
- Liu, W.-C.; Song, R.-F.; Zheng, S.-Q.; Li, T.-T.; Zhang, B.-L.; Gao, X.; Lu, Y.-T. Coordination of plant growth and abiotic stress responses by tryptophan synthase β subunit 1 through modulation of tryptophan and ABA homeostasis in Arabidopsis. Mol. Plant 2022, 15, 973–990. [Google Scholar] [CrossRef] [PubMed]
- Finn, J.; Langevine, C.; Birk, I.; Birk, J.; Nickerson, K.; Rodaway, S. Rational herbicide design by inhibition of tryptophan biosynthesis. Bioorg. Med. Chem. Lett. 1999, 9, 2297–2302. [Google Scholar] [CrossRef]
- Miles, E.W. Tryptophan synthase: A multienzyme complex with an intramolecular tunnel. Chem. Rec. 2001, 1, 140–151. [Google Scholar] [CrossRef]
- Dunn, M.F.; Niks, D.; Ngo, H.; Barends, T.R.; Schlichting, I. Tryptophan synthase: The workings of a channeling nanomachine. Trends Biochem. Sci. 2008, 33, 254–264. [Google Scholar] [CrossRef]
- Wellington, S.; Nag, P.P.; Michalska, K.; E Johnston, S.; Jedrzejczak, R.P.; Kaushik, V.K.; E Clatworthy, A.; Siddiqi, N.; McCarren, P.; Bajrami, B.; et al. A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase. Nat. Chem. Biol. 2017, 13, 943–950. [Google Scholar] [CrossRef]
- Michalska, K.; Chang, C.; Maltseva, N.I.; Jedrzejczak, R.; Robertson, G.T.; Gusovsky, F.; McCarren, P.; Schreiber, S.L.; Nag, P.P.; Joachimiak, A. Allosteric inhibitors of Mycobacterium tuberculosis tryptophan synthase. Protein Sci. 2020, 29, 779–788. [Google Scholar] [CrossRef]
- Dunn, M.F. Allosteric regulation of substrate channeling and catalysis in the tryptophan synthase bienzyme complex. Arch. Biochem. Biophys. 2012, 519, 154–166. [Google Scholar] [CrossRef]
- Niks, D.; Hilario, E.; Dierkers, A.; Ngo, H.; Borchardt, D.; Neubauer, T.J.; Fan, L.; Mueller, L.J.; Dunn, M.F. Allostery and Substrate Channeling in the Tryptophan Synthase Bienzyme Complex: Evidence for Two Subunit Conformations and Four Quaternary States. Biochemistry 2013, 52, 6396–6411. [Google Scholar] [CrossRef][Green Version]
- Buller, A.R.; van Roye, P.; Murciano-Calles, J.; Arnold, F.H. Tryptophan Synthase Uses an Atypical Mechanism To Achieve Substrate Specificity. Biochemistry 2016, 55, 7043–7046. [Google Scholar] [CrossRef]
- Michalska, K.; Gale, J.; Joachimiak, G.; Chang, C.; Hatzos-Skintges, C.; Nocek, B.; Johnston, S.E.; Bigelow, L.; Bajrami, B.; Jedrzejczak, R.P.; et al. Conservation of the structure and function of bacterial tryptophan synthases. IUCrJ 2019, 6, 649–664. [Google Scholar] [CrossRef] [PubMed]
- Burns, D.M.; Horn, V.; Paluh, J.; Yanofsky, C. Evolution of the tryptophan synthetase of fungi. Analysis of Experimentally Fused Escherichia Coli Tryptophan Synthetase Alpha and Beta Chains. J. Biol. Chem. 1990, 265, 2060–2069. [Google Scholar] [CrossRef] [PubMed]
- Sen, K.; Kinoshita, H.; Tazuke, K.; Maki, Y.; Yoshiura, Y.; Yakushi, T.; Shibai, H.; Kurosawa, S.-I. Analysis of the sexual development-promoting region of Schizophyllum commune TRP1 gene. Biosci. Biotechnol. Biochem. 2016, 80, 2033–2044. [Google Scholar] [CrossRef]
- Ireland, C.; Peekhaus, N.; Lu, P.; Sangari, R.; Zhang, A.; Masurekar, P.; An, Z. The tryptophan synthetase gene TRP1 of Nodulisporium sp.: Molecular characterization and its relation to nodulisporic acid A production. Appl. Microbiol. Biotechnol. 2008, 79, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Staats, C.C.; Silva, M.S.N.; Pinto, P.M.; Vainstein, M.H.; Schrank, A. The Metarhizium anisopliae trp1 Gene: Cloning and Regulatory Analysis. Curr. Microbiol. 2004, 49, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Nakai, R.; Sen, K.; Kurosawa, S.