Research on the Interaction Mechanism and Structural Changes in Human Serum Albumin with Hispidin Using Spectroscopy and Molecular Docking
Abstract
1. Introduction
2. Results
2.1. Fluorescence Quenching Analysis
2.2. Synchronous Fluorescence Analysis
2.3. UV/vis Spectroscopy Analysis
2.4. Three-Dimensional Fluorescence Spectroscopy Analysis
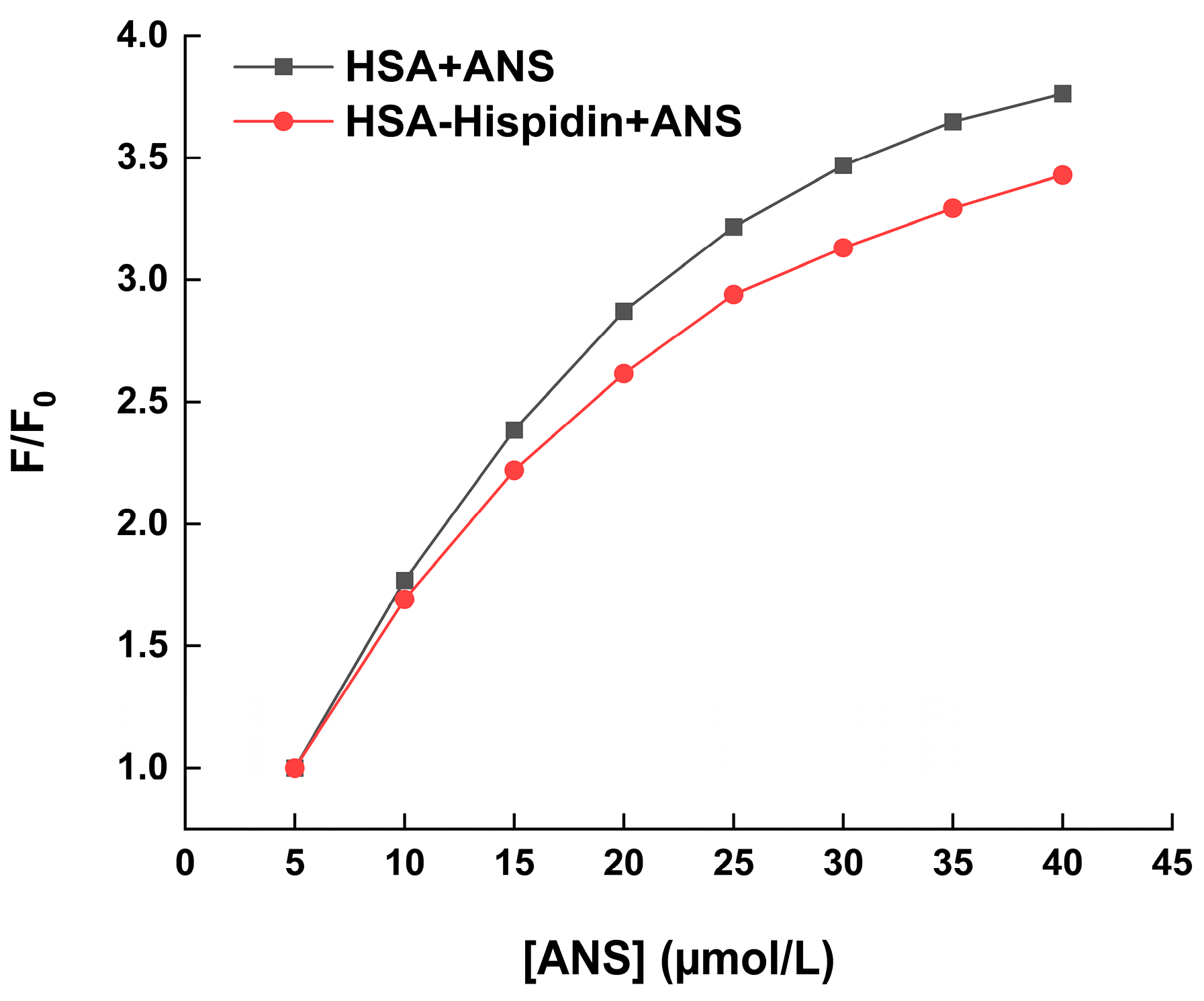
2.5. Hydrophobic Probe Assay and Hydrophobicity
2.6. The Site Competition Analysis
2.7. Molecular Docking Simulation
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of Reaction Solutions
4.3. Fluorescence Spectroscopy Analysis
4.3.1. Identification of the Fluorescent Quenching Process
4.3.2. Ka Measurement and Site Numbering
4.3.3. Determining Evident Thermodynamic Parameters
4.4. Synchronous Fluorescence Analysis
4.5. UV/vis Spectroscopy Analysis
4.6. Three-Dimensional Fluorescence Analysis
4.7. Hydrophobic Probe Assay and Hydrophobicity Measurements
4.8. Binding Site Exploration
4.9. Molecular Docking Simulation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edwards, R.L.; Lewis, D.G.; Wilson, D.V. 983. Constituents of the higher fungi. Part I. Hispidin, a new 4-hydroxy-6-styryl-2-pyrone from polyporus hispidus (Bull.). Fr. J. Chem. Soc. 1961, 4995–5002. [Google Scholar] [CrossRef]
- Edwards, R.L.; Wilson, D.V. 984. Constituents of the higher fungi. Part II. The synthesis of hispidin. J. Chem. Soc. 1961, 5003–5004. [Google Scholar] [CrossRef]
- Li, I.C.; Chang, F.C.; Kuo, C.C.; Chu, H.T.; Li, T.J.; Chen, C.C. Pilot study: Nutritional and preclinical safety investigation of fermented hispidin-enriched Sanghuangporus sanghuang mycelia: A promising functional food material to improve sleep. Front. Nutr. 2021, 8, 788965. [Google Scholar] [CrossRef]
- Lee, I.K.; Cho, S.M.; Seok, S.J.; Yun, B.S. Chemical constituents of Gymnopilus spectabilis and their antioxidant activity. Mycobiology 2008, 36, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Han, J.J.; Bao, L.; He, L.W.; Zhang, X.Q.; Yang, X.L.; Li, S.J.; Yao, Y.J.; Liu, H.W. Phaeolschidins A-E, five hispidin derivatives with antioxidant activity from the fruiting body of Phaeolus schweinitzii collected in the Tibetan Plateau. J. Nat. Prod. 2013, 76, 1448–1453. [Google Scholar] [CrossRef] [PubMed]
