Medicinal Chemistry Strategies for the Modification of Bioactive Natural Products
Abstract
1. Introduction
2. Characteristics of Natural Products
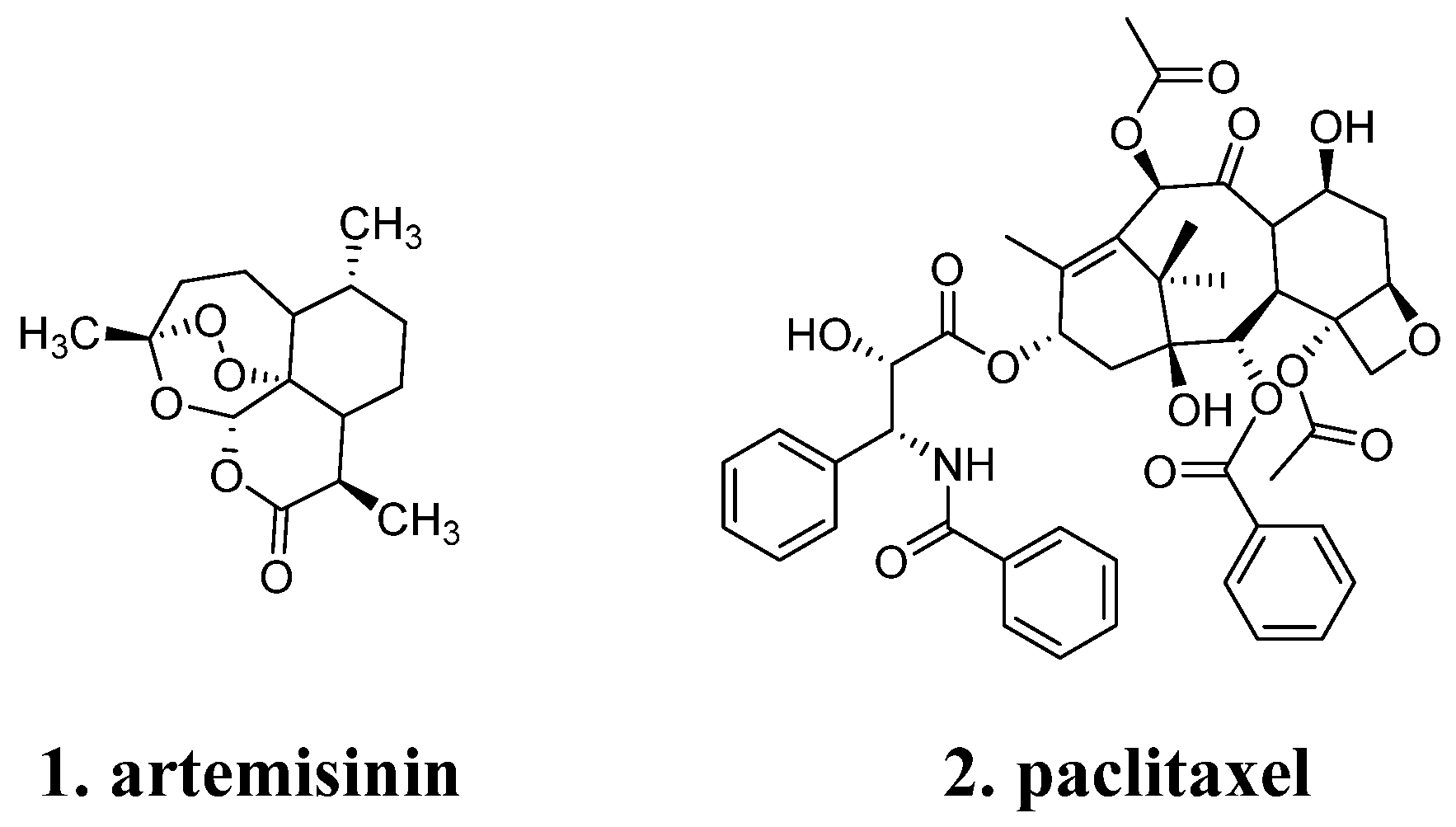
2.1. Diversity and Complexity of Natural Product Structures
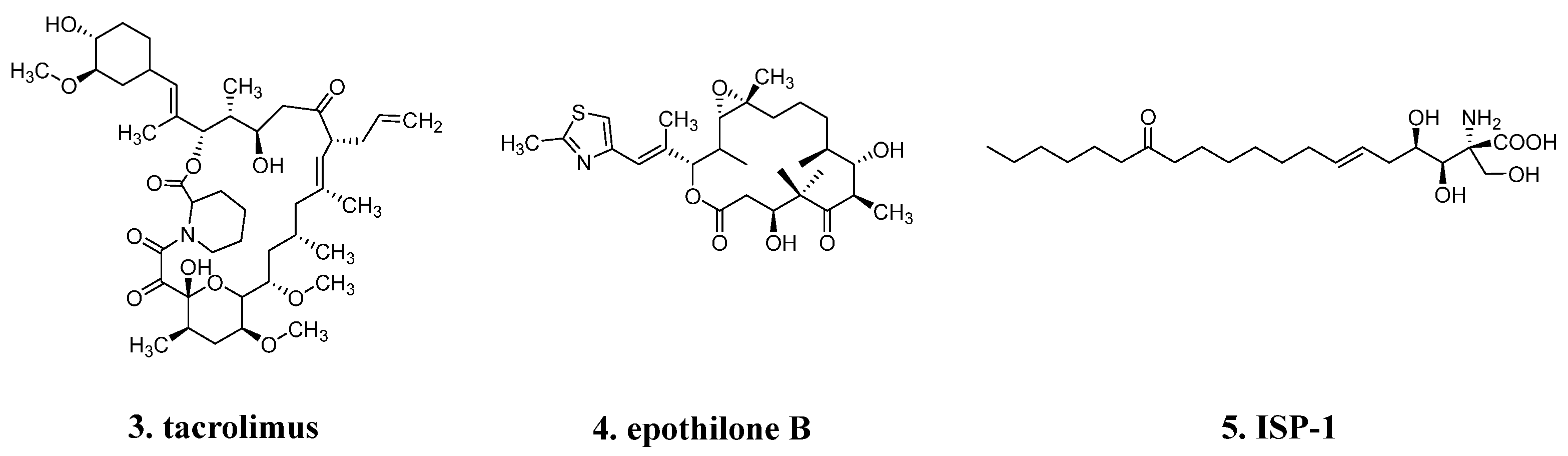
2.2. High sp3 Carbon Content and Few Aromatic Rings
2.3. Low Nitrogen and Halogen Content
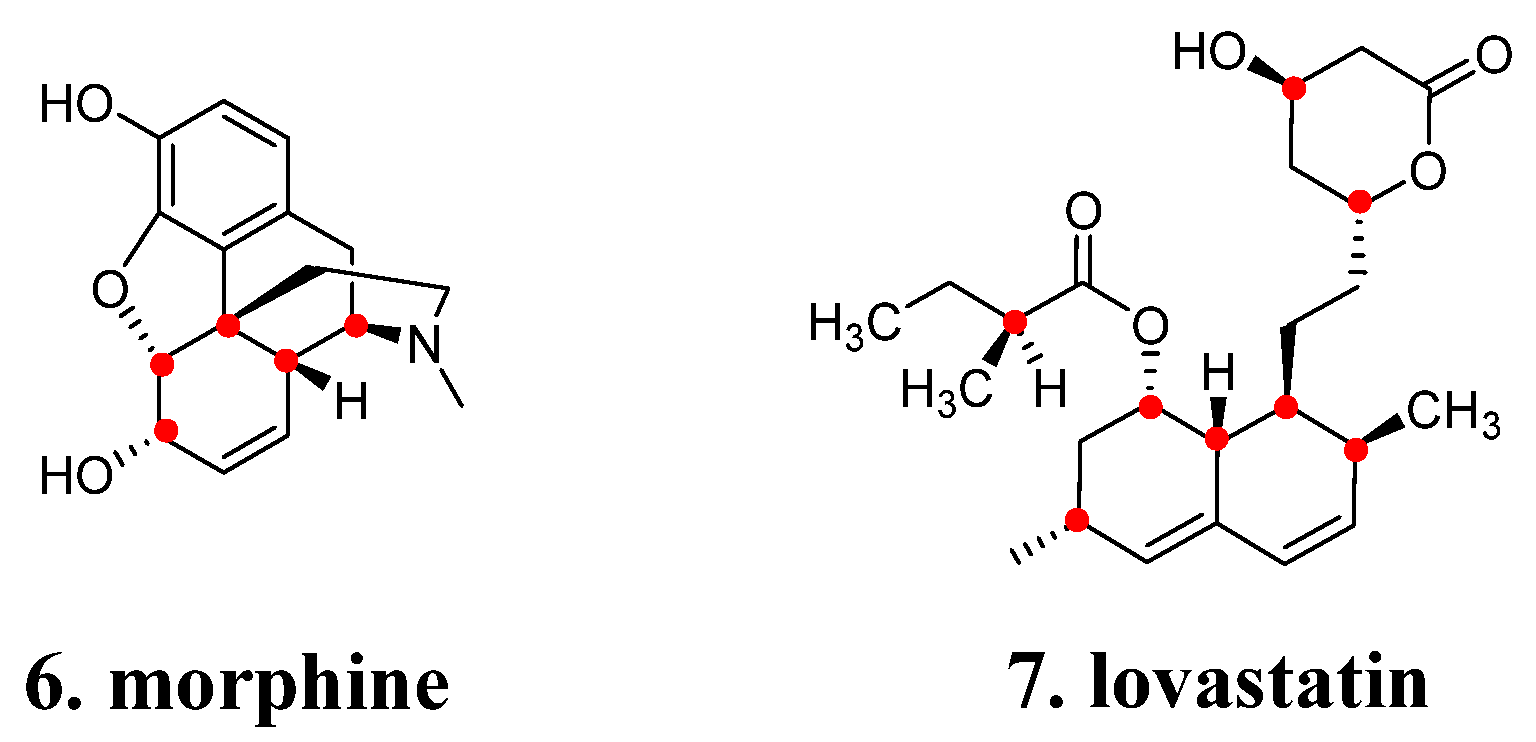
2.4. Chirality and Stereochemistry
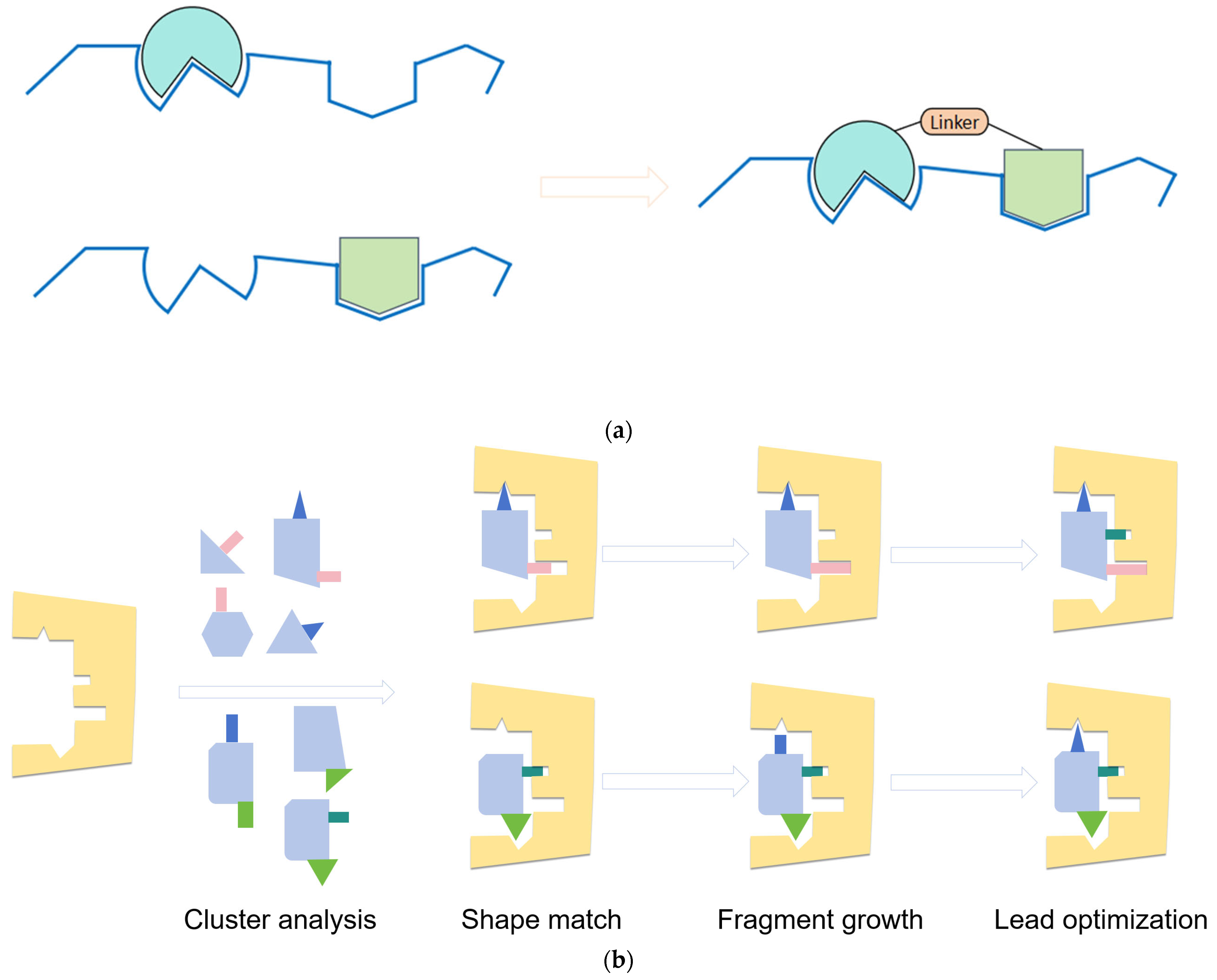
3. Employ Structure-Based Drug Design to Natural Products Optimization
4. Modification Strategies of Successful Examples
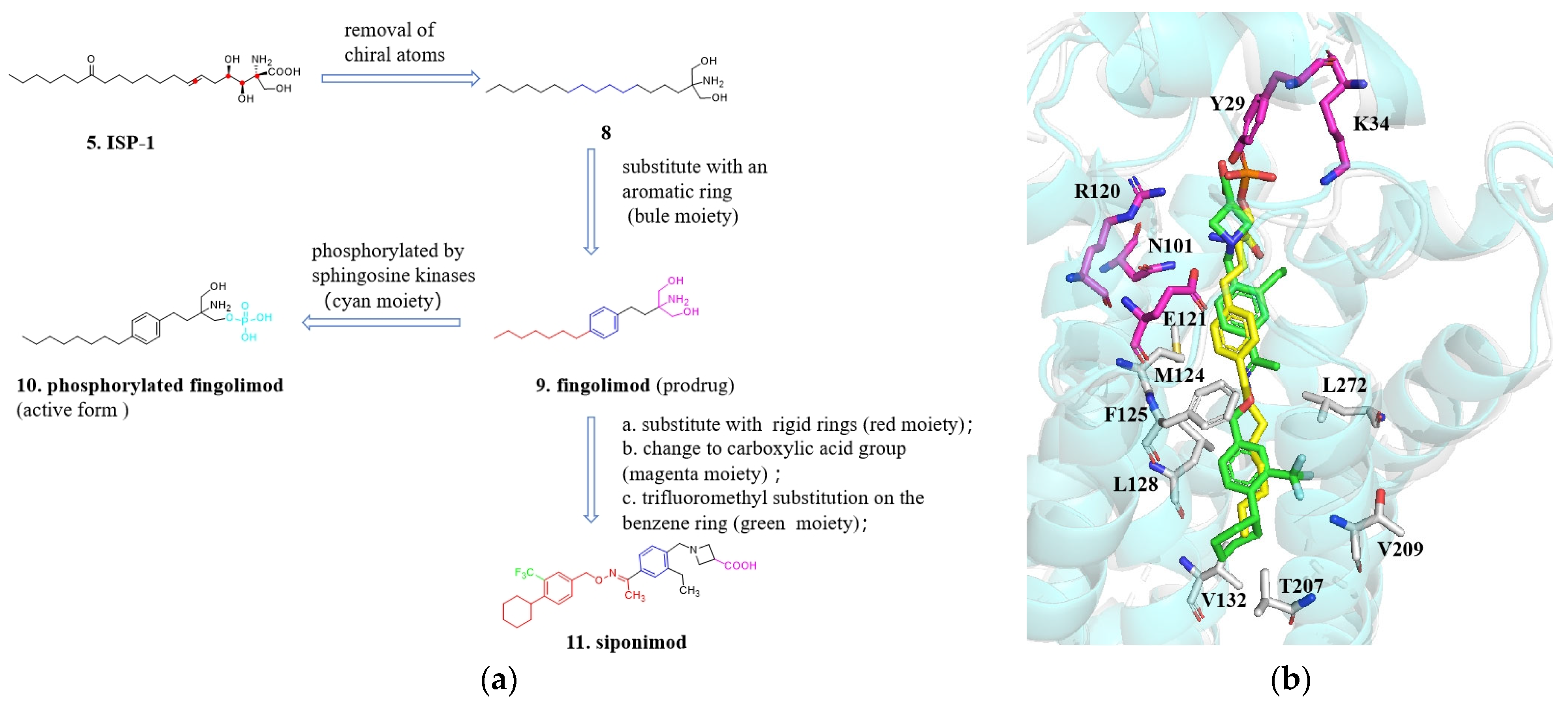
4.1. From ISP-1 to Siponimod
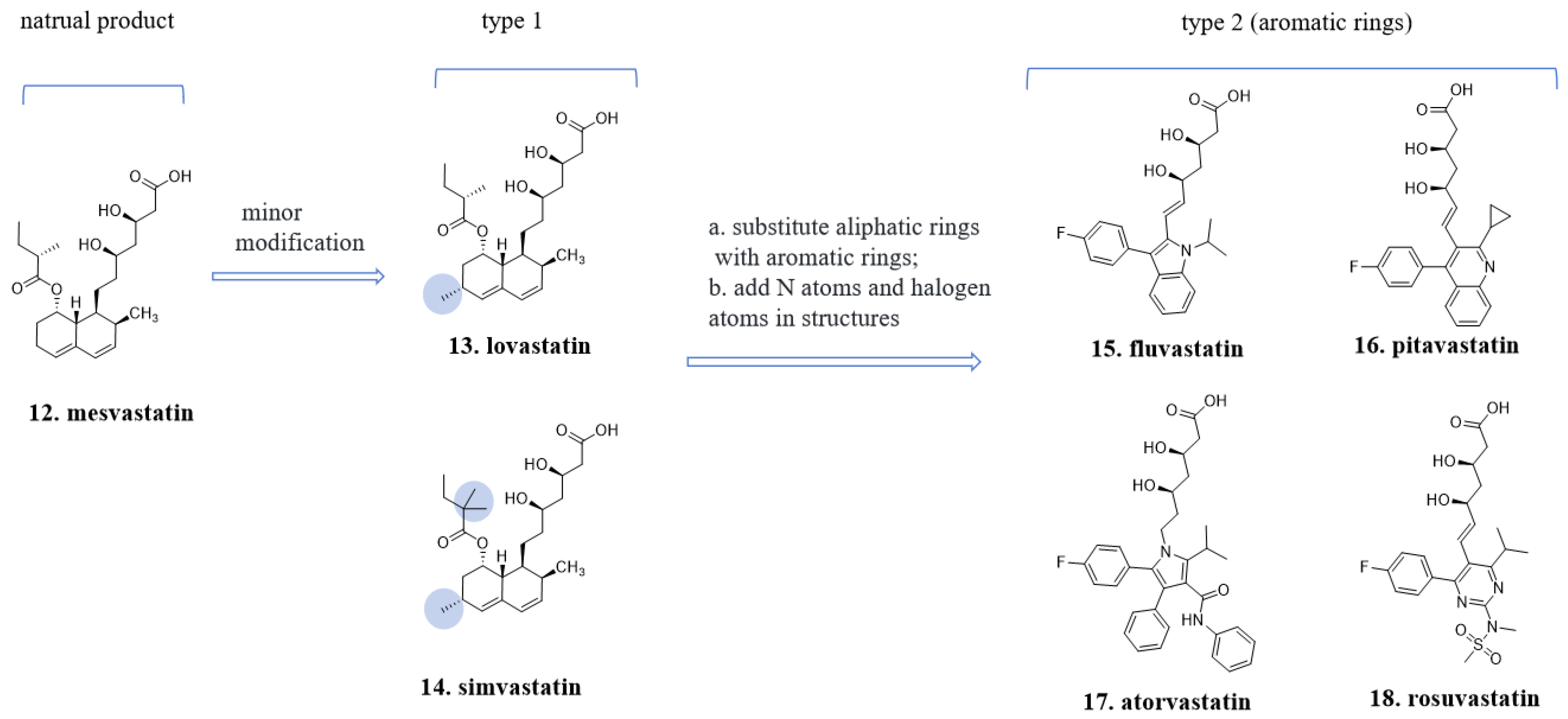
4.2. The “Statin” Drugs
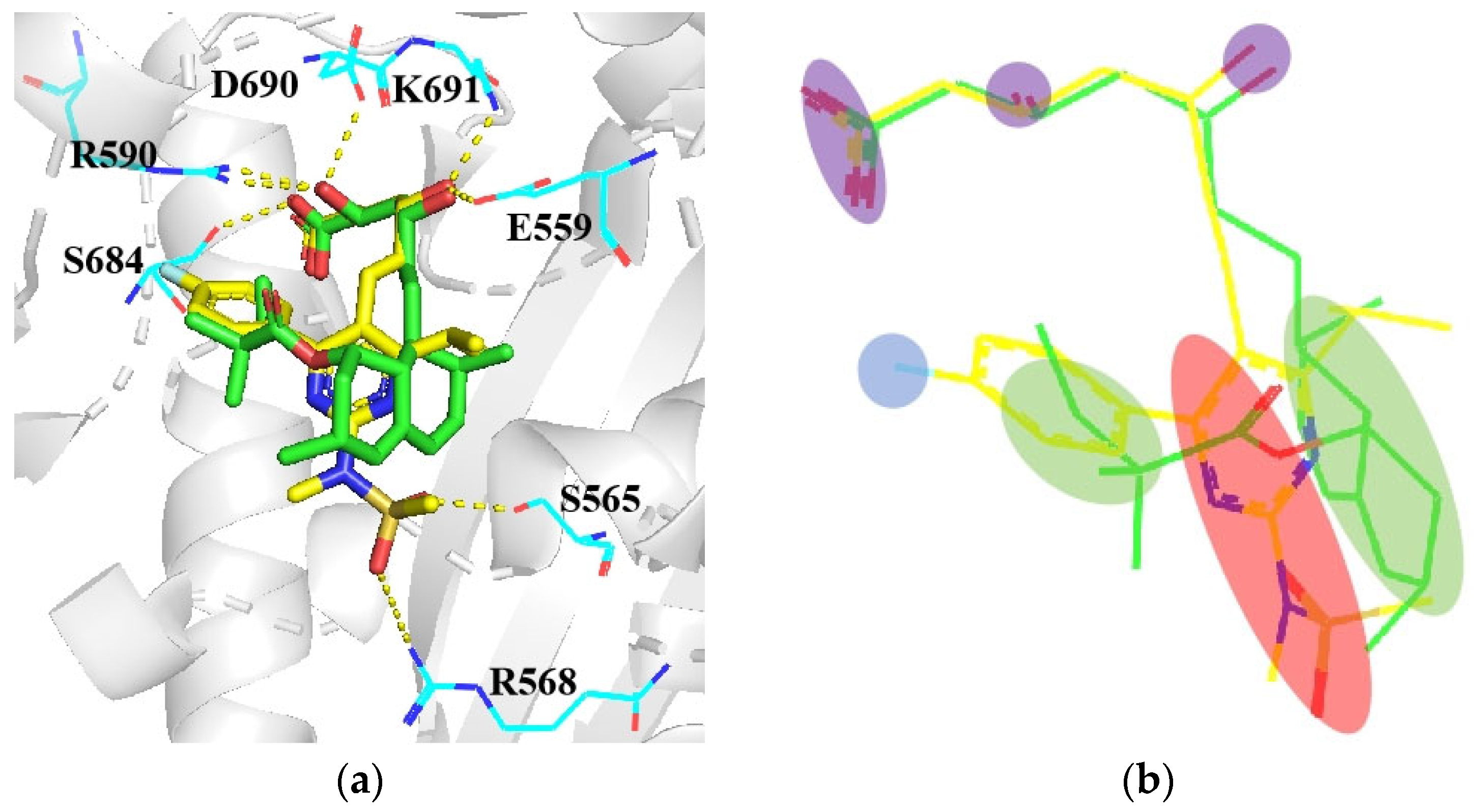
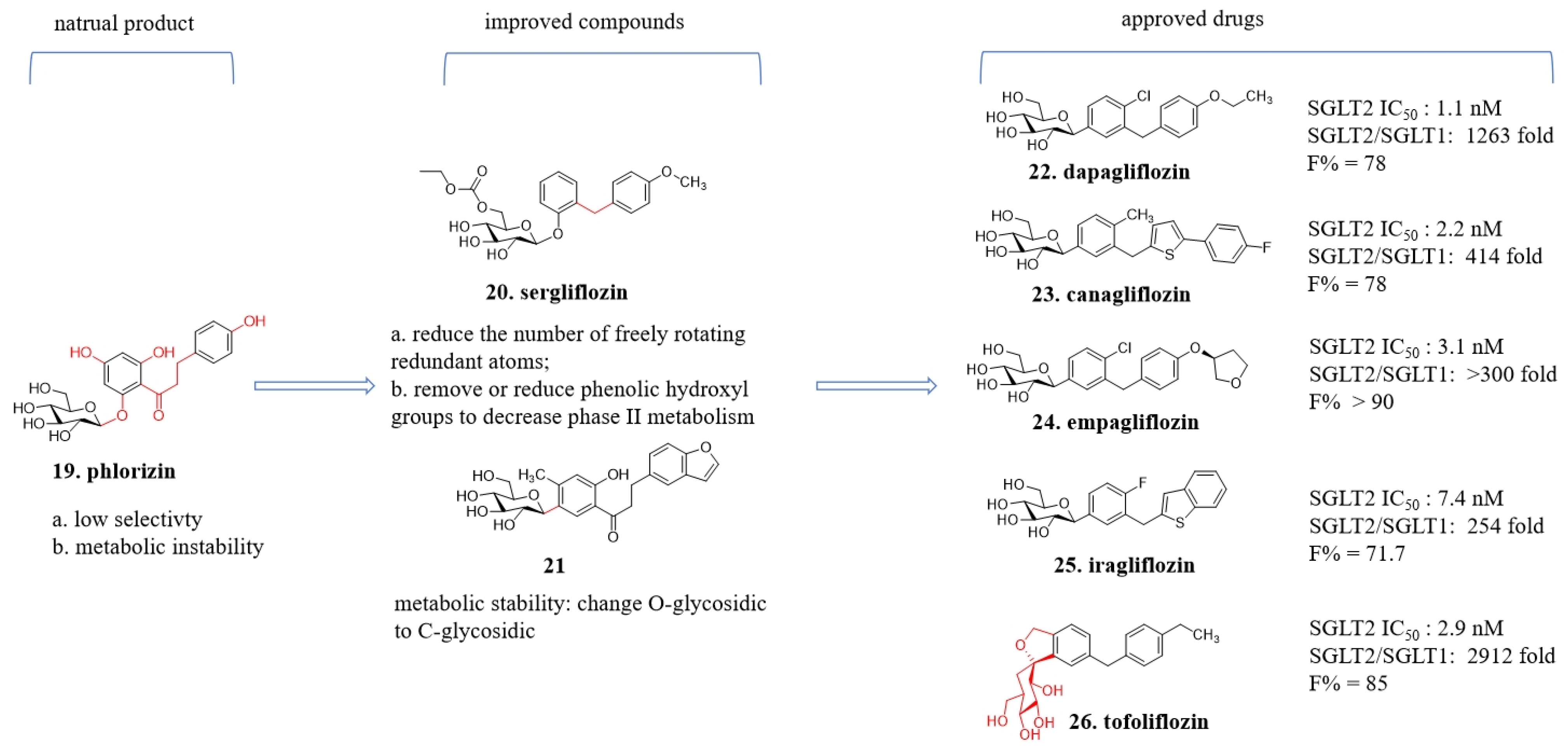
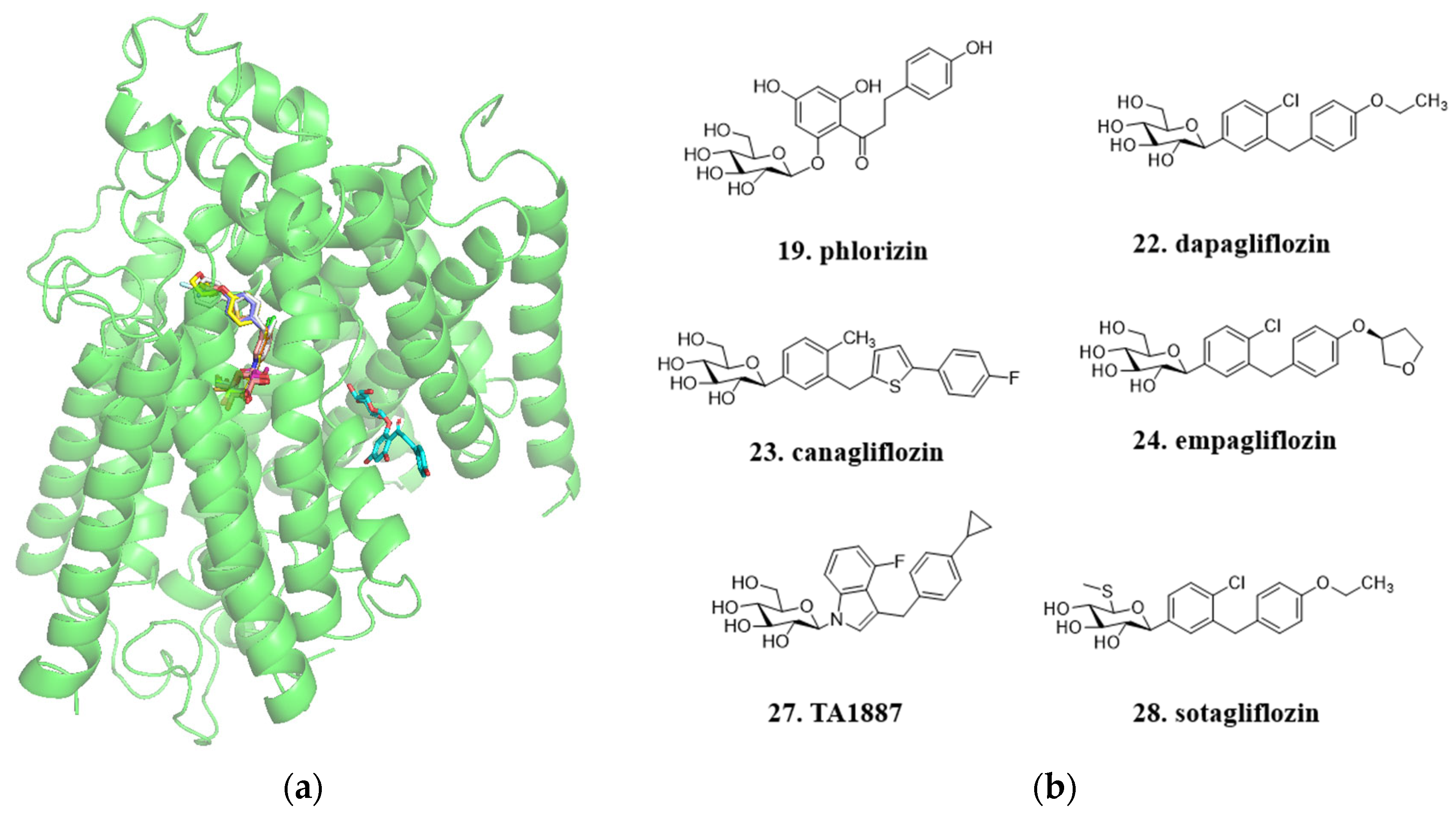
4.3. From Phloridzin to Dapagliflozin
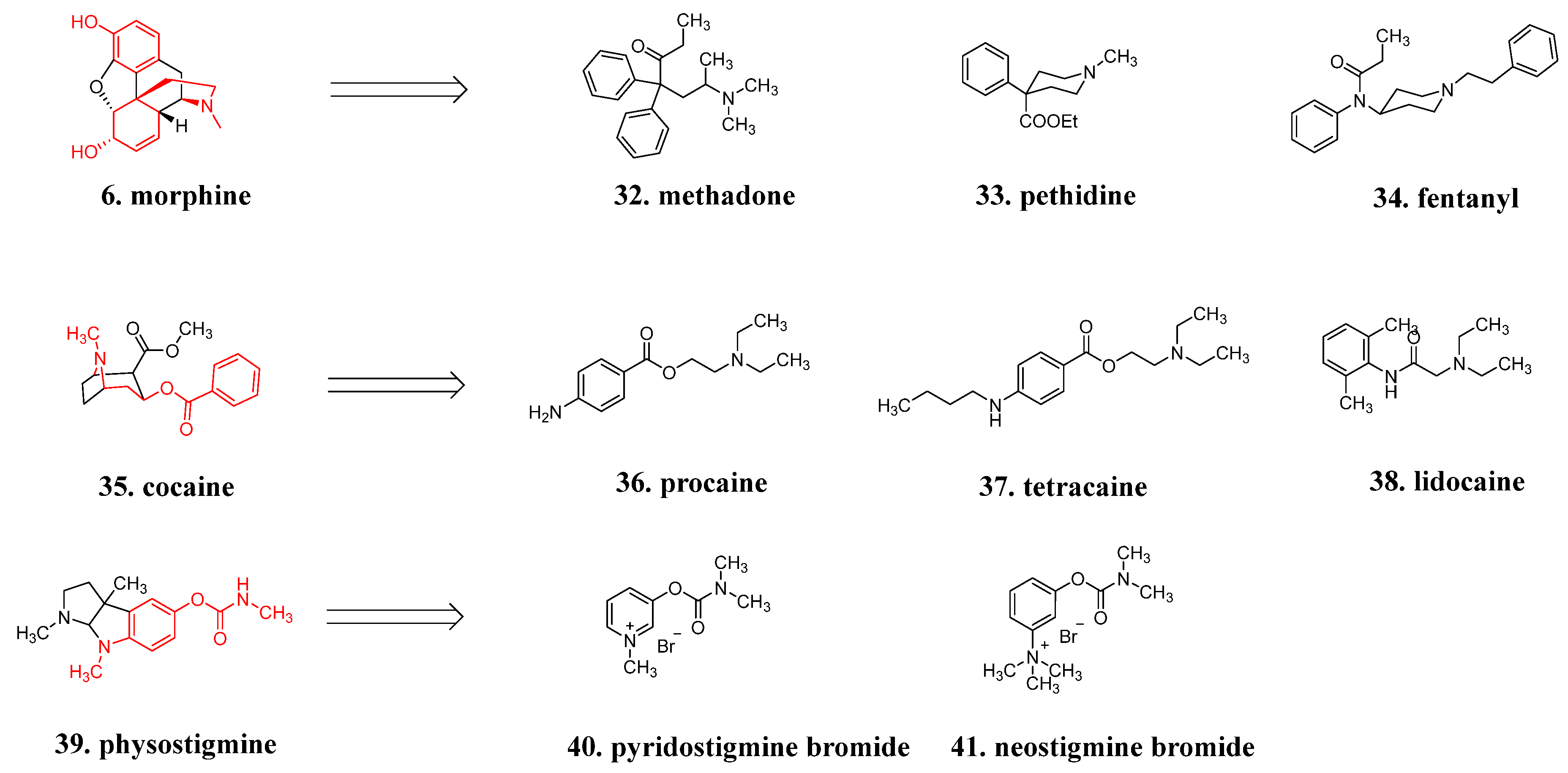
4.4. Structure Simplification
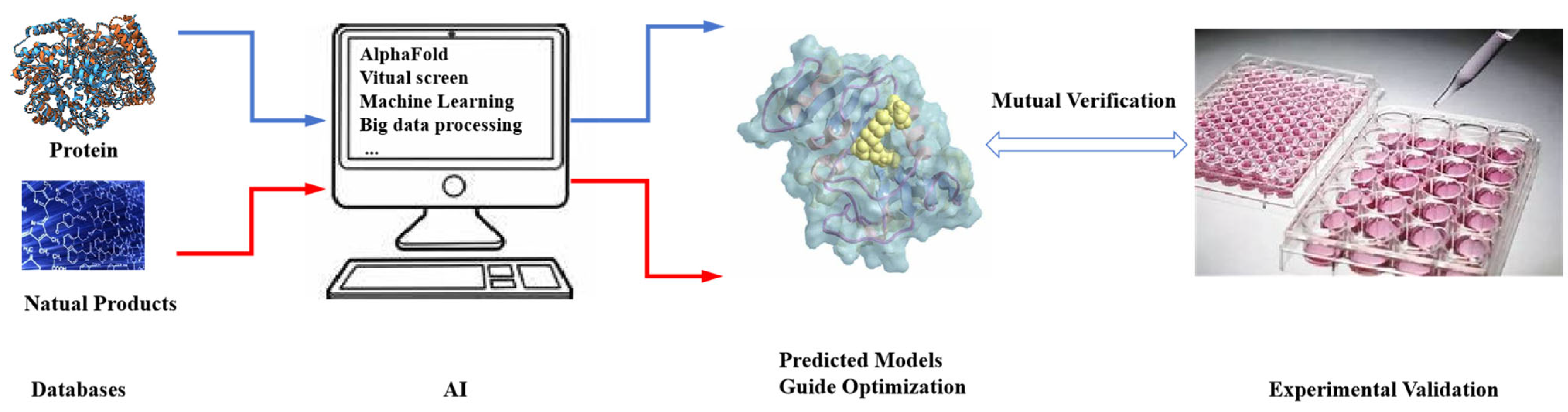
5. Optimization of Bioactive Natural Products Using Computer-Guided and Structure-Based Screening Strategies
6. Brief Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, M.L.; Schneider, G. Scaffold architecture and pharmacophoric properties of natural products and trade drugs: Application in the design of natural product-based combinatorial libraries. J. Comb. Chem. 2001, 3, 284–289. [Google Scholar] [CrossRef]
- Brown, D.G.; Lister, T.; May-Dracka, T.L. New natural products as new leads for antibacterial drug discovery. Bioorg. Med. Chem. Lett. 2014, 24, 413–418. [Google Scholar] [CrossRef]
- Harvey, A.L.; Edrada-Ebel, R.; Quinn, R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [Google Scholar] [CrossRef]
- Malandraki-Miller, S.; Riley, P.R. Use of artificial intelligence to enhance phenotypic drug discovery. Drug Discov. Today 2021, 26, 887–901. [Google Scholar] [CrossRef]
- Singh, S.; Aghdam, S.A.; Lahowetz, R.M.; Brown, A.M.V. Metapangenomics of wild and cultivated banana microbiome reveals a plethora of host-associated protective functions. Environ. Microbiome 2023, 18, 36. [Google Scholar] [CrossRef]
- Ding, J.; Hu, C.X.; Liu, P.Y.; Zhu, X.C.; Du, Z.H. Dynamic mode decomposition of the unsteady flow in a vaneless diffuser. J. Chin. Soc. Power Eng. 2017, 37, 447–453. [Google Scholar]
- Ma, N.; Zhang, Z.Y.; Liao, F.L.; Jiang, T.L.; Tu, Y.Y. The birth of artemisinin. Pharmacol. Ther. 2020, 216, 107658. [Google Scholar] [CrossRef] [PubMed]
- Paller, C.J.; Antonarakis, E.S. Cabazitaxel: A novel second-line treatment for metastatic castration-resistant prostate cancer. Drug Des. Dev. Ther. 2011, 2011, 117–124. [Google Scholar]
- Hopkins, A.L.; Groom, C.R.; Alex, A. Ligand efficiency: A useful metric for lead selection. Drug Discov. Today 2004, 9, 430–431. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.; Reker, D.; Schneider, P.; Schneider, G. Counting on natural products for drug design. Nat. Chem. 2016, 8, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Henkel, T.; Brunne, R.M.; Müller, H.; Reichel, F. Statistical investigation into the structural complementarity of natural products and synthetic compounds. Angew. Chem. Int. Ed. 1999, 38, 643–647. [Google Scholar] [CrossRef]
