High-Resolution Mass Spectrometry Non-Targeted Detection of Per- and Polyfluoroalkyl Substances in Roe Deer (Capreolus capreolus)
Abstract
1. Introduction
2. Results and Discussion
2.1. Realisation of the Strategy for Non-Targeted PFAS Workflow
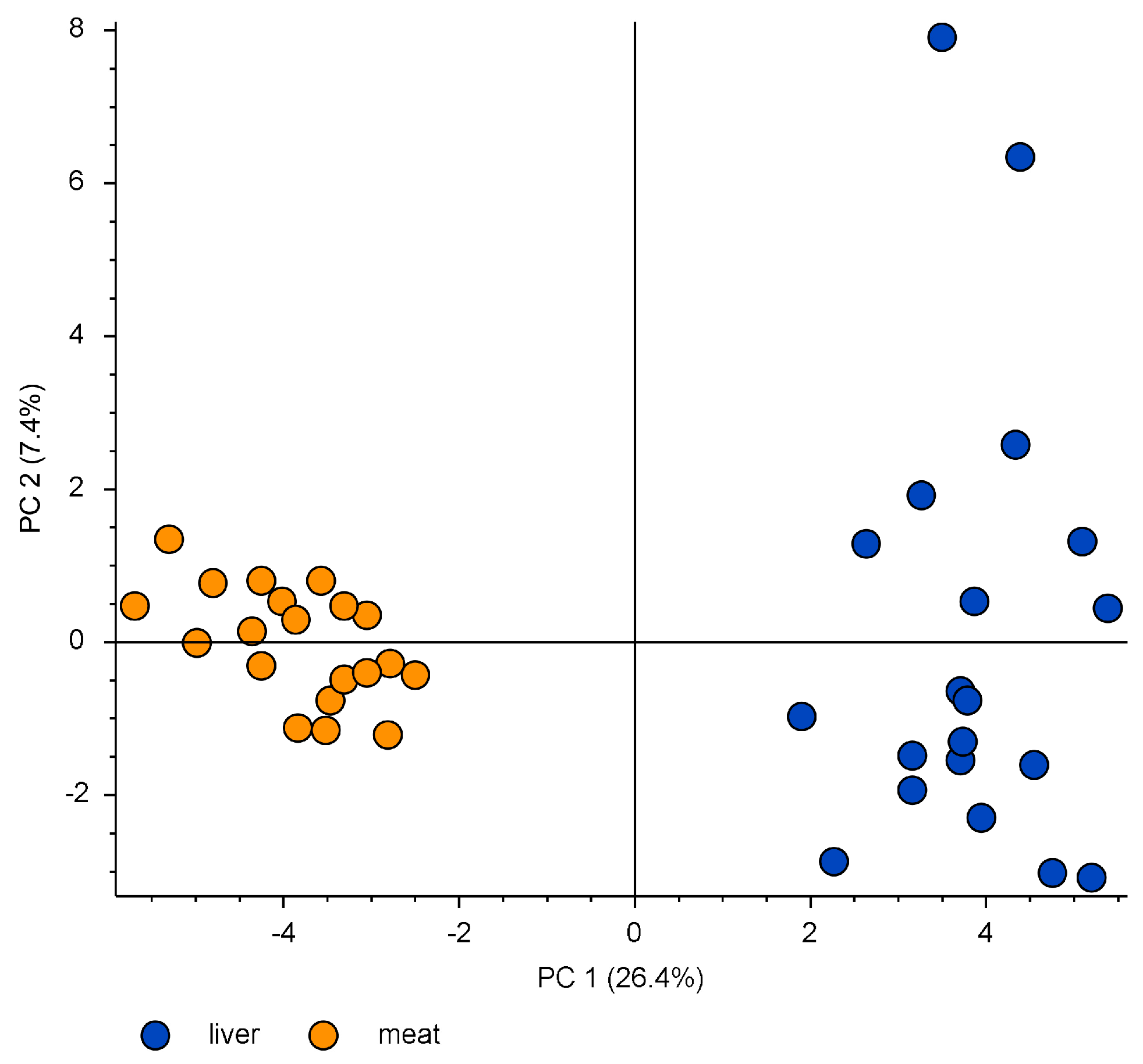
2.2. Investigating the Presence of PFAS in Roe Deer Liver and Muscle
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Sample Collection and Extraction
3.3. HPLC Q-Exactive Orbitrap High-Resolution Mass Spectrometry Analysis
3.4. Non-Targeted PFAS Workflow: Find and Identify Per- and Polyfluoroalkyl Substances
3.5. Statistical Evaluation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Silva, A.O.; Armitage, J.M.; Bruton, T.A.; Dassuncao, C.; Heiger-Bernays, W.; Hu, X.C.; Karrman, A.; Kelly, B.; Ng, C.; Robuck, A.; et al. PFAS exposure pathways for humans and wildlife: A synthesis of current knowledge and key gaps in understanding. Environ. Toxicol. Chem. 2021, 40, 631–657. [Google Scholar] [CrossRef]
- Sznajder-Katarzyńska, K.; Surma, M.; Cieślik, I. A review of perfluoroalkyl acids (PFAAs) in terms of sources, applications, human exposure, dietary intake, toxicity, legal regulation, and methods of determination. J. Chem. 2019, 2019, 2717528. [Google Scholar] [CrossRef]
- Synthesis Paper of per and Poly Fluorinated Chemicals (PFCs). Organisation for Economic Co-Operation and Development and United Nations Environment Program Global PFC Group. Available online: https://web-archive.oecd.org/2017-08-02/250024-synthesis-paper-on-per-and-polyfluorinated-chemicals.htm (accessed on 13 December 2023).
