Cytotoxic and Antibacterial Cyclodepsipeptides from an Endophytic Fungus Fusarium avenaceum W8
Abstract
:1. Introduction
2. Results and Discussion
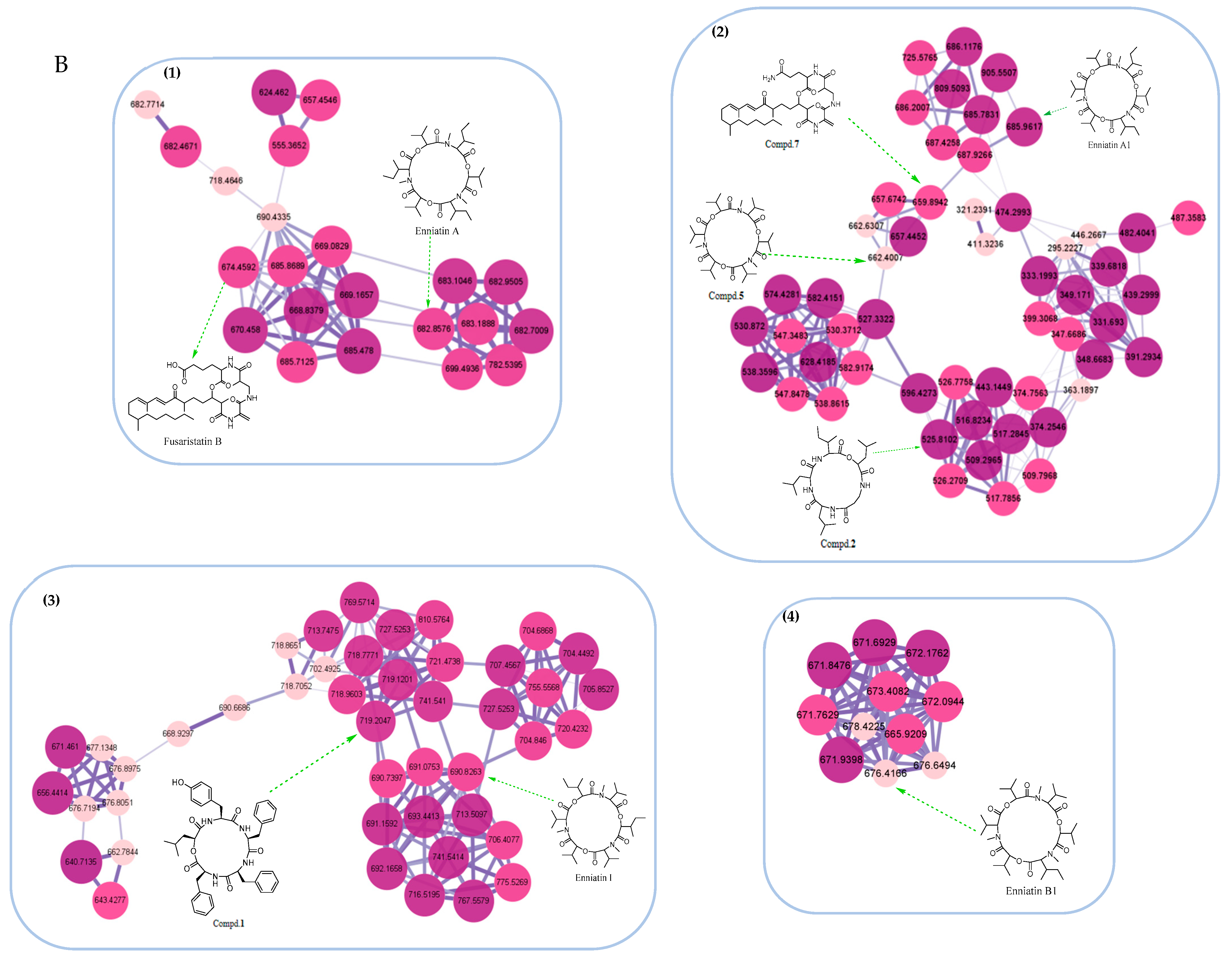
2.1. Bioactivity-Guided Isolation and Molecular Networking Analysis
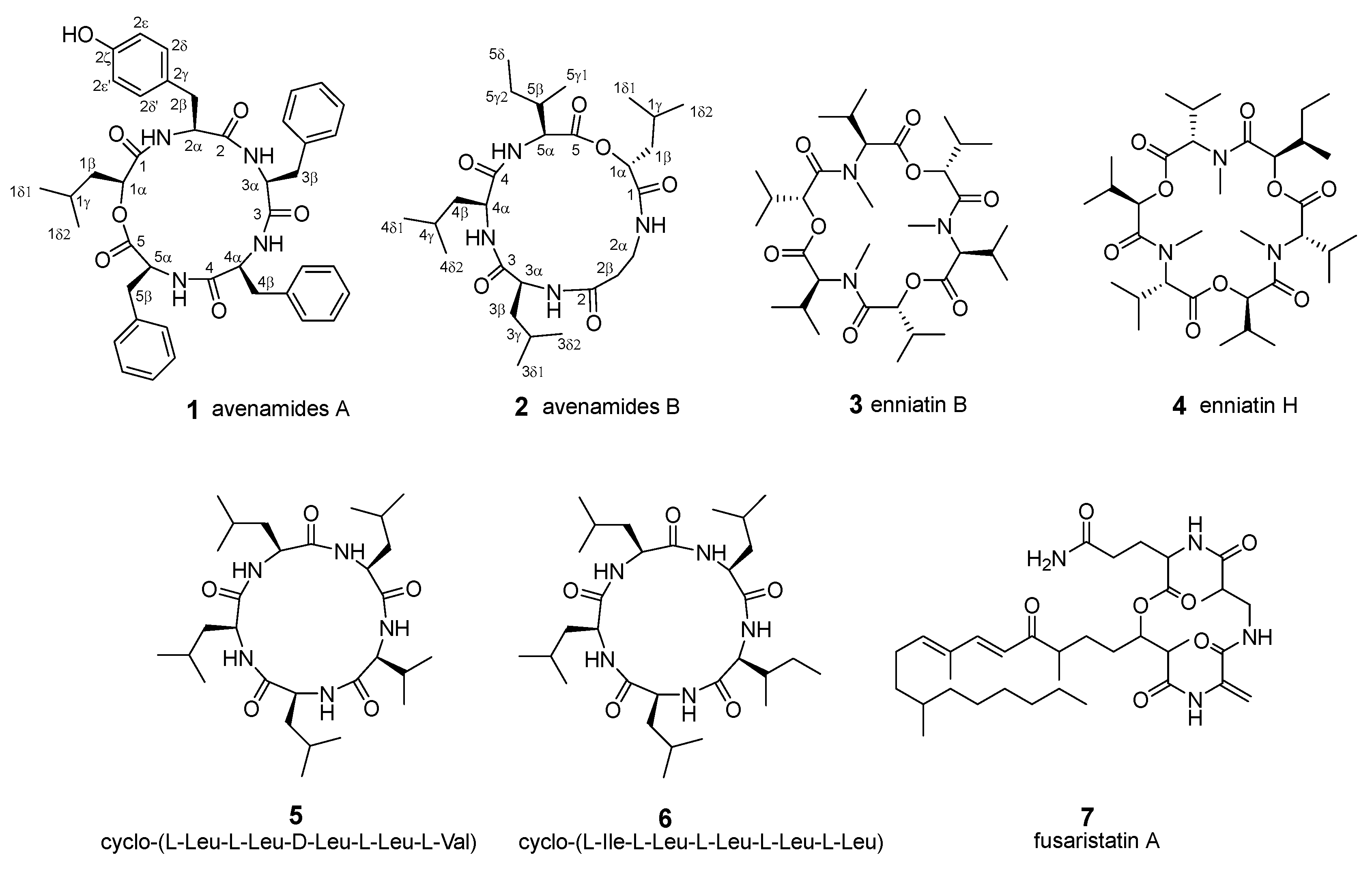
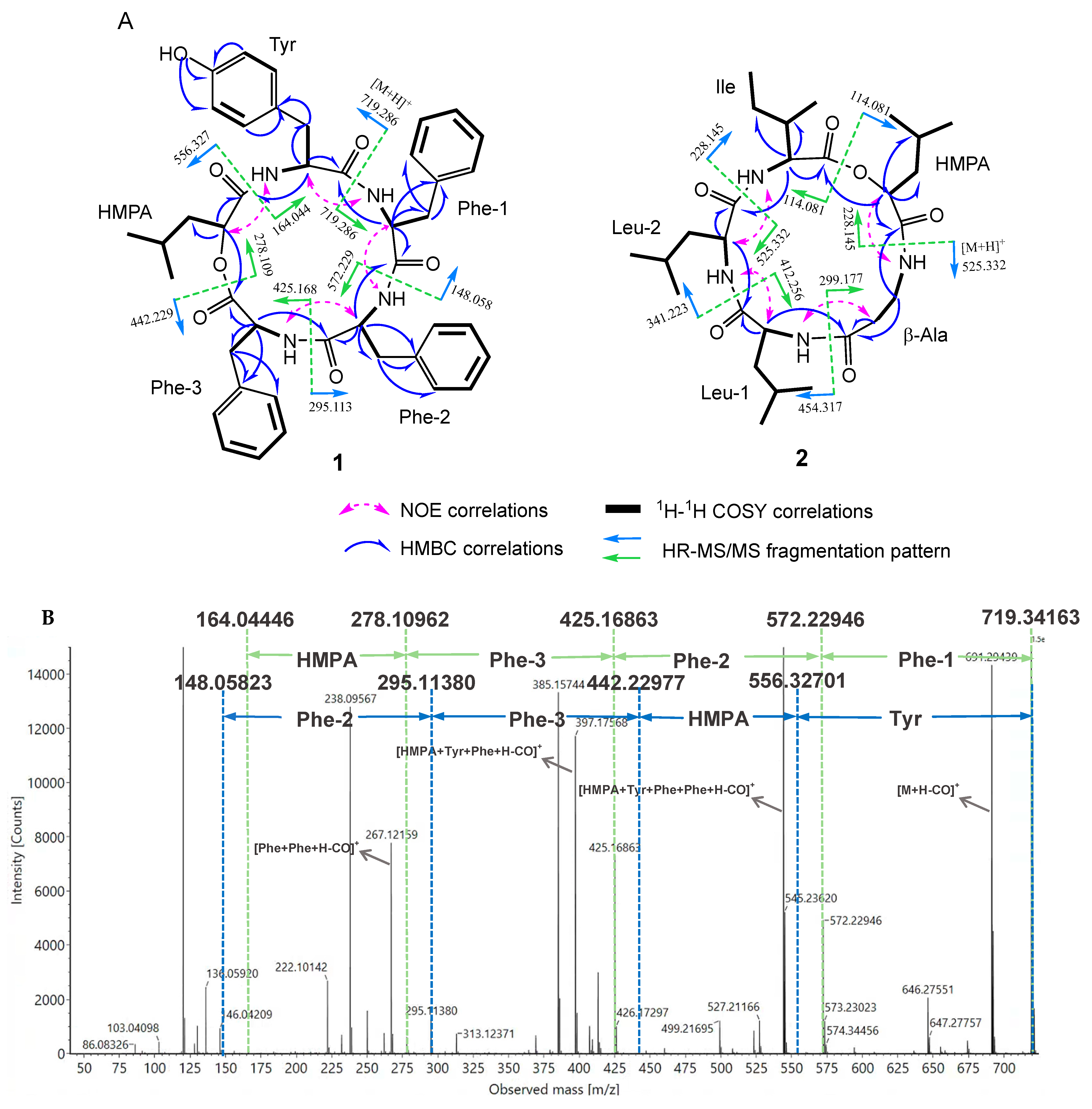
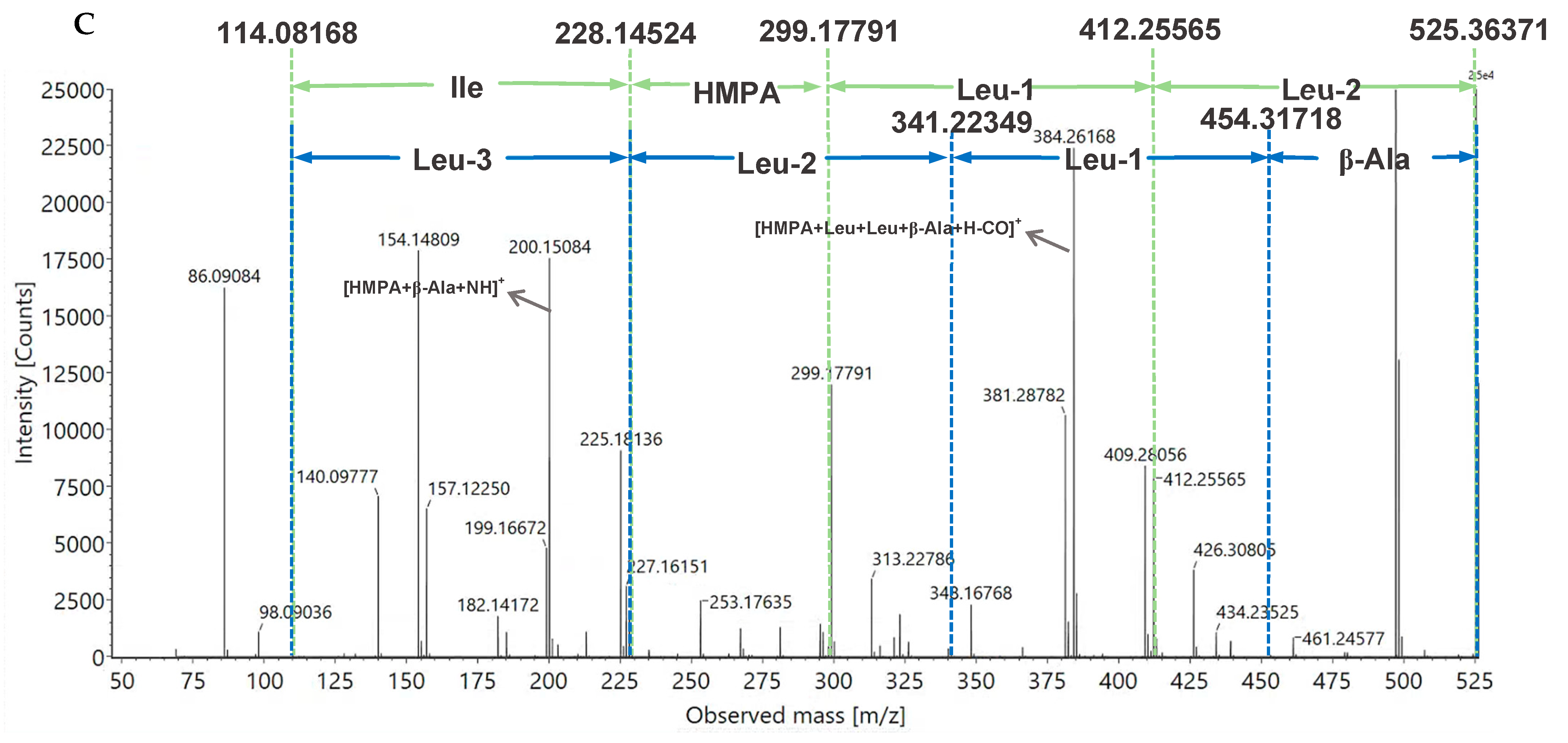
2.2. Structural Elucidation of Avenamides A (1) and B (2)
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation and Isolation
3.4. Advanced Marfey’s Analysis and Chiral HPLC Analysis
3.5. MS/MS Data Conversion and Molecular Network Visualization
3.6. UPLC-HR-MS Data Acquisition
3.7. Cytotoxicity Assay
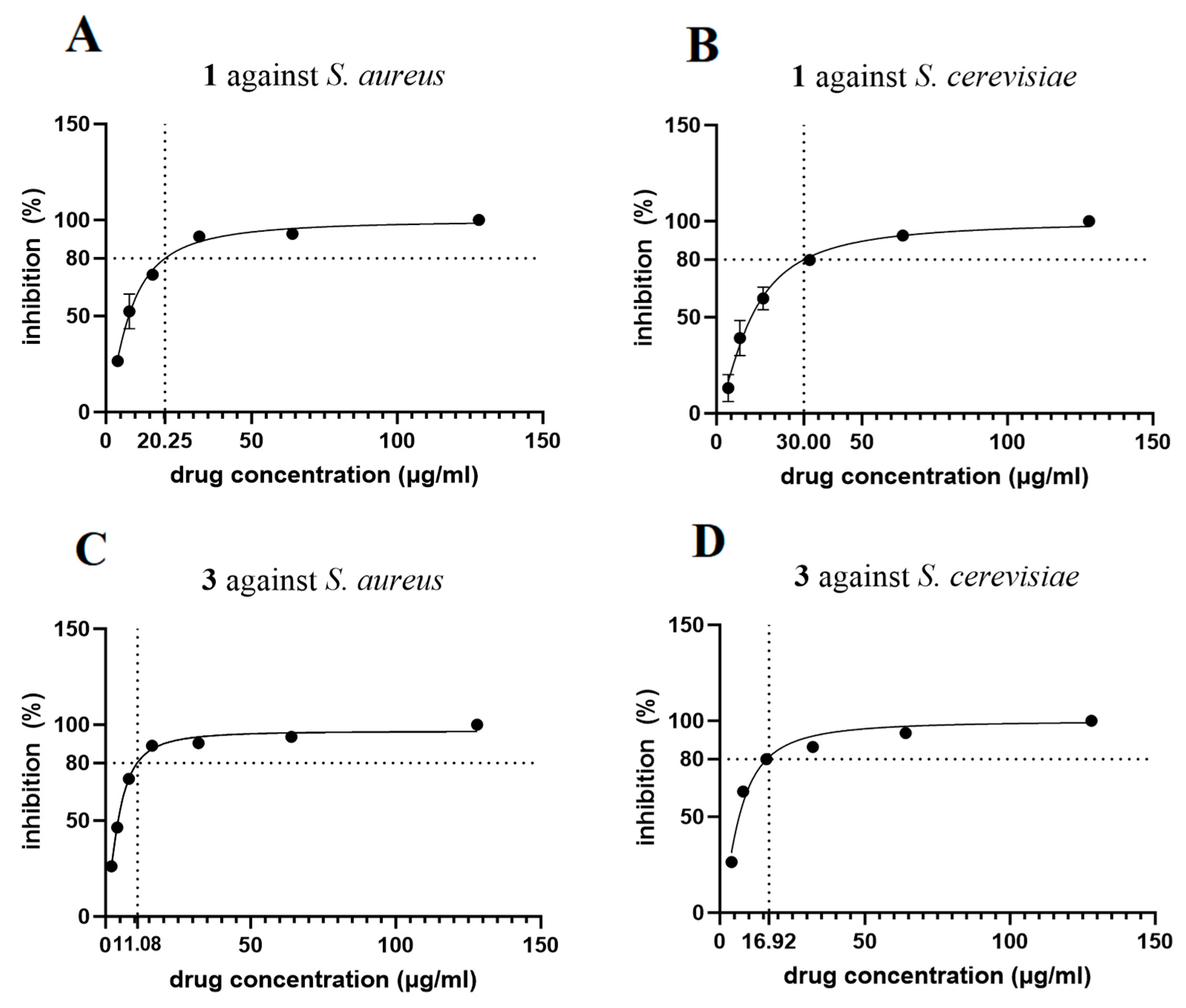
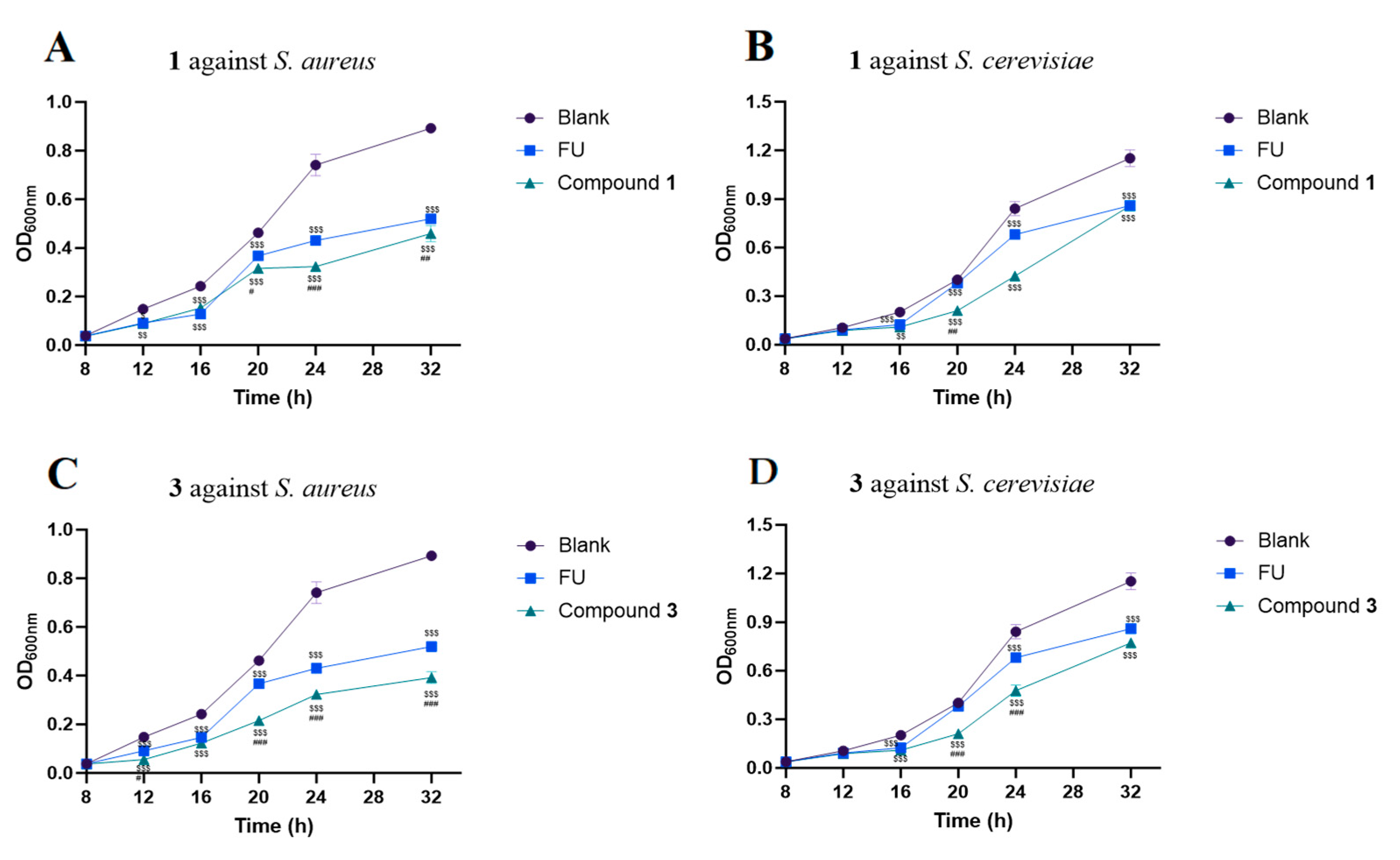
3.8. Antibacterial Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tiwari, P.; Bae, H. Endophytic Fungi: Key Insights, Emerging Prospects, and Challenges in Natural Product Drug Discovery. Microorganisms 2022, 10, 360. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Mahmoud, B.K.; Millán-Aguiñaga, N.; Abdelmohsen, U.R.; Fouad, M.A. The endophytic Fusarium strains: A treasure trove of natural products. RSC Adv. 2023, 13, 1339–1369. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Huang, Z.; Zhu, W.; Liu, Y.; Bai, X.; Zhang, H. Fusarium-Derived Secondary Metabolites with Antimicrobial Effects. Molecules 2023, 28, 3424. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, M.; Waśkiewicz, A.; Stępień, Ł. Fusarium Cyclodepsipeptide Mycotoxins: Chemistry, Biosynthesis, and Occurrence. Toxins 2020, 12, 765. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Sun, Y.; Tang, M.; Sun, P.; Wang, A.; Hao, Y.; Wang, Y.; Pei, Y. Trichodestruxins A-D: Cytotoxic Cyclodepsipeptides from the Endophytic Fungus Trichoderma harzianum. J. Nat. Prod. 2020, 83, 3635–3641. [Google Scholar] [CrossRef] [PubMed]
- Sivanathan, S.; Scherkenbeck, J. Cyclodepsipeptides: A rich source of biologically active compounds for drug research. Molecules 2014, 19, 12368–12420. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gong, X.; Li, P.; Lai, D.; Zhou, L. Structural Diversity and Biological Activities of Cyclic Depsipeptides from Fungi. Molecules 2018, 23, 169. [Google Scholar] [CrossRef] [PubMed]
- Otrubova, K.; Lushington, G.; Velde, D.V.; McGuire, K.L.; McAlpine, S.R. Comprehensive Study of Sansalvamide A Derivatives and their Structure—Activity Relationships against Drug-Resistant Colon Cancer Cell Lines. J. Med. Chem. 2008, 51, 530–544. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Moon, S.-K.; Lee, C. N-methylsansalvamide elicits antitumor effects in colon cancer cells in vitro and in vivo by regulating proliferation, apoptosis, and metastatic capacity. Front. Pharmacol. 2023, 14, 1146966. [Google Scholar] [CrossRef]