- Guo, Z.R. The modification of natural products for medical use. Acta Pharm. Sin. B 2017, 7, 119–136. [Google Scholar] [CrossRef]
- Cavallo, G.; Metrangolo, P.; Milani, R.; Pilati, T.; Terraneo, G. The Halogen Bond. Chem. Rev. 2016, 116, 2478–2601. [Google Scholar] [CrossRef]
- Prabhakara, C.T.; Patil, S.A.; Toragalmath, S.S.; Kinnal, S.M.; Badami, P.S. Synthesis, characterization and biological approach of metal chelates of some first row transition metal ions with halogenated bidentate coumarin Schiff bases containing N and O donor atoms. J. Photochem. Photobiol. B 2016, 157, 1–14. [Google Scholar] [CrossRef]
- Zhao, D.; Chen, C.; Liu, H.Y.; Zheng, L.K.; Tong, Y.; Qu, D.; Han, S.Q. Biological evaluation of halogenated thiazolo [3,2-a]pyrimidin-3-one carboxylic acid derivatives targeting the YycG histidine kinase. Eur. J. Med. Chem. 2014, 87, 500–507. [Google Scholar] [CrossRef]
- Muller, G. Medicinal chemistry of target family-directed masterkeys. Drug Discov. Today 2003, 8, 681–691. [Google Scholar] [CrossRef]
- Bemis, G.W.; Murcko, M.A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 1996, 39, 2887–2893. [Google Scholar] [CrossRef] [PubMed]
- Bemis, G.W.; Murcko, M.A. Properties of known drugs. 2. Side chains. J. Med. Chem. 1999, 42, 5095–5099. [Google Scholar] [CrossRef] [PubMed]
- Lewell, X.Q.; Judd, D.B.; Watson, S.P.; Hann, M.M. RECAP—Retrosynthetic combinatorial analysis procedure: A powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J. Chem. Inf. Comput. Sci. 1998, 38, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Jhoti, H.; Leach, A.R. Structure-Based Drug Discovery; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Fujita, T.; Inoue, K.; Yamamoto, S.; Ikumoto, T.; Sasaki, S.; Toyama, R.; Chiba, K.; Hoshino, Y.; Okumoto, T. Fungal metabolites. Part 11. A potent immunosuppressive activity found in Isaria sinclairii metabolite. J. Antibiot. 1994, 47, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Hirose, R.; Yoneta, M.; Sasaki, S.; Inoue, K.; Kiuchi, M.; Hirase, S.; Chiba, K.; Sakamoto, H.; Arita, M. Potent immunosuppressants, 2-alkyl-2-aminopropane-1,3-diols. J. Med. Chem. 1996, 39, 4451–4459. [Google Scholar] [CrossRef]
- Fujita, T.; Hirose, R.; Hamamichi, N.; Kitao, Y.; Sasaki, S.; Yoneta, M.; Chiba, K. 2-Substituted 2-Aminoethanol—Minimum Essential Structure for Immunosuppressive Activity of Isp-I (Myriocin). Bioorg. Med. Chem. Lett. 1995, 5, 1857–1860. [Google Scholar] [CrossRef]
- Sasaki, S.; Hashimoto, R.; Kiuchi, M.; Inoue, K.; Ikumoto, T.; Hirose, R.; Chiba, K.; Hoshino, Y.; Okumoto, T.; Fujita, T. Fungal Metabolites. 14. novel potent immunosuppressants, mycestericins, produced by mycelia-sterilia. J. Antibiot. 1994, 47, 420–433. [Google Scholar] [CrossRef]