- Oba, Y.; Suzuki, Y.; Martins, G.N.R.; Carvalho, R.P.; Pereira, T.A.; Waldenmaier, H.E.; Kanie, S.; Naito, M.; Oliveira, A.G.; Dörr, F.A.; et al. Identification of hispidin as a bioluminescent active compound and its recycling biosynthesis in the luminous fungal fruiting body. Photochem. Photobiol. Sci. 2017, 16, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Palkina, K.A.; Balakireva, A.V.; Belozerova, O.A.; Chepurnykh, T.V.; Markina, N.M.; Kovalchuk, S.I.; Tsarkova, A.S.; Mishin, A.S.; Yampolsky, I.V.; Sarkisyan, K.S. Domain truncation in hispidin synthase orthologs from non-bioluminescent fungi does not lead to hispidin biosynthesis. Int. J. Mol. Sci. 2023, 24, 1317. [Google Scholar] [CrossRef] [PubMed]
- Beckert, C.; Horn, C.; Schnitzler, J.P.; Lehning, A.; Heller, W.; Veit, M. Styrylpyrone biosynthesis in Equisetum arvense. Phytochemistry 1997, 44, 275–283. [Google Scholar] [CrossRef]
- Yousfi, M.; Djeridane, A.; Bombarda, I.; Chahrazed, H.; Duhem, B.; Gaydou, E.M. Isolation and characterization of a new hispolone derivative from antioxidant extracts of Pistacia atlantica. Phytother. Res. 2009, 23, 1237–1242. [Google Scholar] [CrossRef]
- Fernandes, N.S.; Desoti, V.C.; Dias, A.; da Silva, Y.C.; de Azevedo Dos Santos, A.P.; Passarini, G.M.; Nakamura, C.V.; da Veiga Junior, V.F. Styrylpyrone, isolated from an amazon plant, induces cell cycle arrest and autophagy in Leishmania amazonensis. Nat. Prod. Res. 2021, 35, 4729–4733. [Google Scholar] [CrossRef]
- Abd Wahab, N.Z.; Ibrahim, N. Styrylpyrone derivative (SPD) extracted from Goniothalamus umbrosus binds to dengue virus serotype-2 envelope protein and inhibits early stage of virus replication. Molecules 2022, 27, 4566. [Google Scholar] [CrossRef]
- Avula, S.K.; Das, B.; Csuk, R.; Al-Rawahi, A.; Al-Harrasi, A. Recent advances in the stereoselective total synthesis of natural pyranones having long side chains. Molecules 2020, 25, 1905. [Google Scholar] [CrossRef]
- Gonindard, C.; Bergonzi, C.; Denier, C.; Sergheraert, C.; Klaebe, A.; Chavant, L.; Hollande, E. Synthetic hispidin, a PKC inhibitor, is more cytotoxic toward cancer cells than normal cells in vitro. Cell Biol. Toxicol. 1997, 13, 141–153. [Google Scholar] [CrossRef]
- Kim, D.E.; Kim, B.; Shin, H.S.; Kwon, H.J.; Park, E.S. The protective effect of hispidin against hydrogen peroxide-induced apoptosis in H9c2 cardiomyoblast cells through Akt/GSK-3β and ERK1/2 signaling pathway. Exp. Cell Res. 2014, 327, 264–275. [Google Scholar] [CrossRef]