- All POPs Listed in the Stockholm Convention. Available online: https://www.pops.int/TheConvention/ThePOPs/AllPOPs/tabid/2509/Default.aspx (accessed on 13 December 2023).
- Sunderland, E.M.; Hu, X.C.; Dassuncao, C.; Tokranov, A.K.; Wagner, C.C.; Allen, J.G. A review of the pathways of human exposure to poly-and perfluoroalkyl substances (PFASs) and present understanding of health effects. J. Expo. Sci. Environ. Epidemiol. 2019, 29, 131–147. [Google Scholar] [CrossRef]
- Petrick, L.M.; Wolff, M.S.; Barupal, D.; Teitelbaum, S.L. Comparison of untargeted and targeted perfluoroalkyl acids measured in adolescent girls. Chemosphere 2022, 290, 133303. [Google Scholar] [CrossRef]
- Wang, X.; Yu, N.; Qian, Y.; Shi, W.; Zhang, X.; Geng, J.; Yu, H.; Wei, S. Non-target and suspect screening of per-and polyfluoroalkyl substances in Chinese municipal wastewater treatment plants. Water Res. 2020, 183, 115989. [Google Scholar] [CrossRef] [PubMed]
- Brase, R.A.; Mullin, E.J.; Spink, D.C. Legacy and emerging per-and polyfluoroalkyl substances: Analytical techniques, environmental fate, and health effects. Int. J. Mol. Sci. 2021, 22, 995. [Google Scholar] [CrossRef] [PubMed]
- Kang, Q.; Gao, F.; Zhang, X.; Wang, L.; Liu, J.; Fu, M.; Zhang, S.; Wan, Y.; Shen, H.; Hu, J. Nontargeted identification of per-and polyfluoroalkyl substances in human follicular fluid and their blood-follicle transfer. Environ. Int. 2020, 139, 105686. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, L.M.; Pavlovic, R.; Arioli, F.; Nobile, M.; Di Cesare, F.; Mosconi, G.; Falletta, E.; Malandra, R.; Panseri, S. Presence of perfluoroalkyl substances in Mediterranean sea and North Italian lake fish addressed to Italian consumer. Int. J. Food Sci. Technol. 2022, 57, 1303–1316. [Google Scholar] [CrossRef]
- Draghi, S.; Pavlovic, R.; Pellegrini, A.; Fidani, M.; Riva, F.; Brecchia, G.; Agradi, S.; Arioli, F.; Vigo, D.; Di Cesare, F.; et al. First investigation of the physiological distribution of legacy and emerging perfluoroalkyl substances in raw bovine milk according to the component fraction. Foods 2023, 12, 2449. [Google Scholar] [CrossRef] [PubMed]
- Oró-Nolla, B.; Dulsat-Masvidal, M.; Bertolero, A.; Lopez-Antia, A.; Lacorte, S. Target and untargeted screening of perfluoroalkyl substances in biota using liquid chromatography coupled to quadrupole time of flight mass spectrometry. J. Chromatogr. A 2023, 1701, 464066. [Google Scholar] [CrossRef]