-I.; Shibai, H. Cloning and sequencing analysis ofTRP1gene of Flammulina velutipes. FEMS Microbiol. Lett. 2000, 190, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Karlovsky, P.; Prell, H.H. The TRP1 gene of Phytophthora parasitica encoding indole-3-glycerolphosphate synthase-N-(5′-phosphoribosyl) anthranilate isomerase: Structure and evolutionary distance from homologous fungal genes. Gene 1991, 109, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Skrzynia, C.; Binninger, D.M.; Alspaugh, J., II; Pukkila, P.J. Molecular characterization of TRP1, a gene coding for tryptophan synthetase in the basidiomycete Coprinus cinereus. Gene 1989, 81, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Braus, G.H.; Luger, K.; Paravicini, G.; Schmidheini, T.; Kirschner, K.; Hütter, R. The role of the TRP1 gene in yeast tryptophan biosynthesis. J. Biol. Chem. 1988, 263, 7868–7875. [Google Scholar] [CrossRef] [PubMed]
- Talhinhas, P.; Batista, D.; Diniz, I.; Vieira, A.; Silva, D.N.; Loureiro, A.; Tavares, S.; Pereira, A.P.; Azinheira, H.G.; Guerra-Guimarães, L.; et al. The coffee leaf rust pathogen Hemileia vastatrix: One and a half centuries around the tropics. Mol. Plant Pathol. 2017, 18, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.R.; Fernandes, R.C.; Carvalho, G.M.A.; Barreto, R.W.; Evans, H.C. Cryptosexuality and the Genetic Diversity Paradox in Coffee Rust, Hemileia vastatrix. PLoS ONE 2011, 6, e26387. [Google Scholar] [CrossRef][Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- InterPro. Available online: https://www.ebi.ac.uk/interpro/interproscan.html (accessed on 25 December 2023).
- Cantu, D.; Govindarajulu, M.; Kozik, A.; Wang, M.; Chen, X.; Kojima, K.K.; Jurka, J.; Michelmore, R.W.; Dubcovsky, J. Next Generation Sequencing Provides Rapid Access to the Genome of Puccinia striiformis f. sp. tritici, the Causal Agent of Wheat Stripe Rust. PLoS ONE 2011, 6, e24230. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef]
- Jurcik, A.; Bednar, D.; Byska, J.; Marques, S.M.; Furmanova, K.; Daniel, L.; Kokkonen, P.; Brezovsky, J.; Strnad, O.; Stourac, J.; et al. CAVER Analyst 2.0: Analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics 2018, 34, 3586–3588. [Google Scholar] [CrossRef]
- Schlitter, J.; Engels, M.; Krüger, P. Targeted molecular dynamics: A new approach for searching pathways of conformational transitions. J. Mol. Graph. 1994, 12, 84–89. [Google Scholar] [CrossRef]
- Talhinhas, P.; Azinheira, H.G.; Vieira, B.; Loureiro, A.; Tavares, S.; Batista, D.; Morin, E.; Petitot, A.-S.; Paulo, O.S.; Poulain, J.; et al. Overview of the functional virulent genome of the coffee leaf rust pathogen Hemileia vastatrix with an emphasis on early stages of infection. Front. Plant Sci. 2014, 5, 88. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hilario, E.; Caulkins, B.G.; Huang, Y.-M.M.; You, W.; Chang, C.-E.A.; Mueller, L.J.; Dunn, M.F.; Fan, L. Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2016, 1864, 268–279. [Google Scholar] [CrossRef]
- Cristancho, M.A.; Botero-Rozo, D.O.; Giraldo, W.; Tabima, J.; Riaño-Pachón, D.M.; Escobar, C.; Rozo, Y.; Rivera, L.F.; Durán, A.; Restrepo, S.; et al. Annotation of a hybrid partial genome of the coffee rust (Hemileia vastatrix) contributes to the gene repertoire catalog of the Pucciniales. Front. Plant Sci. 2014, 5, 594. [Google Scholar] [CrossRef] [PubMed]
- Porto, B.N.; Caixeta, E.T.; Mathioni, S.M.; Vidigal, P.M.P.; Zambolim, L.; Zambolim, E.M.; Donofrio, N.; Polson, S.W.; Maia, T.; Chen, C.; et al. Genome sequencing and transcript analysis of Hemileia vastatrix reveal expression dynamics of candidate effectors dependent on host compatibility. PLoS ONE 2019, 14, e0215598. [Google Scholar] [CrossRef]