- Lai, M.C.; Liu, W.Y.; Liou, S.S.; Liu, I.M. Hispidin in the Medicinal Fungus Protects Dopaminergic Neurons from JNK Activation-Regulated Mitochondrial-Dependent Apoptosis in an MPP+-Induced In Vitro Model of Parkinson’s Disease. Nutrients 2023, 15, 549. [Google Scholar] [CrossRef]
- Chien, L.H.; Deng, J.S.; Jiang, W.P.; Chen, C.C.; Chou, Y.N.; Lin, J.G.; Huang, G.J. Study on the potential of Sanghuangporus sanghuang and its components as COVID-19 spike protein receptor binding domain inhibitors. Biomed. Pharmacother. 2022, 153, 113434. [Google Scholar] [CrossRef]
- Wu, F.; Shang, C.; Jin, T.; Shi, L. Hispidin Inhibits Ferroptosis Induced by High Glucose via the miR-15b-5p/GLS2 Axis in Pancreatic Beta Cells. Evid. Based Complement. Alternat. Med. 2023, 2023, 9428241. [Google Scholar]
- Bi, S.; Yan, L.; Sun, Y.; Zhang, H. Investigation of ketoprofen binding to human serum albumin by spectral methods. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2011, 78, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yue, Q.; Gao, B. Probing the molecular mechanism of C. I. Acid red 73 binding to human serum albumin. Environ. Toxicol. Pharmacol. 2010, 30, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, M.; Murakami, K.; Yamada, K.; Kawai, K.; Kunishima, M. Binding of sulforhodamine B to human serum albumin: A spectroscopic study. Dye. Pigment. 2013, 99, 588–593. [Google Scholar] [CrossRef]
- Dong, C.; Lu, N.; Liu, Y. Binding of methacycline to human serum albumin at subdomain IIA using multispectroscopic and molecular modeling methods. Luminescence 2013, 28, 933–941. [Google Scholar] [CrossRef]
- Ali, M.S.; Al-lohedan, H.A. Interaction of human serum albumin with sulfadiazine. J. Mol. Liq. 2014, 197, 124–130. [Google Scholar] [CrossRef]
- Rhodes, A.A.; Swartz, B.L.; Hosler, E.R.; Snyder, D.L.; Benitez, K.M.; Chohan, B.S.; Basu, S. Static quenching of tryptophan fluorescence in proteins by a dioxomolybdenum(VI) thiolate complex. J. Photochem. Photobiol. A Chem. 2014, 293, 81–87. [Google Scholar] [CrossRef]
- Wenskowsky, L.; Wagner, M.; Reusch, J.; Schreuder, H.; Matter, H.; Opatz, T.; Petry, S.M. Resolving Binding Events on the Multifunctional Human Serum Albumin. ChemMedChem 2020, 15, 738–743. [Google Scholar] [CrossRef]