- Chi, L.-P.; Liu, D.; Li, X.-M.; Wan, Y.; Wang, B.-G.; Li, X. Aspertides A-E: Antimicrobial Pentadepsipeptides with a Unique p-Methoxycinnamoyl Amide Group from the Marine Isolates Aspergillus tamarii MA-21 and Aspergillus insuetus SD-512. J. Agric. Food Chem. 2023, 71, 13316–13324. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Kusari, S.; Golz, C.; Strohmann, C.; Spiteller, M. Three cyclic pentapeptides and a cyclic lipopeptide produced by endophytic Fusarium decemcellulare LG53. RSC Adv. 2016, 6, 54092–54098. [Google Scholar] [CrossRef]
- Nilanonta, C.; Isaka, M.; Chanphen, R.; Thong-Orn, N.; Tanticharoen, M.; Thebtaranonth, Y. Unusual enniatins produced by the insect pathogenic fungus Verticillium hemipterigenum: Isolation and studies on precursor-directed biosynthesis. Tetrahedron 2003, 59, 1015–1020. [Google Scholar] [CrossRef]
- Shiono, Y.; Tsuchinari, M.; Shimanuki, K.; Miyajima, T.; Murayama, T.; Koseki, T.; Laatsch, H.; Funakoshi, T.; Takanami, K.; Suzuki, K. Fusaristatins A and B, two new cyclic lipopeptides from an endophytic Fusarium sp. J Antibiot. 