- Fujita, T.; Yoneta, M.; Hirose, R.; Sasaki, S.; Inoue, K.; Kiuchi, M.; Hirase, S.; Adachi, K.; Arita, M.; Chiba, K. Simple compounds, 2-Alkyl-2-amino-1,3-propanediols have potent immunosuppressive activity. Bioorg. Med. Chem. Lett. 1995, 5, 847–852. [Google Scholar] [CrossRef]
- Adachi, K.; Kohara, T.; Nakao, N.; Arita, M.; Chiba, K.; Mishina, T.; Sasaki, S.; Fujita, T. Design, synthesis, and structure-activity-relationships of 2-substituted-2-amino-1,3-propanediols—Discovery of a novel immunosuppressant, Fty720. Bioorg. Med. Chem. Lett. 1995, 5, 853–856. [Google Scholar] [CrossRef]
- Kovarik, J.M.; Hartmann, S.; Bartlett, M.; Riviere, G.J.; Neddermann, D.; Wang, Y.B.; Port, A.; Schmouder, R.L. Oral-intravenous crossover study of fingolimod pharmacokinetics, lymphocyte responses and cardiac effects. Biopharm. Drug Dispos. 2007, 28, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Mandala, S.; Hajdu, R.; Bergstrom, J.; Quackenbush, E.; Xie, J.; Milligan, J.; Thornton, R.; Shei, G.J.; Card, D.; Keohane, C.; et al. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science 2002, 296, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Jia, G.W.; Wu, C.; Wang, W.; Cheng, L.; Li, Q.; Li, Z.Y.; Luo, K.D.; Yang, S.Y.; Yan, W.; et al. Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition. Cell Res. 2021, 31, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.F.; Gray, N.S.; Gao, W.Q.; Mi, Y.; Fan, Y.; Wang, X.; Tuntland, T.; Che, J.W.; Lefebvre, S.; Chen, Y.; et al. Discovery of BAF312 (Siponimod), a potent and selective S1P receptor modulator. ACS Med. Chem. Lett. 2013, 4, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Hanson, M.A.; Roth, C.B.; Jo, E.; Griffith, M.T.; Scott, F.L.; Reinhart, G.; Desale, H.; Clemons, B.; Cahalan, S.M.; Schuerer, S.C.; et al. Crystal structure of a lipid G protein-coupled receptor. Science 2012, 335, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Alberts, A.W.; Chen, J.; Kuron, G.; Hunt, V.; Huff, J.; Hoffman, C.; Rothrock, J.; Lopez, M.; Joshua, H.; Harris, E.; et al. Mevinolin—A highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme—A reductase and a cholesterol-lowering agent. Proc. Natl. Acad. Sci. USA 1980, 77, 3957–3961. [Google Scholar] [CrossRef] [PubMed]
- Istvan, E.S.; Deisenhofer, J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Ojima, I. Use of fluorine in the medicinal chemistry and chemical biology of bioactive compounds—A case study on fluorinated taxane anticancer agents. Chembiochem 2004, 5, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Jencks, W.P. On the attribution and additivity of binding energies. Proc. Natl. Acad. Sci. USA 1981, 78, 4046–4050. [Google Scholar] [CrossRef] [PubMed]
- Hung, A.W.; Silvestre, H.L.; Wen, S.J.; Ciulli, A.; Blundell, T.L.; Abell, C. Application of Fragment Growing and Fragment Linking to the Discovery of Inhibitors of Pantothenate Synthetase. Angew. Chem. Int. Ed. 2009, 48, 8452–8456. [Google Scholar] [CrossRef]