- Rupp, J.; Guckert, M.; Berger, U.; Drost, W.; Mader, A.; Nödler, K.; Nuremberg, G.; Schulze, J.; Sohlmann, R.; Reemtsma, T. Comprehensive target analysis and TOP assay of per-and polyfluoroalkyl substances (PFAS) in wild boar livers indicate contamination hot-spots in the environment. Sci. Total Environ. 2023, 871, 162028. [Google Scholar] [CrossRef] [PubMed]
- Dale, K.; Yadetie, F.; Müller, M.B.; Pampanin, D.M.; Gilabert, A.; Zhang, X.; Tairova, Z.; Haarr, A.; Lille-Langøy, R.; Lyche, J.L.; et al. Proteomics and lipidomics analyses reveal modulation of lipid metabolism by perfluoroalkyl substances in liver of Atlantic cod (Gadus morhua). Aquat. Toxicol. 2020, 227, 105590. [Google Scholar] [CrossRef] [PubMed]
- Bangma, J.; Guillette, T.C.; Bommarito, P.A.; Ng, C.; Reiner, J.L.; Lindstrom, A.B.; Strynar, M.J. Understanding the dynamics of physiological changes, protein expression, and PFAS in wildlife. Environ. Int. 2022, 159, 107037. [Google Scholar] [CrossRef] [PubMed]
- Lettoof, D.C.; Nguyen, T.V.; Richmond, W.R.; Nice, H.E.; Gagnon, M.M.; Beale, D.J. Bioaccumulation and metabolic impact of environmental PFAS residue on wild-caught urban wetland tiger snakes (Notechis scutatus). Sci. Total Environ. 2023, 897, 165260. [Google Scholar] [CrossRef] [PubMed]
- Draghi, S.; Agradi, S.; Riva, F.; Tarhan, D.; Bilgiç, B.; Dokuzeylül, B.; Ercan, A.M.; Or, M.E.; Brecchia, G.; Vigo, D.; et al. Roe Deer (Capreolus capreolus) hair as a bioindicator for the environmental presence of toxic and trace elements. Toxics 2023, 11, 49. [Google Scholar] [CrossRef]
- Cygan-Szczegielniak, D.; Stasiak, K. Effects of age and sex on the content of heavy metals in the hair, liver and the longissimus lumborum muscle of roe deer Capreolus capreolus L. Environ. Sci. Pollut. Res. 2022, 29, 10782–10790. [Google Scholar] [CrossRef]
- Tixier, H.; Duncan, P.; Scehovic, J.; Yant, A.; Gleizes, M.; Lila, M. Food selection by European roe deer (Capreolus Capreolus): Effects of plant chemistry, and consequences for the nutritional value of their diets. J. Zool. 1997, 242, 229–245. [Google Scholar] [CrossRef]
- Koch, A.; Aro, R.; Wang, T.; Yeung, L.W.Y. Towards a comprehensive analytical workflow for the chemical characterisation of organofluorine in consumer products and environmental samples. TrAC—Trends Anal. Chem. 2020, 123, 115423. [Google Scholar] [CrossRef]
- Jia, S.; Marques Dos Santos, M.; Li, C.; Snyder, S.A. Recent advances in mass spectrometry analytical techniques for per- and polyfluoroalkyl substances (PFAS). Anal. Bioanal. Chem. 2022, 414, 2795–2807. [Google Scholar] [CrossRef]
- Nason, S.L.; Koelmel, J.; Zuverza-Mena, N.; Stanley, C.; Tamez, C.; Bowden, J.A.; Godri Pollitt, K.J. Software comparison for nontargeted analysis of PFAS in AFFF-contaminated soil. J. Am. Soc. Mass Spectrom. 2020, 32, 840–846. [Google Scholar] [CrossRef]