2007, 60, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Talontsi, F.M.; Facey, P.; Tatong, M.D.K.; Islam, M.T.; Frauendorf, H.; Draeger, S.; von Tiedemann, A.; Laatsch, H. Zoosporicidal metabolites from an endophytic fungus Cryptosporiopsis sp. of Zanthoxylum leprieurii. Phytochemistry 2012, 83, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Barret, M.O.; Boyd, K.G.; Adams, D.R.; Boyd, A.S.; Burgess, J.G. JM47, a cyclic tetrapeptide HC-toxin analogue from a marine Fusarium species. Phytochemistry 2002, 60, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Lee, C. Structural analysis of a new cytotoxic demethylated analogue of Neo-N-methylsansalvamide with a different peptide sequence produced by Fusarium solani isolated from potato. J. Agri. Food Chem. 2012, 60, 4342–4347. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Phat, C.; Choi, S.-U.; Lee, C. Synergistic effect of a novel cyclic pentadepsipetide, neoN-methylsansalvamide, and paclitaxel on human multidrug resistance cancer cell lines. Anti-Cancer Drugs 2013, 24, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Hamano, K.; Kinoshita, M.; Tanzawa, K.; Yoda, K.; Ohki, Y.; Nakamura, T. Leualacin, a novel calcium blocker from Hapsidospora irregularis: II. Structure deremination. J. Antibiot. 1992, 45, 906–913. [Google Scholar] [CrossRef] [PubMed]

| 1 | 2 | ||||
|---|---|---|---|---|---|
| No. | δC | δH, mult. (J in Hz) | No. | δC | δH, mult. (J in Hz) |
| OLeu | OLeu | ||||
| 1 | 168.5 | − | 1 | 168.9 | - |
| 2 | 73.9 | 4.85 (dd, J = 8.3, 5.8 Hz, 1H) | 2 | 72.9 | 5.1 (dd, J = 7.3, 4.6 Hz, 1H) |
| 3 | 40.2 | 1.58 (dd, J = 14.3, 7.5 Hz, 1H); 1.42 (dd, J = 14.3, 5.8 Hz, 1H) | 3 | 40.7 | 1.54–1.65 (m, 2H) |
| 4 | 23.7 | 1.30 (m, 1H) | 4 | 24.1 | 1.62 (m, 1H) |
| 5 | 22.5 | 0.83 (d, J = 6.5 Hz, 3H) | 5 | 23.4 | 0.81 (d, J = 6.7 Hz, 3H) |
| 6 | 21.9 | 0.79 (d, J = 6.5 Hz, 3H) | 6 | 23.4 | 0.86 (d, J = 6.8 Hz, 3H) |