- Miyaura, N.; Suzuki, A. Palladium-Catalyzed Cross-Coupling Reactions of Organoboron Compounds. Chem. Rev. 1995, 95, 2457–2483. [Google Scholar] [CrossRef]
- Driver, M.S.; Hartwig, J.F. A second-generation catalyst for aryl halide amination: Mixed secondary amines from aryl halides and primary amines catalyzed by (DPPF)PdCl2. J. Am. Chem. Soc. 1996, 118, 7217–7218. [Google Scholar] [CrossRef]
- Beletskaya, I.P.; Cheprakov, A.V. Copper in cross-coupling reactions—The post-Ullmann chemistry. Coord. Chem. Rev. 2004, 248, 2337–2364. [Google Scholar] [CrossRef]
- Collaborators, C. Efficacy and safety of cholesterol-lowering treatment: Prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 2005, 366, 1358. [Google Scholar]
- Akizawa, T.; Yamaguchi, Y.; Otsuka, T.; Reusch, M. A Phase 3, multicenter, randomized, two-Arm, open-label study of intermittent oral dosing of roxadustat for the treatment of anemia in Japanese erythropoiesis-stimulating agent-naive chronic kidney disease patients not on dialysis. Nephron 2020, 144, 372–382. [Google Scholar] [CrossRef]
- Sheridan, R.P. The most common chemical replacements in drug-like compounds. J. Chem. Inf. Comp. Sci. 2002, 42, 103–108. [Google Scholar] [CrossRef]
- de Kloe, G.E.; Bailey, D.; Leurs, R.; de Esch, I.J.P. Transforming fragments into candidates: Small becomes big in medicinal chemistry. Drug Discov. Today 2009, 14, 630–646. [Google Scholar] [CrossRef] [PubMed]
- Chasis, H.; Jolliffe, N.; Smith, H.W. The action of phlorizin on the excretion of glucose, xylose, sucrose, creatinine and urea by man. J. Clin. Investig. 1933, 12, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Tsujihara, K.; Hongu, M.; Saito, K.; Inamasu, M.; Arakawa, K.; Oku, A.; Matsumoto, M. Na+-glucose cotransporter inhibitors as antidiabetics. I. Synthesis and pharmacological properties of 4’-dehydroxyphlorizin derivatives based on a new concept. Chem. Pharm. Bull. 1996, 44, 1174–1180. [Google Scholar] [CrossRef] [PubMed]
- Tsujihara, K.; Hongu, M.; Saito, K.; Kawanishi, H.; Kuriyama, K.; Matsumoto, M.; Oku, A.; Ueta, K.; Tsuda, M.; Saito, A. Na-glucose cotransporter (SGLT) inhibitors as antidiabetic agents: 4: Synthesis and pharmacological properties of 4′-dehydroxyphlorizin derivatives substituted on the B ring. J. Med. Chem. 1999, 42, 5311–5324. [Google Scholar] [CrossRef] [PubMed]
- Oku, A.; Ueta, K.; Arakawa, K.; Ishihara, T.; Nawano, M.; Kuronuma, Y.; Matsumoto, M.; Saito, A.; Tsujihara, K.; Anai, M.; et al. T-1095, an inhibitor of renal Na-glucose cotransporters, may provide a novel approach to treating diabetes. Diabetes 1999, 48, 1794–1800. [Google Scholar] [CrossRef] [PubMed]
