Characterizing Interactions Between Small Peptides and Dimethyl Sulfoxide Using Infrared Spectroscopy and Computational Methods
Abstract
1. Introduction
2. Results and Discussion
2.1. Direct Interactions Between Model Peptides and DMSO–ATR-FTIR Studies
2.2. FTIR Investigation the Water Structure Around Peptides in the Presence of DMSO
2.2.1. Experimental vs. Theoretical Spectra of Affected Water in Peptide–DMSO Systems
2.2.2. Analysis of the Spectral Shares of Affected Water in the Experimental Spectra in Peptide–DMSO Systems
2.2.3. Structural and Energetic Analysis of Changed-Affected Water Molecules
2.3. Interactions of Solute Hydration Shells–DFT/ONIOM Studies
2.3.1. Water Around Diglycine and NAGMA
2.3.2. Water in Diglycine—DMSO Systems
2.3.3. Water in NAGMA–DMSO Systems
3. Materials and Methods
3.1. Chemicals and Solutions
3.2. ATR-FTIR Studies of Weak Peptide–DMSO Interactions
3.3. FTIR Investigation of Water Structure–HDO Difference Spectra Method
3.3.1. Sample Preparations
3.3.2. FTIR Measurements
3.3.3. Analysis of HDO Spectral Data
3.4. DFT/ONIOM Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATR | Attenuated Total Reflectance |
| DFT | Density Functional Theory |
| DMSO | dimethyl sulfoxide |
| FTIR | Fourier transform infrared |
| fwhh | Full width at the half-height |
| NAGMA | N-acetyl-glycine-methylamide |
| NALMA | N-acetyl-leucine-methylamide |
| NMA | N-methylacetamide |
| ONIOM | Our own N-layered Integrated Molecular Orbital and Molecular Mechanics |
References
- Rupley, J.A.; Careri, G. Protein Hydration and Function. Adv. Protein Chem. 1991, 41, 37–172. [Google Scholar] [CrossRef] [PubMed]