| Tyr | β-Ala | ||||
| 1 | 170.6 | − | 1 | 171.1 | - |
| 2 | 54.9 | 4.39 (dd, J = 8.8, 6.6 Hz, 1H) | 2 | 35.4 | 3.49 (m, 1H); 3.01 (m, 1H) |
| 3 | 36.3 | 2.79 (m, 1H); 2.85 (m, 1H) | 3 | 34.9 | 2.4 (t, J = 13.7 Hz, 1H); 2.12 (dt, J = 14.9, 3.1 Hz, 1H) |
| 4 | 127.1 | − | NH | 7.48 (brs, 1H) | |
| 5 | 129.9 | 6.93 (d, J = 8.0 Hz, 1H) | |||
| 6 | 115.0 | 6.63 (d, J = 8.0 Hz, 1H) | |||
| 7 | 155.9 | − | |||
| 8 | 115.0 | 6.63 (d, J = 8.0 Hz, 1H) | |||
| 9 | 129.9 | 6.93 (d, J = 8.0 Hz, 1H) | |||
| NH | 7.91 (d, J = 9.3 Hz, 1H) | ||||
| OH | 9.22 (s, 1H) | ||||
| Phe1 | Leu1 | ||||
| 1 | 171.0 | - | 1 | 171.2 | - |
| 2 | 57.1 | 4.00 (m, 1H) | 2 | 51.1 | 4.23 (m, 1H) |
| 3 | 36.0 | 2.85 (m, 1H); 3.02 (m, 1H) | 3 | 40.1 | 1.57–1.66 (m, 2H) |
| 4 | 137.5 | - | 4 | 24.5 | 1.50–1.60 (m, 1H) |
| 5 | 129.0 | 7.12 (d, J = 7.4 Hz, 1H) | 5 | 22.3 | 0.83 (d, J = 6.8 Hz, 3H) |
| 6 | 128.3 | 7.28 (m, 1H) | 6 | 23.3 | 0.91 (d, J = 6.8 Hz, 3H) |
| 7 | 126.5 | 7.19 (m, 1H) | NH | 8.12 (brs, 1H) | |