- Cheng, Z. Studies on the interaction between scopoletin and two serum albumins by spectroscopic methods. J. Lumin. 2012, 132, 2719–2729. [Google Scholar] [CrossRef]
- Patra, D.; Barakat, C.; Tafech, R.M. Study on effect of lipophilic curcumin on sub-domain IIA site of human serum albumin during unfolded and refolded states: A synchronous fluorescence spectroscopic study. Colloids Surf. B Biointerfaces 2012, 94, 354–361. [Google Scholar] [CrossRef]
- Cohen, L.H.; Nicoll-Griffith, D.A. Plasma protein binding methods in drug discovery and development: Bioanalysis. In Encyclopedia of Drug Metabolism and Interactions; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 1–18. [Google Scholar]
- Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Unraveling the mysteries of serum albumin-more than just a serum protein. Front. Physiol. 2014, 5, 299. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, G.D.; Birkett, D.J.; Wade, D.N. The characterization of two specific drug binding sites on HSA. Mol. Pharmacol. 1975, 11, 824–832. [Google Scholar] [PubMed]
- Petitpas, I.; Bhattacharya, A.A.; Twine, S.; East, M.; Curry, S. Crystal structure analysis of warfarin binding to human serum albumin: Anatomy of drug site I. J Biol Chem. 2001, 276, 22804–22809. [Google Scholar] [CrossRef]
- De Wolf, F.A.; Brett, G.M. Ligand-binding proteins: Their potential for application in systems for controlled delivery and uptake of ligands. Pharmacol. Rev. 2000, 52, 207–236. [Google Scholar] [PubMed]
- Krenzel, E.S.; Chen, Z.; Hamilton, J.A. Correspondence of fatty acid and drug binding sites on human serum albumin: A two-dimensional nuclear magnetic resonance study. Biochemistry 2013, 52, 1559–1567. [Google Scholar] [CrossRef]
- Xie, Y.; Yang, W.; Chen, X.; Xiao, J. Inhibition of flavonoids on acetylcholine esterase: Binding and structure-activity relationship. Food Funct. 2014, 5, 2582–2589. [Google Scholar] [CrossRef]
- Lu, Q.; Chen, C.; Zhao, S.; Ge, F.; Liu, D. Investigation of the Interaction Between Gallic Acid and α-Amylase by Spectroscopy. Int. J. food Prop. 2015, 19, 2481–2494. [Google Scholar] [CrossRef]