- Martins, N.F.; Bresso, E.; Togawa, R.C.; Urban, M.; Antoniw, J.; Maigret, B.; Hammond-Kosack, K. Searching for Novel Targets to Control Wheat Head Blight Disease—I-Protein Identification, 3D Modeling and Virtual Screening. Adv. Microbiol. 2016, 06, 811–830. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank, 1999–. In International Tables for Crystallography; International Union of Crystallography: Chester, UK, 2006; pp. 675–684. [Google Scholar]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Protein Structure Modeling with MODELLER. In Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 39–54. [Google Scholar]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- di Luccio, E.; Koehl, P. The H-factor as a novel quality metric for homology modeling. J. Clin. Bioinform. 2012, 2, 18. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- de Souza, J.V.P.; Kioshima, E.S.; Murase, L.S.; Lima, D.d.S.; Seixas, F.A.V.; Maigret, B.; Cardoso, R.F. Identification of new putative inhibitors of Mycobacterium tuberculosis 3-dehydroshikimate dehydratase from a combination of ligand- and structure-based and deep learning in silico approaches. J. Biomol. Struct. Dyn. 2023, 41, 2971–2980. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Falsafi-Zadeh, S.; Karimi, Z.; Galehdari, H. VMD DisRg: New User-Friendly Implement for calculation distance and radius of gyration in VMD program. Bioinformation 2012, 8, 341–343. [Google Scholar] [CrossRef] [PubMed]
- Hünenberger, P.; Mark, A.; van Gunsteren, W. Fluctuation and Cross-correlation Analysis of Protein Motions Observed in Nanosecond Molecular Dynamics Simulations. J. Mol. Biol. 1995, 252, 492–503. [Google Scholar] [CrossRef]
- Grant, B.J.; Rodrigues, A.P.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef]
- Brown, D.K.; Penkler, D.L.; Amamuddy, O.S.; Ross, C.; Atilgan, A.R.; Atilgan, C.; Bishop, Ö.T. MD-TASK: A software suite for analyzing molecular dynamics trajectories. Bioinformatics 2017, 33, 2768–2771. [Google Scholar] [CrossRef]
- Weyand, M.; Schlichting, I. Crystal Structure of Wild-Type Tryptophan Synthase Complexed with the Natural Substrate Indole-3-glycerol Phosphate. Biochemistry 1999, 38, 16469–16480. [Google Scholar] [CrossRef]
- Haider, K.; Cruz, A.; Ramsey, S.; Gilson, M.K.; Kurtzman, T. Solvation Structure and Thermodynamic Mapping (SSTMap): An Open-Source, Flexible Package for the Analysis of Water in Molecular Dynamics Trajectories. J. Chem. Theory Comput. 2017, 14, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Ray, N.; Cavin, X.; Paul, J.-C.; Maigret, B. Intersurf: Dynamic interface between proteins. J. Mol. Graph. Model. 2005, 23, 347–354. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pavelka, A.; Sebestova, E.; Kozlikova, B.; Brezovsky, J.; Sochor, J.; Damborsky, J. CAVER: Algorithms for Analyzing Dynamics of Tunnels in Macromolecules. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015, 13, 505–517. [Google Scholar] [CrossRef] [PubMed]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, N.F.; Viana, M.J.A.; Maigret, B. Fungi Tryptophan Synthases: What Is the Role of the Linker Connecting the α and β Structural Domains in Hemileia vastatrix TRPS? A Molecular Dynamics Investigation. Molecules 2024, 29, 756. https://doi.org/10.3390/molecules29040756
Martins NF, Viana MJA, Maigret B. Fungi Tryptophan Synthases: What Is the Role of the Linker Connecting the α and β Structural Domains in Hemileia vastatrix TRPS? A Molecular Dynamics Investigation. Molecules. 2024; 29(4):756. https://doi.org/10.3390/molecules29040756
Chicago/Turabian StyleMartins, Natália F., Marcos J. A. Viana, and Bernard Maigret. 2024. "Fungi Tryptophan Synthases: What Is the Role of the Linker Connecting the α and β Structural Domains in Hemileia vastatrix TRPS? A Molecular Dynamics Investigation" Molecules 29, no. 4: 756. https://doi.org/10.3390/molecules29040756
APA StyleMartins, N. F., Viana, M. J. A., & Maigret, B. (2024). Fungi Tryptophan Synthases: What Is the Role of the Linker Connecting the α and β Structural Domains in Hemileia vastatrix TRPS? A Molecular Dynamics Investigation. Molecules, 29(4), 756. https://doi.org/10.3390/molecules29040756