- Levy, Y.; Onuchic, J.N. Water Mediation in Protein Folding and Molecular Recognition. Annu. Rev. Biophys. 2006, 35, 389–415. [Google Scholar] [CrossRef] [PubMed]
- Canchi, D.R.; García, A.E. Cosolvent Effects on Protein Stability. Annu. Rev. Phys. Chem. 2013, 64, 273–293. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.; Boikess, R.S.; Schwartz, R.A.; Cohen, P.J. Dimethyl sulfoxide (DMSO): A solvent that may solve selected cutaneous clinical challenges. Arch. Dermatol. Res. 2023, 315, 1465–1472. [Google Scholar] [CrossRef] [PubMed]
- Zalar, M.; Svilenov, H.L.; Golovanov, A.P. Binding of excipients is a poor predictor for aggregation kinetics of biopharmaceutical proteins. Eur. J. Pharm. Biopharm. 2020, 151, 127–136. [Google Scholar] [CrossRef]
- Clare, R. Trevitt, D.R. Yashwanth Kumar, N.J.F.; Williamson, M.P. Interactions between the protein barnase and co-solutes studied by NMR. Commun. Chem. 2024, 7, 44. [Google Scholar] [CrossRef]
- Anderson, C.F.; Courtenay, E.S.; Record, M.T. Thermodynamic Expressions Relating Different Types of Preferential Interaction Coefficients in Solutions Containing Two Solute Components. J. Phys. Chem. B 2002, 106, 418–433. [Google Scholar] [CrossRef]
- Scherer, T.M. Role of Cosolute–Protein Interactions in the Dissociation of Monoclonal Antibody Clusters. J. Phys. Chem. B 2015, 119, 13027–13038. [Google Scholar] [CrossRef]
- Canchi, D.R.; Paschek, D.; Garcia, A.E. Equilibrium study of protein denaturation by urea. J. Am. Chem. Soc. 2010, 132, 2338–2344. [Google Scholar] [CrossRef]
- Bolen, D.W.; Baskakov, I.V. The osmophobic effect: Natural selection of a thermodynamic force in protein folding. J. Mol. Biol. 2001, 310, 955–963. [Google Scholar] [CrossRef]