- Jacob, P.; Barzen-Hanson, K.A.; Helbling, D.E. Target and nontarget analysis of per-and polyfluoralkyl substances in wastewater from electronics fabrication facilities. Environ. Sci. Technol. 2021, 55, 2346–2356. [Google Scholar] [CrossRef]
- Hensema, T.J.; Berendsen, B.J.; van Leeuwen, S.P. Non-targeted identification of per-and polyfluoroalkyl substances at trace level in surface water using fragment ion flagging. Chemosphere 2021, 265, 128599. [Google Scholar] [CrossRef]
- de Vega, R.G.; Cameron, A.; Clases, D.; Dodgen, T.M.; Doble, P.A.; Bishop, D.P. Simultaneous targeted and non-targeted analysis of per-and polyfluoroalkyl substances in environmental samples by liquid chromatography-ion mobility-quadrupole time of flight-mass spectrometry and mass defect analysis. J. Chromatogr. A 2021, 1653, 462423. [Google Scholar] [CrossRef] [PubMed]
- Death, C.; Bell, C.; Champness, D.; Milne, C.; Reichman, S.; Hagen, T. Per-and polyfluoroalkyl substances (PFAS) in livestock and game species: A review. Sci. Total Environ. 2021, 774, 144795. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Allen, D.; Tian, S.; Oler, E.; Gautam, V.; Greiner, R.; Metz, T.O.; Wishart, D.S. CFM-ID 4.0–a web server for accurate MS-based metabolite identification. Nucleic Acids Res. 2022, 50, W165–W174. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Schmidt, S.; Müller-Hannemann, M.; Neumann, S. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinform. 2010, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Schymanski, E.L.; Singer, H.P.; Longrée, P.; Loos, M.; Ruff, M.; Stravs, M.A.; Ripolles Vidal, C.; Hollender, J. Strategies to characterize polar organic contamination in wastewater: Exploring the capability of high resolution mass spectrometry. Environ. Sci. Technol. 2014, 48, 1811–1818. [Google Scholar] [CrossRef] [PubMed]
- Barzen-Hanson, K.A.; Roberts, S.C.; Choyke, S.; Oetjen, K.; McAlees, A.; Riddell, N.; McCrindle, R.; Lee Ferguson, P.; Higgins, C.P.; Field, J.A. Discovery of 40 classes of per-and polyfluoroalkyl substances in historical aqueous film-forming foams (AFFFs) and AFFF-impacted groundwater. Environ. Sci. Technol. 2017, 51, 2047–2057. [Google Scholar] [CrossRef]
- Chiesa, L.M.; Pavlovic, R.; Nobile, M.; Di Cesare, F.; Malandra, R.; Pessina, D.; Panseri, S. Discrimination between fresh and frozen-thawed fish involved in food safety and fraud protection. Foods 2020, 9, 1896. [Google Scholar] [CrossRef]