- Ehrenkranz, J.R.L.; Lewis, N.G.; Kahn, C.R.; Roth, J. Phlorizin: A review. Diabetes-Metab. Res. 2005, 21, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Katsuno, K.; Fujimori, Y.; Takemura, Y.; Hiratochi, M.; Itoh, F.; Komatsu, Y.; Fujikura, H.; Isaji, M. Sergliflozin, a novel selective inhibitor of low-affinity sodium glucose cotransporter (SGLT2), validates the critical role of SGLT2 in renal glucose reabsorption and modulates plasma glucose level. J. Pharmacol. Exp. Ther. 2007, 320, 323. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.; Ellsworth, B.A.; Nirschl, A.A.; Mccann, P.J.; Patel, M.; Girotra, R.N.; Wu, G.; Sher, P.M.; Morrison, E.P.; Biller, S.A. Discovery of dapagliflozin: A potent, selective renal sodium-dependent glucose cotransporter 2 (SGLT2) inhibitor for the treatment of type 2 diabetes. J. Med. Chem. 2008, 51, 1145–1149. [Google Scholar] [CrossRef]
- Nomura, S.; Sakamaki, S.; Hongu, M.; Kawanishi, E.; Koga, Y.; Sakamoto, T.; Yamamoto, Y.; Ueta, K.; Kimata, H.; Nakayama, K.; et al. Discovery of Canagliflozin, a novel C-glucoside with thiophene ring, as sodium-dependent glucose cotransporter 2 inhibitor for the treatment of type 2 diabetes mellitus. J. Med. Chem. 2010, 53, 6355–6360. [Google Scholar] [CrossRef]
- Schernthaner, G.; Gross, J.L.; Rosenstock, J.; Guarisco, M.; Fu, M.; Yee, J.; Kawaguchi, M.; Canovatchel, W.; Meininger, G. Canagliflozin compared with sitagliptin for patients with type 2 diabetes who do not have adequate glycemic control with metformin plus sulfonylurea: A 52-week randomized trial. Diabetes Care 2013, 36, 2508–2515. [Google Scholar] [CrossRef] [PubMed]
- Imamura, M.; Nakanishi, K.; Suzuki, T.; Ikegai, K.; Shiraki, R.; Ogiyama, T.; Murakami, T.; Kurosaki, E.; Noda, A.; Kobayashi, Y.; et al. Discovery of ipragliflozin (ASP1941): A novel C-glucoside with benzothiophene structure as a potent and selective sodium glucose co-transporter 2 (SGLT2) inhibitor for the treatment of type 2 diabetes mellitus. Bioorgan Med. Chem. 2012, 20, 3263–3279. [Google Scholar] [CrossRef]
- Ohtake, Y.; Sato, T.; Kobayashi, T.; Nishimoto, M.; Taka, N.; Takano, K.; Yamamoto, K.; Ohmori, M.; Yamaguchi, M.; Takami, K.; et al. Discovery of Tofogliflozin, a novel C-arylglucoside with an O-spiroketal ring system, as a highly selective sodium glucose cotransporter 2 (SGLT2) inhibitor for the treatment of type 2 diabetes. J. Med. Chem. 2012, 55, 7828–7840. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Liu, R.; Guan, C.; Zhang, Y.; Chen, Z.; Hoerer, S.; Nar, H.; Chen, L. Structural basis of inhibition of the human SGLT2-MAP17 glucose transporter. Nature 2022, 601, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Li, A.R.; Zhang, J.; Greenberg, J.; Lee, T.; Liu, J. Discovery of non-glucoside SGLT2 inhibitors. Bioorg. Med. Chem. Lett. 2011, 21, 2472–2475. [Google Scholar] [CrossRef]
- Wu, J.S.; Peng, Y.H.; Wu, J.M.; Hsieh, C.J.; Wu, S.H.; Coumar, M.S.; Song, J.S.; Lee, J.C.; Tsai, C.H.; Chen, C.T.; et al. Discovery of non-glycoside sodium-dependent glucose co-transporter 2 (SGLT2) inhibitors by ligand-based virtual screening. J. Med. Chem. 2010, 53, 8770–8774. [Google Scholar] [CrossRef]
- Wu, S.H.; Yao, C.H.; Hsieh, C.J.; Liu, Y.W.; Chao, Y.S.; Song, J.S.; Lee, J.C. Development and application of a fluorescent glucose uptake assay for the high-throughput screening of non-glycoside SGLT2 inhibitors. Eur. J. Pharm. Sci. 2015, 74, 40–44. [Google Scholar] [CrossRef]