- Panuszko, A.; Pieloszczyk, M.; Kuffel, A.; Jacek, K.; Biernacki, K.; Demkowicz, S.; Stangret, J.; Bruździak, P. Hydration of Simple Model Peptides in Aqueous Osmolyte Solutions. Int. J. Mol. Sci. 2021, 22, 9350. [Google Scholar] [CrossRef] [PubMed]
- Panuszko, A.; Bruździak, P.; Kaczkowska, E.; Stangret, J. General Mechanism of Osmolytes’ Influence on Protein Stability Irrespective of the Type of Osmolyte Cosolvent. J. Phys. Chem. B 2016, 120, 11159–11169. [Google Scholar] [CrossRef]
- Timasheff, S.N. Protein-solvent preferential interactions, protein hydration, and the modulation of biochemical reactions by solvent components. Proc. Natl. Acad. Sci. USA 2002, 99, 9721–9726. [Google Scholar] [CrossRef]
- Zou, Q.; Bennion, B.J.; Daggett, V.; Murphy, K.P. The molecular mechanism of stabilization of proteins by TMAO and its ability to counteract the effects of urea. J. Am. Chem. Soc. 2002, 124, 1192–1202. [Google Scholar] [CrossRef]
- Sukenik, S.; Sapir, L.; Gilman-Politi, R.; Harries, D. Diversity in the mechanisms of cosolute action on biomolecular processes. Faraday Discuss. 2013, 160, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.K.; Bates, S.P.; Crain, J.; Martyna, G.J. Solution Structure of the Aqueous Model Peptide N-Methylacetamide. J. Phys. Chem. B 2006, 110, 21319–21326. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer-Stenner, R. Visible and UV-resonance Raman spectroscopy of model peptides. J. Raman Spectrosc. 2001, 32, 711–732. [Google Scholar] [CrossRef]