- National Institute of Standards and Technology (NIST). Science Data Portal. Suspect List of Possible per- and Polyfluoroalkyl Substances. Available online: https://data.nist.gov/od/id/mds2-2387 (accessed on 13 December 2023).
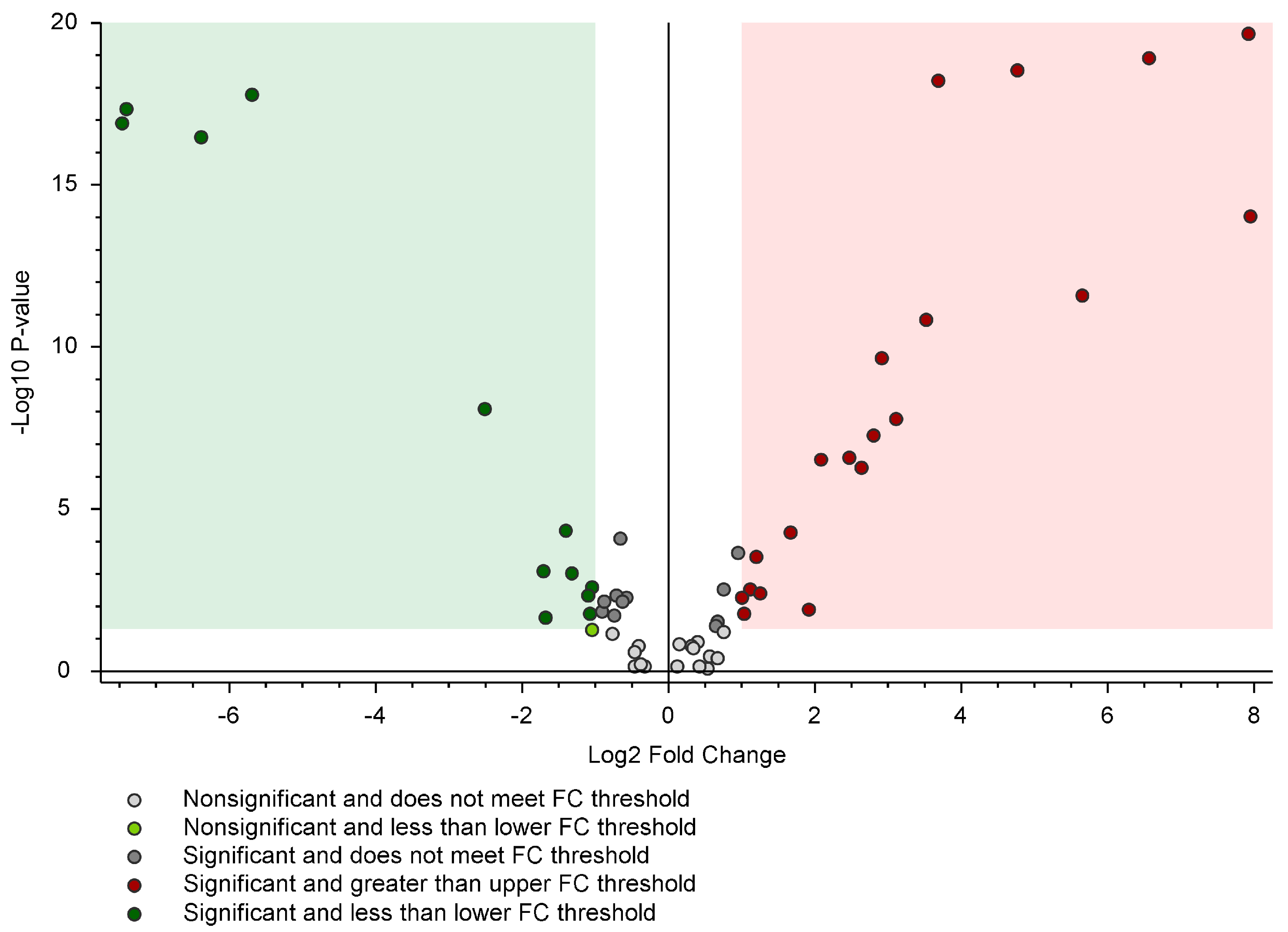

| Code | IUPAC Name | Formula | m/z [M-H]− | Mass Error [ppm] | Diagnostic Ions in DDA | RT [min] | Area (Max.) | Conf Level |
|---|---|---|---|---|---|---|---|---|
| 1 | 1,1,1,2,2,3,3,4-Octafluoro-7,7-dimethyl-4-octene | C10H12F8 | 283.0731 | −1.46 | 71.012; 115.247; 169.001 | 12.5 | 2.7 × 106 | 3 |
| 2 | 1,1,1,2,2,3,3,4,4,5,5,6,6-Tridecafluoro-7-pentadecene | C15H17F13 | 443.1031 | −4.2 | 85.011; 318.98; 343.976 | 12.64 | 3.9 × 107 | 3 |
| 3 | 1,1,1,2,2,3,3,4,4,5,5,6,6-Tridecafluorohexadecane | C16H21F13 | 459.1341 | −4.67 | 141.204; 318.979; 331.100 | 13.62 | 2.5 × 106 | 2 |
| 4 | 1,1,1,2,2,3,3,4,4,5,5-Undecafluorononadecane | C19H29F11 | 465.2001 | −4.27 | 181.201; 268.974 | 8.19 | 7.2 × 106 | 3 |
| 5 | 1,1,1,2,2,3,3,4,4,5,5-Undecafluoropentane | C5HF11 | 268.9836 | 2.19 | 118.978; 169.001 | 13.61 | 4.3 × 107 | 2 |
| 6 | 1,1,1,2,2,3,3,4,4-Nonafluorodocosane | C22H37F9 | 471.2660 | −3.89 | 71.089; 387.1833 | 10.40 | 4.9 × 107 | 3 |
| 7 | 1,1,1,2,2,3,3-Heptafluorododecane | C12H19F7 | 295.1306 | 1.34 | 275.128; 168.987 | 9.74 | 4.1 × 108 | 3 |
| 8 | 1,1,1,3,3,3-Hexafluoro-2-(trifluoromethyl)propan-2-yl heptafluorobutanoate | C8F16O2 | 430.9587 | 3.86 | 118.96; 212.978; 218.78 | 8.46 | 4.9 × 106 | 2 |
| 9 | 1-((3-(Dimethylamino)propyl)amino)-4,4,5,5,6,6,7,7,8,8,9,9,10,10,11,11,12,13,13,13-icosafluoro-12-(trifluoromethyl)tridecan-1-ol | C19H19F23N2O | 727.1064 | 0.88 | 115.087; 129.112; 568.963; 695.043 | 11.65 | 1.0 × 107 | 3 |
| 10 | 1-(2-chloro-1,1,2,3,3,3-hexafluoropropoxy)-1,1,2,3,3,3-hexafluoropropan-2-ol | C6HClF12O2 | 366.9409 | 2.15 | 116.996; 166.995; 232.987; 348.949; 366.944 | 17.86 | 7.4 × 107 | 2 |
| 11 | 1-(Difluoromethoxy)-1,1,2,2-tetrafluoro-2-(trifluoromethoxy)ethane | C4HF9O2 | 250.9767 | 2.74 | 84.981; 134.986; 184.984 | 18.77 | 3.4 × 107 | 2 |
| 12 | (Pentafluoroethyl)-(trifluoromethyl)cyclohexane | C9H10F8 | 269.0577 | −2.04 | 118.989; 151.097; 201.091 | 11.82 | 3.9 × 108 | 2 |
| 13 | 1-(Perfluoro-n-hexyl)dodecane | C18H25F13 | 487.1654 | −4.16 | 318.979 | 12.46 | 3.1 × 107 | 3 |
| 14 | 1-(Tetradec-1-en-1-yl)perfluorohexane | C20H27F13 | 513.1809 | −4.21 | 318.979; 455.117; 493.178 | 13.63 | 3.0 × 107 | 2 |
| 15 | 1-hydro-pentadecafluoroheptane | C7HF15 | 368.9772 | 1.64 | 168.987; 268.982 | 16.78 | 1.6 × 107 | 2 |
| 16 | 1-sec-Butyl-2-(1,1,2,2-tetrafluoroethoxy)benzene | C12H14F4O | 249.0904 | −1.49 | 116.996; 133.065; 219.043; 221.061; 233.061 | 12.98 | 4.5 × 107 | 3 |