- Stanley, T.H. The Fentanyl Story. J. Pain. 2014, 15, 1215–1226. [Google Scholar] [CrossRef]
- Calatayud, J.; González, A. History of the development and evolution of local anesthesia since the coca leaf. Anesthesiology 2003, 98, 1503–1508. [Google Scholar] [CrossRef]
- Koch, M.A.; Schuffenhauer, A.; Scheck, M.; Wetzel, S.; Casaulta, M.; Odermatt, A.; Ertl, P.; Waldmann, H. Charting biologically relevant chemical space: A structural classification of natural products (SCONP). Proc. Natl. Acad. Sci. USA 2005, 102, 17272–17277. [Google Scholar] [CrossRef]
- Over, B.; Wetzel, S.; Grütter, C.; Nakai, Y.; Renner, S.; Rauh, D.; Waldmann, H. Natural-product-derived fragments for fragment-based ligand discovery. Nat. Chem. 2013, 5, 21–28. [Google Scholar] [CrossRef]
- Wetzel, S.; Klein, K.; Renner, S.; Rauh, D.; Oprea, T.I.; Mutzel, P.; Waldmann, H. Interactive exploration of chemical space with Scaffold Hunter. Nat. Chem. Biol. 2009, 5, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.; van Otterlo, W.A.L.; Seoane, M.D.; Möcklinghoff, S.; Hofmann, B.; Wetzel, S.; Schuffenhauer, A.; Ertl, P.; Oprea, T.I.; Steinhilber, D.; et al. Bioactivity-guided mapping and navigation of chemical space. Nat. Chem. Biol. 2009, 5, 585–592. [Google Scholar] [CrossRef]
- Cherezov, V.; Rosenbaum, D.M.; Hanson, M.A.; Rasmussen, S.G.; Thian, F.S.; Kobilka, T.S.; Choi, H.J.; Kuhn, P.; Weis, W.I.; Kobilka, B.K.; et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 2007, 318, 1258–1265. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, Y.; Ye, S.; Jiang, Y. Structure of the gating ring from the human large-conductance Ca2+-gated K+ channel. Nature 2010, 466, 393–397. [Google Scholar] [CrossRef]
- Liao, J.; Li, H.; Zeng, W.; Sauer, D.B.; Belmares, R.; Jiang, Y. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science 2012, 335, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.S.; Lee, H.M.; Yang, F.; Che, C.M.; Wong, C.C.; Abagyan, R.; Leung, C.H.; Ma, D.L. Structure-based discovery of natural-product-like TNF-alpha inhibitors. Angew. Chem. Int. Ed. Engl. 2010, 49, 2860–2864. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, Z.B.; Yao, Z.; Zheng, S.; Li, Y.; Zhu, W.; Tan, X.; Luo, X.; Shen, J.; Chen, K.; et al. Discovering potassium channel blockers from synthetic compound database by using structure-based virtual screening in conjunction with electrophysiological assay. J. Med. Chem. 2007, 50, 83–93. [Google Scholar] [CrossRef]
- Giege, R. A historical perspective on protein crystallization from 1840 to the present day. FEBS J. 2013, 280, 6456–6497. [Google Scholar] [CrossRef]
- Kendrew, J.C.; Dickerson, R.E.; Strandberg, B.E.; Hart, R.G.; Davies, D.R.; Phillips, D.C.; Shore, V.C. Structure of myoglobin: A three-dimensional Fourier synthesis at 2 A. resolution. Nature 1960, 185, 422–427. [Google Scholar] [CrossRef]