- Verheijen, M.; Lienhard, M.; Schrooders, Y.; Clayton, O.; Nudischer, R.; Boerno, S.; Timmermann, B.; Selevsek, N.; Schlapbach, R.; Gmuender, H.; et al. DMSO induces drastic changes in human cellular processes and epigenetic landscape in vitro. Sci. Rep. 2019, 9, 4641. [Google Scholar] [CrossRef]
- Notman, R.; Noro, M.; O’Malley, B.; Anwar, J. Molecular Basis for Dimethylsulfoxide (DMSO) Action on Lipid Membranes. J. Am. Chem. Soc. 2006, 128, 13982–13983. [Google Scholar] [CrossRef]
- Arakawa, T.; Kita, Y.; Timasheff, S.N. Protein precipitation and denaturation by dimethyl sulfoxide. Biophys. Chem. 2007, 131, 62–70. [Google Scholar] [CrossRef]
- Voets, I.K.; Cruz, W.A.; Moitzi, C.; Lindner, P.; Arêas, E.P.G.; Schurtenberger, P. DMSO-Induced Denaturation of Hen Egg White Lysozyme. J. Phys. Chem. B 2010, 114, 11875–11883. [Google Scholar] [CrossRef] [PubMed]
- Tjernberg, A.; Markova, N.; Griffiths, W.J.; HalléN, D. DMSO-Related Effects in Protein Characterization. SLAS Discov. 2006, 11, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.E.; Malardier-Jugroot, C.; Head-Gordon, T. Effects of co-solvents on peptide hydration water structure and dynamics. Phys. Chem. Chem. Phys. 2010, 12, 393–405. [Google Scholar] [CrossRef]
- Panuszko, A.; Stangret, J.; Nowosielski, B.; Bruździak, P. Interactions between hydration spheres of two different solutes in solution: The least squares fitting with constraints as a tool to determine water properties in ternary systems. J. Mol. Liq. 2020, 310, 113181. [Google Scholar] [CrossRef]
- Panuszko, A.; Bruździak, P.; Śmiechowski, M.; Stasiulewicz, M.; Stefaniak, J.; Stangret, J. DMSO hydration redefined: Unraveling the hydrophobic hydration of solutes with a mixed hydrophilic–hydrophobic characteristic. J. Mol. Liquids 2019, 294, 111661. [Google Scholar] [CrossRef]