| 17 | 12,12,13,13,13-Pentafluorotridecanoic acid | C13H21F5O2 | 303.1390 | 0.19 | 175.058; 213.071; 283.138 | 9.18 | 7.9 × 107 | 3 |
| 18 | 1H-Benzimidazole, 5,6-dimethyl-2-(pentafluoroethyl)- | C11H9F5N2 | 263.0600 | −4.8 | 118.069; 243.051 | 6.85 | 4.5 × 108 | 2 |
| 19 | 1H-Perfluorohexane | C6HF13 | 318.9804 | 1.89 | 101.056; 118.987; 168.1987 | 15.65 | 3.0 × 107 | 2 |
| 20 | 2,2,3,3,4,4,5,5,6,6,7,7,8,9,9,9-hexadecafluoro-8-(trifluoromethyl)nonanoic acid | C10HF19O2 | 512.9610 | 1.89 | 168.987; 344.987 | 18.29 | 1.6 × 107 | 2 |
| 21 | 2-(3-(2-chloro-1,1,2,3,3,3-hexafluoropropoxy)-1,1,2,2,3,3-hexafluoropropoxy)-1,1,2,2-tetrafluoroethan-1-ol | C8HClF16O3 | 482.9300 | 2.8 | 67.001; 96.991; 116.998; 182.989; 298.977 | 19.76 | 9.9 × 107 | 3 |
| 22 | 2-(Nonafluorobutyl)benzoic acid | C11H5F9O2 | 339.0085 | 3.52 | 295.0189; 339.007 | 10.22 | 3.2 × 107 | 3 |
| 23 | 2-Ethyl-4-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-3-[(2-methylpropyl)sulfanyl]benzoic acid | C16H17F7O2S | 405.0771 | 1.49 | 89.055; 207.028; 281.044 | 9.09 | 4.5 × 106 | 3 |
| 24 | 2-Vinylperfluorobutane | C6H3F9 | 245.0030 | 4.71 | 224.998 | 8.97 | 1.8 × 107 | 2 |
| 25 | 2H-Nonafluorobutane | C4HF9 | 218.9863 | 0.38 | 101.977; 118.997; 151.087 | 9.94 | 2.1 × 107 | 2 |
| 26 | 3,4,5,5,6,6,7,7,8,8,9,9,10,10,10-Pentadecafluoro-3-decen-2-one | C10H3F15O | 422.9880 | 2.01 | 380.9765; 404.9765 | 18.3 | 3.9 × 108 | 2 |
| 27 | 3-((2-chloro-1,1,2,3,3,3-hexafluoropropoxy)difluoromethoxy)-1,1,2,2,3,3-hexafluoropropan-1-ol | C7HClF14O3 | 432.9323 | 1.25 | 67.0001; 116.997; 166.993; 182.999; 232.977 | 18.89 | 4.2 × 107 | 3 |
| 28 | 3-(12,12,13,13,14,14,15,15,15-Nonafluoropentadecyl)-1,2-benzenediol | C21H27F9O2 | 481.1809 | 2.94 | 215.063; 323.100; 389.072 | 23.27 | 3.6 × 106 | 3 |
| 29 | 3-(3-(2-chloro-1,1,2,3,3,3-hexafluoropropoxy)-1,1,2,2,3,3-hexafluoropropoxy)-1,1,2,2,3,3-hexafluoropropan-1-ol | C9HClF18O3 | 532.9271 | 3.08 | 182.99; 466.933 | 20.41 | 2.4 × 108 | 3 |
| 30 | 3-(Butylsulfanyl)-2-ethyl-4-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)benzoic acid | C16H17F7O2S | 405.0771 | 1.54 | 181.033; 235.0798; 305.024; 315.026; 361.086 | 9.43 | 3.1 × 106 | 2 |
| 31 | 3-[Ethyl(perfluoro-1-oxopentyl)amino]-2-hydroxypropyl heptanoate | C17H24F9NO4 | 476.1503 | 2.9 | 109.057; 218.986; 304.039; 346.095; 364.061; 448.115 | 11.79 | 3.2E × 106 | 2 |
| 32 | 3-Fluoro-4-{(E)-[4’-(heptafluoropropyl)-4-biphenylyl]diazenyl}phenol | C21H12F8N2O | 459.0763 | 2.93 | 109.012; 373.099; 439.069 | 13.73 | 5.6 × 107 | 3 |