- Craik, D.J.; Wilce, J.A. Studies of protein-ligand interactions by NMR. Methods Mol. Biol. 1997, 60, 195–232. [Google Scholar] [PubMed]
- Guntert, P.; Mumenthaler, C.; Wuthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997, 273, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.A.; Glaeser, R.M. Electron microscopy of frozen hydrated biological specimens. J. Ultrastruct. Res. 1976, 55, 448–456. [Google Scholar] [CrossRef] [PubMed]
- De Rosier, D.J.; Klug, A. Reconstruction of three dimensional structures from electron micrographs. Nature 1968, 217, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Bammes, B.E.; Rochat, R.H.; Jakana, J.; Chen, D.H.; Chiu, W. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. J. Struct. Biol. 2012, 177, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. What’s next for AlphaFold and the AI protein-folding revolution. Nature 2022, 604, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.B.; Hendrickson, W.A.; Henderson, R.; Brunger, A.T. The protein-folding problem: Not yet solved. Science 2022, 375, 507. [Google Scholar] [CrossRef]
- Chen, S.J.; Hassan, M.; Jernigan, R.L.; Jia, K.; Kihara, D.; Kloczkowski, A.; Kotelnikov, S.; Kozakov, D.; Liang, J.; Liwo, A.; et al. Opinion: Protein folds vs. protein folding: Differing questions, different challenges. Proc. Natl. Acad. Sci. USA 2023, 120, e2214423119. [Google Scholar] [CrossRef]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003, 10, 980. [Google Scholar] [CrossRef]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Erehman, J.; Gohlke, B.O.; Wilhelm, T.; Preissner, R.; Dunkel, M. Super Natural II—A database of natural products. Nucleic Acids Res. 2015, 43, D935–D939. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.W.; Tsai, T.Y.; Chen, K.C.; Yang, S.C.; Huang, H.J.; Chang, T.T.; Sun, M.F.; Chen, H.Y.; Tsai, F.J.; Chen, C.Y. iSMART: An integrated cloud computing web server for traditional Chinese medicine for online virtual screening, de novo evolution and drug design. J. Biomol. Struct. Dyn. 2011, 29, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.Y.; Chang, K.W.; Chen, C.Y. iScreen: World’s first cloud-computing web server for virtual screening and de novo drug design based on TCM database@Taiwan. J. Comput. Aided Mol. Des. 2011, 25, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Wienken, C.J.; Baaske, P.; Rothbauer, U.; Braun, D.; Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010, 1, 100. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Chen, X.; Tong, Q.; Zhu, H.; He, Y.; Lei, L.; Xue, Y.; Yao, G.; Luo, Z.; Wang, J.; et al. Novel small molecule 11beta-HSD1 inhibitor from the endophytic fungus Penicillium commune. Sci. Rep. 2016, 6, 26418. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.A.; Murphy, K.P. Isothermal titration calorimetry. Methods Mol. Biol. 2005, 305, 1–16. [Google Scholar]
- Freyer, M.W.; Lewis, E.A. Isothermal titration calorimetry: Experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 2008, 84, 79–113. [Google Scholar]