- Biernacki, K.A.; Kaczkowska, E.; Bruździak, P. Aqueous solutions of NMA, Na2HPO4, and NaH2PO4 as models for interaction studies in phosphate–protein systems. J. Mol. Liq. 2018, 265, 361–371. [Google Scholar] [CrossRef]
- Kaczkowska, E.; Panuszko, A.; Bruździak, P. Interactions in Ternary Aqueous Solutions of NMA and Osmolytes—PARAFAC Decomposition of FTIR Spectra Series. Int. J. Mol. Sci. 2021, 22, 1684. [Google Scholar] [CrossRef]
- Godlewska, J.; Cieśla, B.; Wawer, J.; Bruździak, P. DMSO and TMAO—Differences in Interactions in Aqueous Solutions of the K-Peptide. Int. J. Mol. Sci. 2022, 23, 1872. [Google Scholar] [CrossRef]
- Bruździak, P. Vapor correction of FTIR spectra—A simple automatic least squares approach. Spectrochim. Acta Part Mol. Biomol. Spectrosc. 2019, 223, 117373. [Google Scholar] [CrossRef]
- Duplan, J.C.; Mahi, L.; Brunet, J.L. NMR determination of the equilibrium constant for the liquid H2O–D2O mixture. Chem. Phys. Lett. 2005, 413, 400–403. [Google Scholar] [CrossRef]
- Stangret, J. Solute-affected vibrational spectra of water in Ca[Cl04)2 aqueous solutions. Spectrosc. Lett. 1988, 21, 369–381. [Google Scholar] [CrossRef]
- Stangret, J.; Gampe, T. Hydration sphere of tetrabutylammonium cation. FTIR studies of HDO spectra. J. Phys. Chem. B 1999, 103, 3778–3783. [Google Scholar] [CrossRef]
- Śmiechowski, M.; Stangret, J. Vibrational spectroscopy of semiheavy water (HDO) as a probe of solute hydration. Pure Appl. Chem. 2010, 82, 1869–1887. [Google Scholar] [CrossRef]