| 33 | 3-Pyridinecarboxamide, N-[4-(nonafluorobutoxy)phenyl]- | C16H9F9N2O2 | 431.0450 | 0.66 | 78.034; 104.014; 121.041; 181.041; 403.013; 431.044 | 10.87 | 5.6 × 106 | 2 |
| 34 | 4,4-Bis(trifluoromethyl)-2H,4H-1,3-benzodioxine | C10H6F6O2 | 271.0192 | −2.65 | 245.004 | 10.20 | 5.0 × 108 | 3 |
| 35 | 4-[Ethyl(2,2,2-trifluoroethyl)amino]-2-(trifluoromethyl)benzonitrile | C12H10F6N2 | 295.0685 | 3.3 | 168.989; 211.995; 293.053; 295.068 | 7.61 | 5.7 × 107 | 2 |
| 36 | 5,5,6,6,7,7,8,8,9,9,10,10,10-Tridecafluoro-1-decanol | C10H9F13O | 427.0153 | 3.5 | 361.026; 391.0397 | 15.06 | 2.7 × 106 | 3 |
| 37 | 5,5,6,6,7,7,8,8,9,9,10,10,11,11,12,12,12-Heptadecafluoro-2-methyl-2-dodecanol | C13H11F17O | 505.0463 | −0.63 | 485.040 | 9.03 | 9.7 × 106 | 3 |
| 38 | 6-(1,1,1,3,3,3-Hexafluoro-2-hydroxypropan-2-yl)-3,5,5-trimethylcyclohex-2-en-1-ol | C12H16F6O2 | 305.0974 | −2.66 | 123.087; 166.997; 287.087; 303.082; | 13.55 | 2.7 × 107 | 2 |
| 39 | 6-Chloro-1,1,2,2,3,3,4,4,5,5,6,7-dodecafluoroheptane | C7H3ClF12 | 348.9671 | 1.70 | 328.9596; 348.966; | 9.59 | 6.8 × 107 | 3 |
| 40 | 9,9,10,10,11,11,12,12,12-Nonafluorododecan-1-ol | C12H17F9O | 347.1080 | 4.86 | 218.986; 273.033; 327.101 | 9.37 | 3.2 × 107 | 2 |
| 41 | Diethyl (3,3,4,4,5,5,6,6,6-nonafluorohexyl)phosphonate | C10H14F9O3P | 383.0449 | −4.03 | 106.991; 326.943; 337.075; 355.067 | 9.26 | 2.5 × 107 | 2 |
| 42 | Hexadecyl 2,3,3,3-tetrafluoropropanoate | C19H34F4O2 | 369.2405 | −4.69 | 129.128; 144.991; 185.023; 349.236 | 11.41 | 2.1 × 107 | 3 |
| 43 | N-[3-(Dimethylamino)propyl]perfluoro-4-(methyl)cyclohexanecarboxamide N-oxide | C13H13F13N2O2 | 475.0713 | 3.4 | 115.087; 117.1033; 143.085; 145.098 | 12.14 | 8.7 × 106 | 3 |
| 44 | N-[3-(Dimethylamino)propyl]perfluorobutanamide | C9H13F7N2O | 297.0843 | 0.02 | 168.992; 211.995; 240.026; 252.027; 277.078 | 11.92 | 1.5 × 108 | 2 |
| 45 | N-[3-(Dimethyloxidoamino)propyl]perfluoro-3,7-dioxaoctanamide | C11H13F11N2O4 | 445.0608 | −4.23 | 115.092; 143.089; 211.087; 359.067; 384.014; 429.037 | 10.86 | 4.6 × 107 | 2 |
| 46 | N-dihydroxyethyl amino propyl-perfluorodecane amide | C17H17F19N2O3 | 657.0890 | 4.16 | 169.098; 468.977; 511.978; 554.0229; 613.147; 637.097 | 12.49 | 6.8 × 106 | 2 |
| 47 | N-dimethylammoniocarboxypropyl-perfluoropropane sulfonamide | C9H13F7N2O4S | 377.0405 | −1.84 | 143.083; 168.986; 232.951; 275.993; 315.004; 333.051 | 13.80 | 9.2 × 106 | 2 |
| 48 | Perfluoro-2,2,3,3-tetramethyl butanoic acid | C8HF15O2 | 412.9674 | 2.24 | 193.997; 368.978 | 16.79 | 2.4 × 107 | 3 |
| 49 | Perfluoro-6-methylheptanesulfonate | C8HF17O3S | 498.9319 | 3.35 | 168.954; 330.158 | 17.91 | 8.7 × 107 | 2 |