- Morgan, H.; Taylor, D.M. A surface plasmon resonance immunosensor based on the streptavidin-biotin complex. Biosens. Bioelectron. 1992, 7, 405–410. [Google Scholar] [CrossRef]

| Dataset | Number of Compounds | Description | Web Site | Ref |
|---|---|---|---|---|
| ZINC | 224,205 | A free database of commercially-available compounds for virtual screening | https://zinc15.docking.org/substances/subsets/natural-products/ (accessed on 26 January 2024) | [82] |
| Super Natural III | 790,096 | A freely available database of natural products and natural product-based derivatives. Information on pathways, mechanism of action, toxicity, vendor information. | https://bioinf-applied.charite.de/supernatural_3/index.php?site=home (accessed on 26 January 2024) | [83] |
| TCM Database | 37,170 | It is currently the world largest and most comprehensive free down small molecular database on traditional Chinese medicine for virtual screening. | http://ismart.cmu.edu.tw (accessed on 26 January 2024) | [84,85] |
| CMNPD(Marine Natural Product Database) | 32,000 | CMNPD is a manually curated open access knowledge base dedicated to marine natural products research. It provides information on chemical entities with various physicochemical and pharmacokinetic properties, standardized biological activity data, systematic taxonomy and geographical distribution of source organisms, and detailed literature citations | https://www.cmnpd.org/ (accessed on 26 January 2024) | 1 |
| TimTec NPL | 800 | Tap natural bioactivity potential with molecules that are primarily sourced from plants with the remaining samples from bacteria, fungus, and animal sources. | http://www.timtec.net/ (accessed on 26 January 2024) | 1 |
| COCONUT(COlleCtion of Open Natural ProdUcTs) | 407,270 | One of the biggest and best annotated resources for NPs available free of charge and without any restriction. | https://coconut.naturalproducts.net/ (accessed on 26 January 2024) | 1 |
| TriForC | 266 | A pipeline for the discovery, sustainable production and commercial utilization of known and novel high-value triterpenes with new or superior biological activities | http://bioinformatics.psb.ugent.be/triforc/#/home (accessed on 26 January 2024) | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Xue, X. Medicinal Chemistry Strategies for the Modification of Bioactive Natural Products. Molecules 2024, 29, 689. https://doi.org/10.3390/molecules29030689
Ding Y, Xue X. Medicinal Chemistry Strategies for the Modification of Bioactive Natural Products. Molecules. 2024; 29(3):689. https://doi.org/10.3390/molecules29030689
Chicago/Turabian StyleDing, Yuyang, and Xiaoqian Xue. 2024. "Medicinal Chemistry Strategies for the Modification of Bioactive Natural Products" Molecules 29, no. 3: 689. https://doi.org/10.3390/molecules29030689
APA StyleDing, Y., & Xue, X. (2024). Medicinal Chemistry Strategies for the Modification of Bioactive Natural Products. Molecules, 29(3), 689. https://doi.org/10.3390/molecules29030689