- Berglund, B.; Lindgren, J.; Tegenfeldt, J. O–H and O–D Stretching Vibrations in Isotopically Dilute HDO Molecules in Some Solid Hydrates. J. Mol. Struct. 1978, 43, 169–177. [Google Scholar] [CrossRef]
- Walker, M.; Harvey, A.J.A.; Sen, A.; Dessent, C.E.H. Performance of M06, M06-2X, and M06-HF Density Functionals for Conformationally Flexible Anionic Clusters: M06 Functionals Perform Better than B3LYP for a Model System with Dispersion and Ionic Hydrogen-Bonding Interactions. J. Phys. Chem. A 2013, 117, 12590–12600. [Google Scholar] [CrossRef]
- Chung, L.W.; Sameera, W.M.C.; Ramozzi, R.; Page, A.J.; Hatanaka, M.; Petrova, G.P.; Harris, T.V.; Li, X.; Ke, Z.; Liu, F.; et al. The ONIOM Method and Its Applications. Chem. Rev. 2015, 115, 5678–5796. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 2012, 4, 17. [Google Scholar] [CrossRef]
- Frisch, M.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.; et al. Gaussian 09, Revision D.01; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]

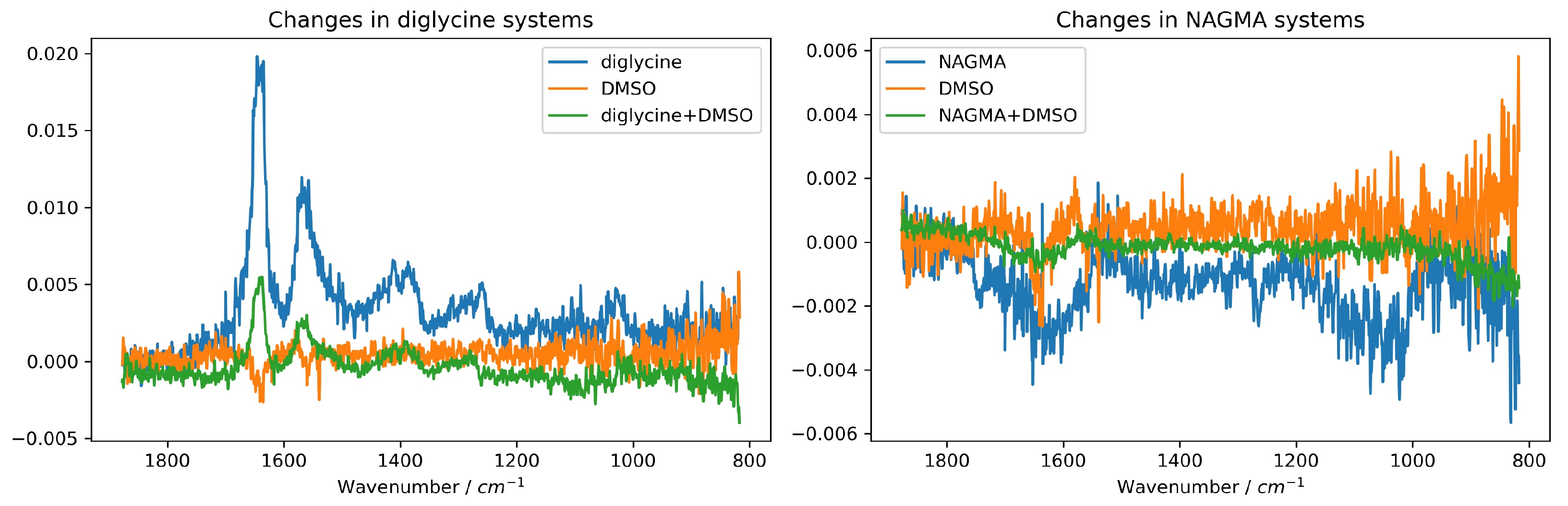
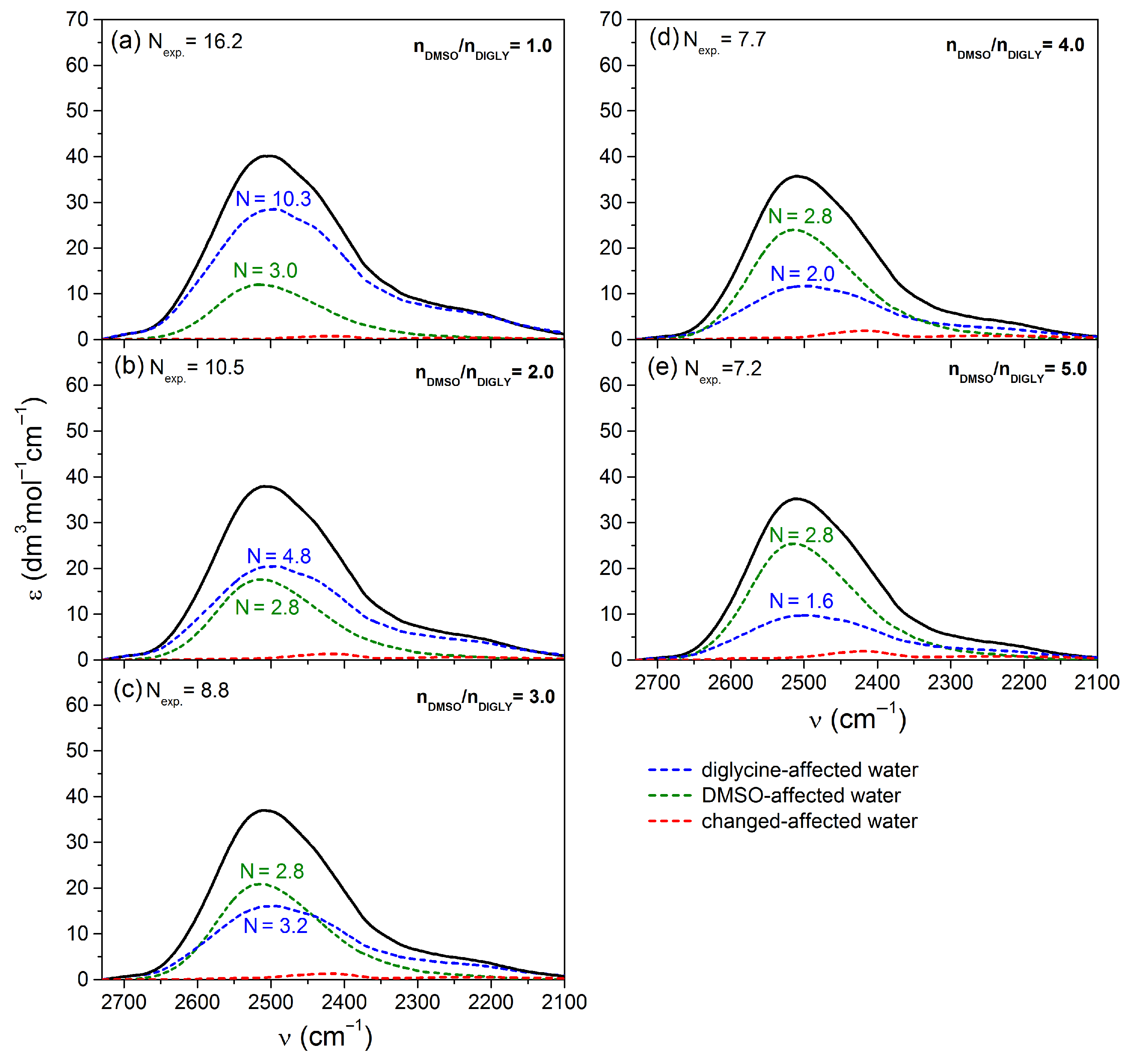

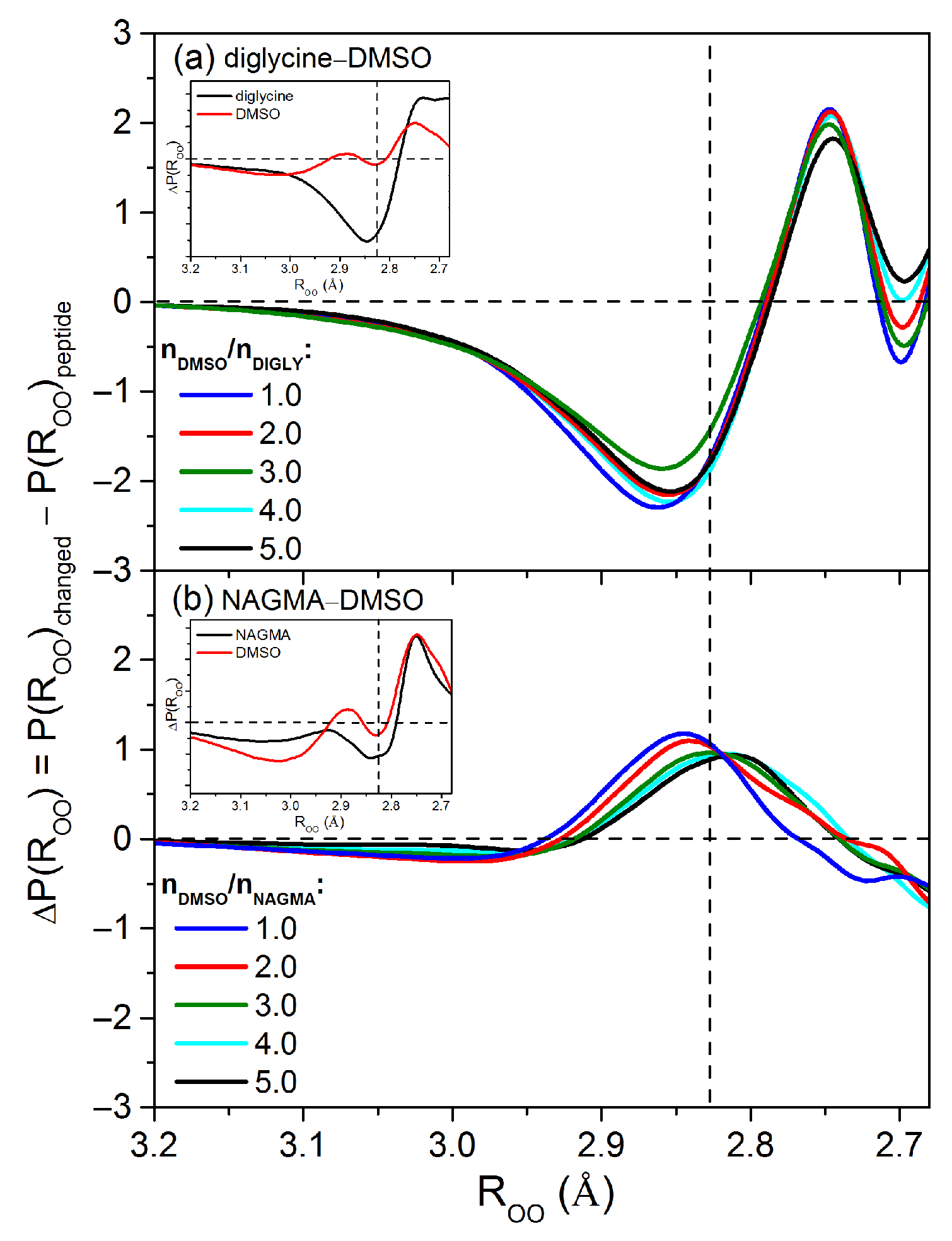
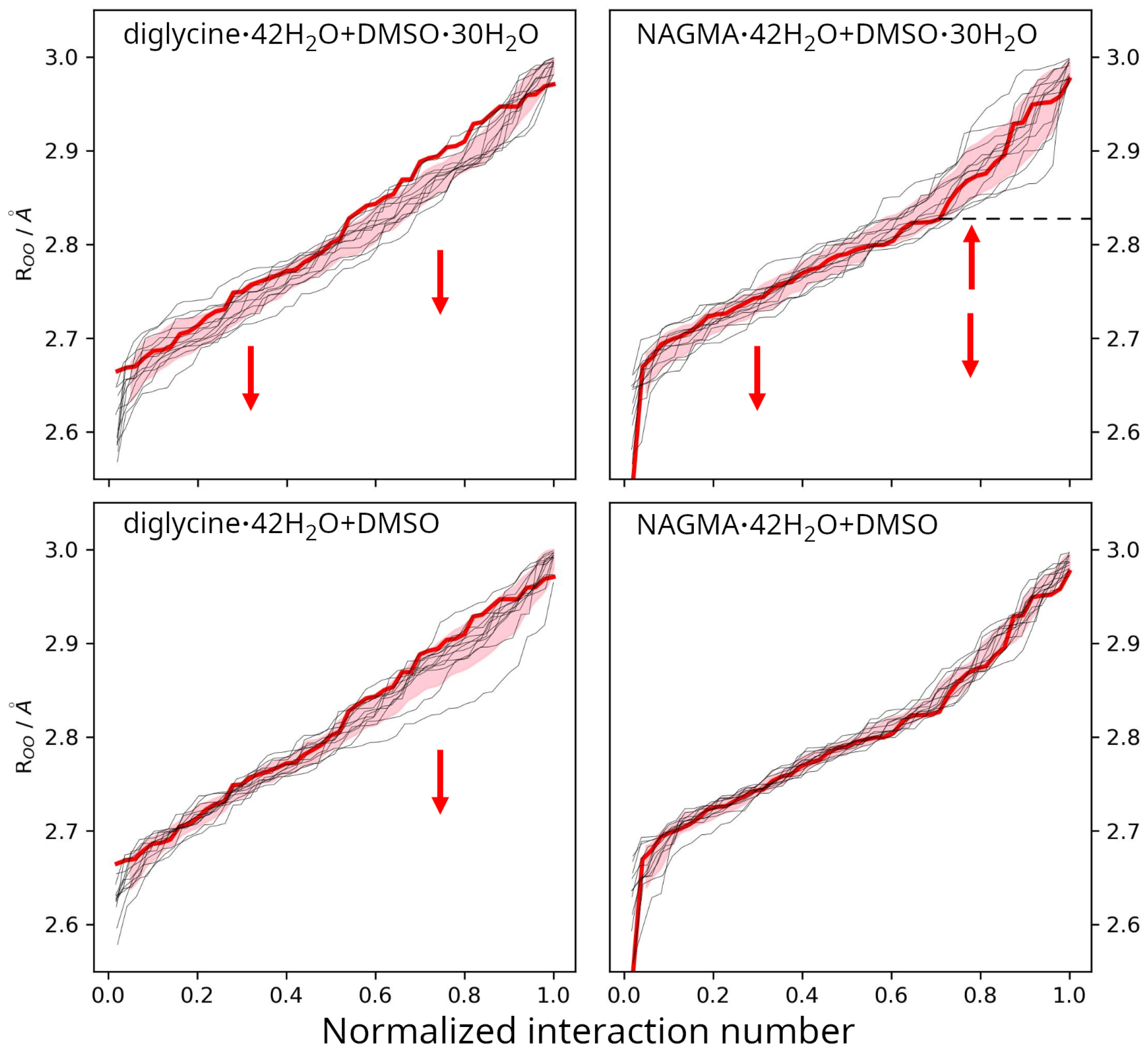
| / a | b | c | d | e |
|---|---|---|---|---|
| Pure Solutes | ||||
| diglycine | 2496 ± 2 | 2449 ± 2 | 2.778 ± 0.003 | 2.802 ± 0.003 |
| NAGMA | 2505 ± 2 | 2488 ± 2 | 2.827 ± 0.003 | 2.834 ± 0.003 |
| DMSO | 2514 ± 2 | 2483 ± 2 | 2.821 ± 0.003 | 2.831 ± 0.003 |
| bulk water | 2509 ± 2 | 2497 ± 2 | 2.827 ± 0.003 | 2.844 ± 0.003 |
| Changed-Affected Water in Diglycine–DMSO System | ||||
| 1.0 | 2415 ± 2 | 2349 ± 2 | 2.744 ± 0.003 | 2.729 ± 0.003 |
| 2.0 | 2415 ± 2 | 2356 ± 2 | 2.744 ± 0.003 | 2.732 ± 0.003 |