| 50 | Perfluoro-n-hexanesulfonate | C6HF13O3S | 398.9372 | 1.38 | 118.472, 168.278 | 15.75 | 6.3 × 107 | 1 |
| 51 | Perfluoro-n-octanesulfonate | C8HF17O3S | 498.9321 | 3.77 | 118.232, 168.145; 218.784 | 18.26 | 1.5 × 108 | 1 |
| 52 | Perfluoroheptanesulphonyl fluoride | C7F16O2S | 450.9282 | −2.02 | 82.960; 368.980; 182.970 | 11.66 | 1.0 × 107 | 2 |
| 53 | Perfluoro-n-heptanoic acid | C7HF13O2 | 362.9704 | 2.27 | 96.974; 318.978 | 15.65 | 3.2 × 107 | 1 |
| 54 | Perfluoro-n-hexanoic acid | C6HF11O2 | 312.9735 | 2.3 | 193.97; 218.941; 268.983 | 13.61 | 3.2 × 107 | 1 |
| 55 | Perfluoro-n-nonanoic acid | C9HF17O2 | 462.9648 | 3.46 | 118.997; 298.048; 318.974 | 18.28 | 3.3 × 107 | 1 |
| 56 | Perfluoropropyl formate | C4HF7O2 | 212.9790 | −0.9 | 94.990; 184.984 | 4.53 | 3.4 × 106 | 2 |
| 57 | Perfluoro-n-pentanoic acid | C5HF9O2 | 262.9766 | 2.15 | 118.950; 143.910; 218.990 | 9.94 | 1.9 × 107 | 1 |
| 58 | Perfluoro-n-unidecanoic acid | C11HF21O2 | 562.9583 | 2.63 | 116.97; 243.97; 318.89 | 20.21 | 2.0 × 107 | 1 |
| 59 | Pyrifluquinazon | C19H15F7N4O2 | 463.1013 | 0.55 | 103.030; 298.047; 313.024; 443.097 | 15.49 | 7.8 × 106 | 3 |
| 60 | 2,2,3,3,4,4,5,5,6-nonafluoro-6-oxohexanoic acid | C6HF9O3 | 290.9723 | 4.87 | 168.972; 246.971 | 7.28 | 3.9 × 106 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlovic, R.; Draghi, S.; Pellegrini, A.; Fornesi Silva, C.; Di Cesare, F.; Curone, G.; Arioli, F.; Fidani, M. High-Resolution Mass Spectrometry Non-Targeted Detection of Per- and Polyfluoroalkyl Substances in Roe Deer (Capreolus capreolus). Molecules 2024, 29, 617. https://doi.org/10.3390/molecules29030617
Pavlovic R, Draghi S, Pellegrini A, Fornesi Silva C, Di Cesare F, Curone G, Arioli F, Fidani M. High-Resolution Mass Spectrometry Non-Targeted Detection of Per- and Polyfluoroalkyl Substances in Roe Deer (Capreolus capreolus). Molecules. 2024; 29(3):617. https://doi.org/10.3390/molecules29030617
Chicago/Turabian StylePavlovic, Radmila, Susanna Draghi, Alberto Pellegrini, Claudia Fornesi Silva, Federica Di Cesare, Giulio Curone, Francesco Arioli, and Marco Fidani. 2024. "High-Resolution Mass Spectrometry Non-Targeted Detection of Per- and Polyfluoroalkyl Substances in Roe Deer (Capreolus capreolus)" Molecules 29, no. 3: 617. https://doi.org/10.3390/molecules29030617
APA StylePavlovic, R., Draghi, S., Pellegrini, A., Fornesi Silva, C., Di Cesare, F., Curone, G., Arioli, F., & Fidani, M. (2024). High-Resolution Mass Spectrometry Non-Targeted Detection of Per- and Polyfluoroalkyl Substances in Roe Deer (Capreolus capreolus). Molecules, 29(3), 617. https://doi.org/10.3390/molecules29030617