- Rajabi, M.; Farhadian, S.; Shareghi, B.; Asgharzadeh, S.; Momeni, L. Noncovalent interactions of bovine trypsin with curcumin and effect on stability, structure, and function. Colloids Surf. B Biointerfaces 2019, 183, 110287. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Liu, M.; Chen, S.; Chen, X.; Wang, J. New insight into molecular interactions of imidazolium ionic liquids with bovine serum albumin. J. Phys. Chem. B 2011, 115, 12306–12314. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.; Daniel, E.; Weber, G.; Daniel, E. Cooperative effects in binding by bovine serum albumin. II. The binding of 1-anilino-8-naphthalenesulfonate. Polarization of the ligand fluorescence and quenching of the protein fluorescence. Biochemistry 1966, 5, 1900–1907. [Google Scholar] [CrossRef]
- Alexandri, E.; Primikyri, A.; Papamokos, G.; Venianakis, T.; Gkalpinos, V.G.; Tzakos, A.G.; Karydis-Messinis, A.; Moschovas, D.; Avgeropoulos, A.; Gerothanassis, I.P. NMR and computational studies reveal novel aspects in molecular recognition of unsaturated fatty acids with non-labeled serum albumin. FEBS J. 2022, 289, 5617–5636. [Google Scholar] [CrossRef] [PubMed]
- Alexandri, E.; Venianakis, T.; Primikyri, A.; Papamokos, G.; Gerothanassis, I.P. Molecular basis for the selectivity of DHA and EPA in Sudlow’s drug binding sites in human serum albumin with the combined use of NMR and docking calculations. Molecules 2023, 28, 3724. [Google Scholar] [CrossRef] [PubMed]
- Venianakis, T.; Primikyri, A.; Opatz, T.; Petry, S.; Papamokos, G.; Gerothanassis, I.P. NMR and Docking Calculations Reveal Novel Atomistic Selectivity of a Synthetic High-Affinity Free Fatty Acid vs. Free Fatty Acids in Sudlow’s Drug Binding Sites in Human Serum Albumin. Molecules 2023, 28, 7991. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Xu, D.; Saeeduddin, M.; Riaz, A.; Zeng, X. Characterization of molecular structures of theaflavins and the interactions with bovine serum albumin. J. Food Sci. Technol. 2017, 54, 3421–3432. [Google Scholar] [CrossRef]
- Wang, L.; Li, H.; Yang, Y.; Zhang, D.; Wu, M.; Pan, B.; Xing, B. Identifying structural characteristics of humic acid to static and dynamic fluorescence quenching of phenanthrene, 9-phenanthrol, and naphthalene. Water Res. 2017, 122, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Yang, Q.; Mao, X.; Chai, W.M.; Peng, Y. Study on the interaction between 4-(1H-indol-3-yl)-2-(p-tolyl)quinazoline-3-oxide and human serum albumin. Bioorg. Med. Chem. 2020, 28, 115720. [Google Scholar] [CrossRef]
- Peng, X.; Sun, Y.; Qi, W.; Su, R.; He, Z. Study of the Interaction Between Coenzyme Q10 and Human Serum Albumin: Spectroscopic Approach. J. Solution Chem. 2014, 43, 585–607. [Google Scholar] [CrossRef]