| 3.0 | 2419 ± 2 | 2363 ± 2 | 2.744 ± 0.003 | 2.739 ± 0.003 |
| 4.0 | 2417 ± 2 | 2354 ± 2 | 2.744 ± 0.003 | 2.729 ± 0.003 |
| 5.0 | 2417 ± 2 | 2361 ± 2 | 2.744 ± 0.003 | 2.732 ± 0.003 |
| Changed-Affected Water in NAGMA–DMSO System | ||||
| 1.0 | 2511 ± 2 | 2496 ± 2 | 2.836 ± 0.003 | 2.839 ± 0.003 |
| 2.0 | 2507 ± 2 | 2493 ± 2 | 2.836 ± 0.003 | 2.836 ± 0.003 |
| 3.0 | 2505 ± 2 | 2493 ± 2 | 2.827 ± 0.003 | 2.834 ± 0.003 |
| 4.0 | 2504 ± 2 | 2494 ± 2 | 2.822 ± 0.003 | 2.834 ± 0.003 |
| 5.0 | 2502 ± 2 | 2495 ± 2 | 2.819 ± 0.003 | 2.836 ± 0.003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panuszko, A.; Pastwa, P.; Gajewski, J.; Bruździak, P. Characterizing Interactions Between Small Peptides and Dimethyl Sulfoxide Using Infrared Spectroscopy and Computational Methods. Molecules 2024, 29, 5869. https://doi.org/10.3390/molecules29245869
Panuszko A, Pastwa P, Gajewski J, Bruździak P. Characterizing Interactions Between Small Peptides and Dimethyl Sulfoxide Using Infrared Spectroscopy and Computational Methods. Molecules. 2024; 29(24):5869. https://doi.org/10.3390/molecules29245869
Chicago/Turabian StylePanuszko, Aneta, Przemysław Pastwa, Jacek Gajewski, and Piotr Bruździak. 2024. "Characterizing Interactions Between Small Peptides and Dimethyl Sulfoxide Using Infrared Spectroscopy and Computational Methods" Molecules 29, no. 24: 5869. https://doi.org/10.3390/molecules29245869
APA StylePanuszko, A., Pastwa, P., Gajewski, J., & Bruździak, P. (2024). Characterizing Interactions Between Small Peptides and Dimethyl Sulfoxide Using Infrared Spectroscopy and Computational Methods. Molecules, 29(24), 5869. https://doi.org/10.3390/molecules29245869