- Bahri, A.; Henriquet, C.; Pugnière, M.; Marchesseau, S.; Chevalier-Lucia, D. Binding analysis between monomeric β-casein and hydrophobic bioactive compounds investigated by surface plasmon resonance and fluorescence spectroscopy. Food Chem. 2019, 286, 289–296. [Google Scholar] [CrossRef]
- Linani, A.; Benarous, K.; Bou-Salah, L.; Yousfi, M. Hispidin, harmaline, and harmine as potent inhibitors of bovine xanthine oxidase: Gout treatment, in vitro, ADMET prediction, and SAR studies. Bioorg Chem. 2021, 112, 104937. [Google Scholar] [CrossRef]
- Rohn, S. Possibilities and limitations in the analysis of covalent interactions between phenolic compounds and proteins. Food Res. Int. 2014, 65, 13–19. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Dong, L.; Li, J.; He, W.; Chen, X.; Hu, Z. Investigation of the interaction between naringin and human serum albumin. J. Mol. Struct. 2008, 875, 1–8. [Google Scholar] [CrossRef]
- Bian, Q.; Liu, J.; Tian, J.; Hu, Z. Binding of genistein to human serum albumin demonstrated using tryptophan fluorescence quenching. Int. J. Biol. Macromol. 2004, 34, 333–337. [Google Scholar] [CrossRef]
- Boye, J.I.; Ma, C.Y.; Ismail, A. Thermal stability of beta-lactoglobulins A and B: Effect of SDS, urea, cysteine and N-ethylmaleimide. J. Dairy Res. 2004, 71, 207–215. [Google Scholar] [CrossRef]
- Ota, C.; Tanaka, S.I.; Takano, K. Revisiting the rate-limiting step of the ANS-protein binding at the protein surface and inside the hydrophobic cavity. Molecules 2021, 26, 420. [Google Scholar] [CrossRef]
- Guliyeva, A.J.; Gasymov, O.K. ANS fluorescence: Potential to discriminate hydrophobic sites of proteins in solid states. Biochem. Biophys. Rep. 2020, 24, 100843. [Google Scholar] [CrossRef] [PubMed]
- Dufour, C.; Dangles, O. Flavonoid-serum albumin complexation: Determination of binding constants and binding sites by fluorescence spectroscopy. Biochim. Biophys. Acta 2005, 1721, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.K.; Han, M.S.; Lee, M.S.; Kim, Y.S.; Yun, B.S. Styrylpyrones from the medicinal fungus Phellinus baumii and their antioxidant properties. Bioorg. Med. Chem. Lett. 2010, 20, 5459–5461. [Google Scholar] [CrossRef] [PubMed]
- Zsila, F.; Bikádi, Z.; Simonyi, M. Probing the binding of the flavonoid, quercetin to human serum albumin by circular dichroism, electronic absorption spectroscopy and molecular modelling methods. Biochem. Pharmacol. 2003, 65, 447–456. [Google Scholar] [CrossRef]
- Mandeville, J.S.; Froehlich, E.; Tajmir-Riahi, H.A. Study of curcumin and genistein interactions with human serum albumin. J. Pharm. Biomed. Anal. 2009, 49, 468–474. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, L.; Li, J.; Chen, X. Studies on the interaction of gallic acid with human serum albumin in membrane mimetic environments. Talanta 2008, 76, 246–253. [Google Scholar] [CrossRef]
- Gentili, P.L.; Ortica, F.; Favaro, G. Static and dynamic interaction of a naturally occurring photochromic molecule with bovine serum albumin studied by UV-visible absorption and fluorescence spectroscopy. J. Phys. Chem. B 2008, 112, 16793–16801. [Google Scholar] [CrossRef]
- Li, T.; Hu, P.; Dai, T.; Li, P.; Ye, X.; Chen, J.; Liu, C. Comparing the binding interaction between β-lactoglobulin and flavonoids with different structure by multi-spectroscopy analysis and molecular docking. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 201, 197–206. [Google Scholar] [CrossRef]
- Wang, Y.F.; Wang, X.Y. Binding, stability, and antioxidant activity of quercetin with soy protein isolate particles. Food Chem. 2015, 188, 24–29. [Google Scholar] [CrossRef]
- Zhao, Y.; Cao, Y.; Han, F.M.; Chen, Y. Study on the interaction between cinnamic acid and human serum albumin by fluorescence quenching method. Guang Pu Xue Yu Guang Pu Fen Xi 2008, 28, 904–907. [Google Scholar]
- Mohanty, D.P.; Mohapatra, S.; Misra, S.; Sahu, P.S. Milk derived bioactive peptides and their impact on human health—A review. Saudi J. Biol. Sci. 2016, 23, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cheng, Z.; Wang, Y.; Fu, L. Dietary protein-phenolic interactions: Characterization, biochemical-physiological consequences, and potential food applications. Crit. Rev. Food Sci. Nutr. 2021, 61, 3589–3615. [Google Scholar] [CrossRef] [PubMed]
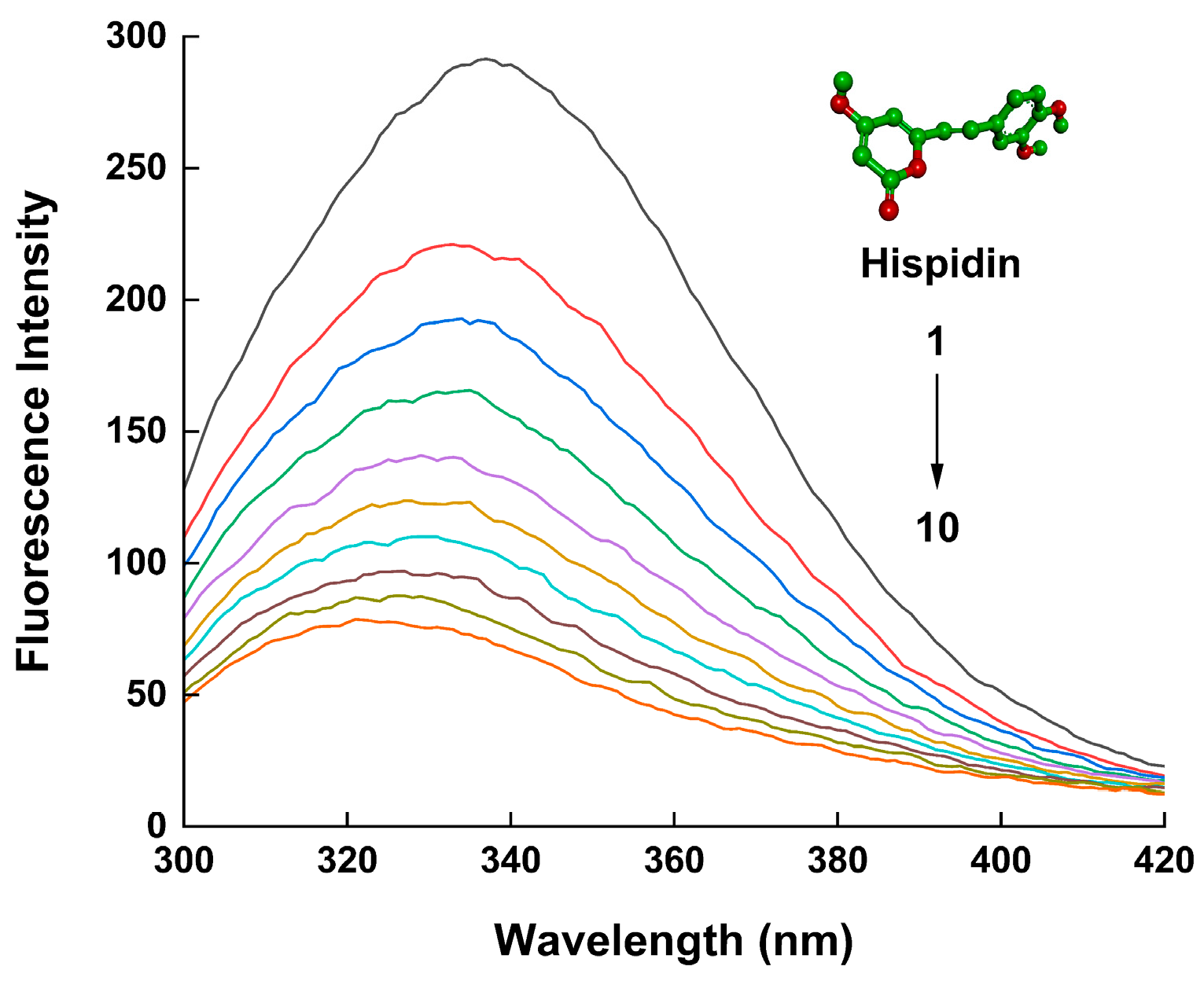
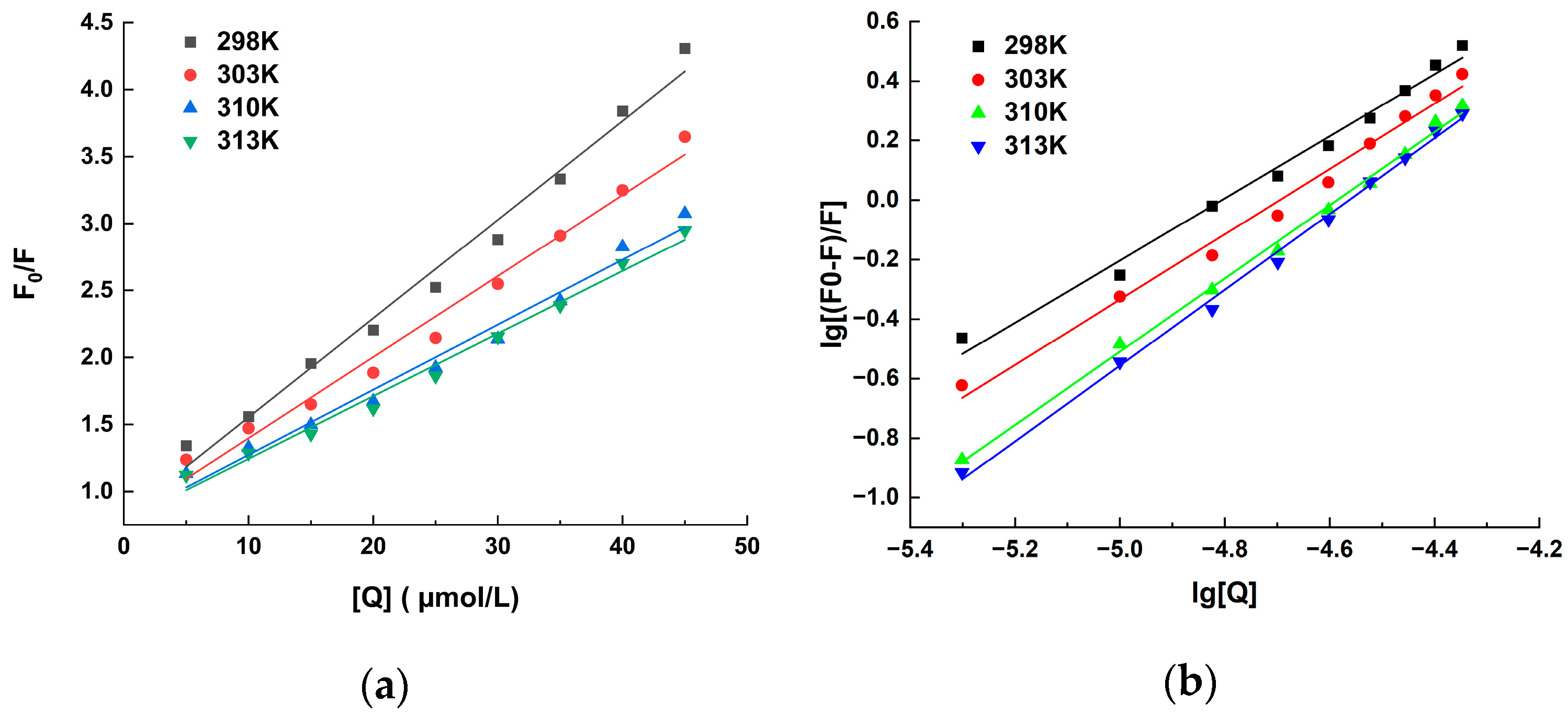
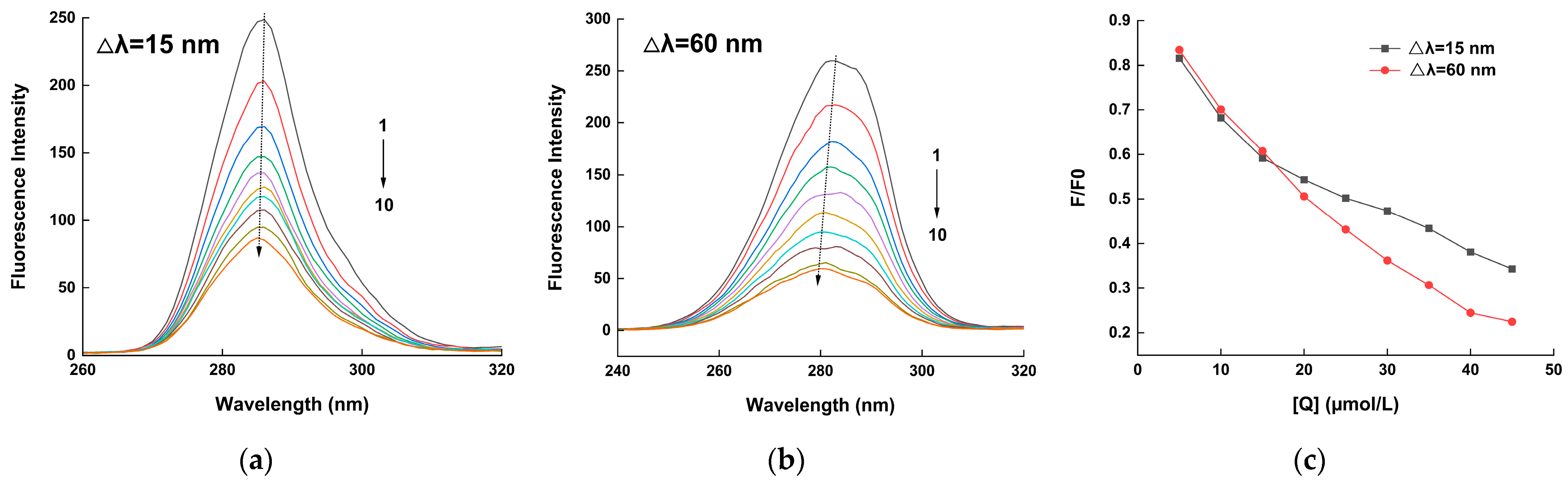

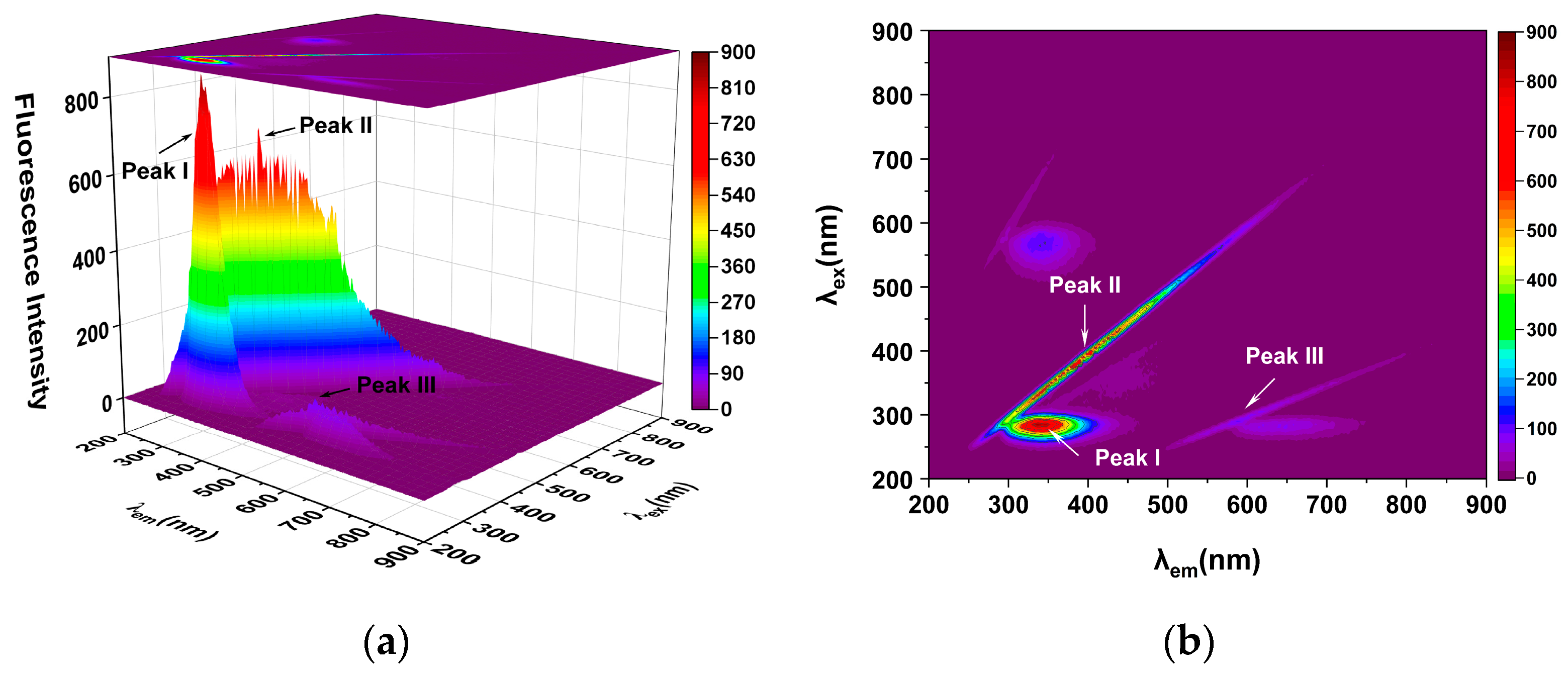


| T (k) | Equation | Ksv (104 L/mol) | Kq (1012 L/(mol·s)) | R2 |
|---|---|---|---|---|
| 298 | Y = 0.0737Q + 0.8165 | 7.37 | 7.37 | 0.98 |
| 303 | Y = 0.0605Q + 0.7929 | 6.05 | 6.05 | 0.98 |
| 310 | Y = 0.0485Q + 0.7882 | 4.85 | 4.85 | 0.98 |
| 313 | Y = 0.0467Q + 0.7773 | 4.67 | 4.67 | 0.98 |
| T (k) | Ka (105 L/mol) | n | ∆H (kJ/mol) | ∆S (J/(mol·K)) | ∆G (kJ/mol) |
|---|---|---|---|---|---|
| 298 | 1.03 | 1.04 | 98.75 | 426.29 | −28.60 |
| 303 | 1.42 | 1.09 | −30.00 | ||
| 310 | 4.36 | 1.22 | −33.47 | ||
| 313 | 6.43 | 1.27 | −34.80 |
| Protein Complex | Ka (L/mol) |
|---|---|
| HSA + Hispidin | 10.30 × 104 |
| HSA + Hispidin + SDS | 2.84 × 104 |
| HSA + Hispidin + Urea | 3.09 × 104 |
| System | Parameter | Peak II | Peak I |
|---|---|---|---|
| HSA | Peak position Δλex/Δλem (nm/nm) | 350/350 | 280/340 |
| Intensity F | 410.7 | 780.6 | |
| Hispidin–HSA | Peak position Δλex/Δλem (nm/nm) | 350/350 | 280/332 |
| Intensity F | 207.6 | 397.9 |
| Molar Ratio [Hispidin]/[HSA] | S0 | R2 |
|---|---|---|
| 0:1 | 15.72 | 0.96 |
| 1:1 | 14.87 | 0.97 |
| 3:1 | 10.03 | 0.98 |
| System | Ka (105 L/mol) | R2 |
|---|---|---|
| HSA + hispidin | 1.299 | 0.9932 |
| HSA + hispidin + ibuprofen | 0.877 | 0.9971 |
| HSA + hispidin + warfarin | 0.745 | 0.9901 |
| HSA + hispidin + digitoxin | 1.576 | 0.9960 |
| HSA + hispidin + hemin | 2.495 | 0.9965 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.-H.; Wang, W.-Q.; Zhou, Y.-W.; Gao, X.-J.; Zhang, Q.; Zhang, M.-H. Research on the Interaction Mechanism and Structural Changes in Human Serum Albumin with Hispidin Using Spectroscopy and Molecular Docking. Molecules 2024, 29, 655. https://doi.org/10.3390/molecules29030655
Fan S-H, Wang W-Q, Zhou Y-W, Gao X-J, Zhang Q, Zhang M-H. Research on the Interaction Mechanism and Structural Changes in Human Serum Albumin with Hispidin Using Spectroscopy and Molecular Docking. Molecules. 2024; 29(3):655. https://doi.org/10.3390/molecules29030655
Chicago/Turabian StyleFan, Si-Hua, Wen-Qiang Wang, Yu-Wen Zhou, Xue-Jun Gao, Qiang Zhang, and Ming-Hui Zhang. 2024. "Research on the Interaction Mechanism and Structural Changes in Human Serum Albumin with Hispidin Using Spectroscopy and Molecular Docking" Molecules 29, no. 3: 655. https://doi.org/10.3390/molecules29030655
APA StyleFan, S.-H., Wang, W.-Q., Zhou, Y.-W., Gao, X.-J., Zhang, Q., & Zhang, M.-H. (2024). Research on the Interaction Mechanism and Structural Changes in Human Serum Albumin with Hispidin Using Spectroscopy and Molecular Docking. Molecules, 29(3), 655. https://doi.org/10.3390/molecules29030655