| 8 | 128.3 | 7.28 (m, 1H) | |||
| 9 | 129.0 | 7.12 (d, J = 7.4 Hz, 1H) | |||
| NH | 8.29 (s, 1H) | ||||
| Phe2 | Leu2 | ||||
| 1 | 170.8 | - | 1 | 172.9 | - |
| 2 | 56.2 | 4.27 (m, 1H) | 2 | 52.0 | 4.29 (m, 1H) |
| 3 | 37.1 | 2.88 (m, 2H) | 3 | 41.1 | 1.46–1.50 (m, 2H) |
| 4 | 137.4 | - | 4 | 24.3 | 1.60–1.66 (m, 1H) |
| 5 | 128.9 | 6.99 (d, J = 7.0 Hz, 1H) | 5 | 21.6 | 0.89 (d, J = 6.6 Hz, 3H) |
| 6 | 128.3 | 7.20 (m, 1H) | 6 | 22.0 | 0.88 (d, J = 6.8 Hz, 3H) |
| 7 | 126.4 | 7.21 (m, 1H) | NH | 8.46 (brs, 1H) | |
| 8 | 128.3 | 7.20 (m, 1H) | |||
| 9 | 128.9 | 6.99 (d, J = 7.0 Hz, 1H) | |||
| NH | 8.05 (s, 1H) | ||||
| Phe3 | Ile | ||||
| 1 | 168.8 | - | 1 | 169.2 | - |
| 2 | 54.2 | 4.61 (q, J = 8.0 Hz, 1H) | 2 | 57.9 | 4.03 (m, 1H) |
| 3 | 36.5 | 3.09 (dd, J = 13.8, 9.0 Hz, 1H); 3.18 (dd, J = 13.8, 6.5 Hz, 1H) | 3 | 34.3 | 2.04 (m, 1H) |
| 4 | 136.8 | - | 4 | 24.2 | 1.03 (dq, J = 17.4, 9.9, 8.4 Hz, 1H); 1.44 (m, 1H) |
| 5 | 129.2 | 7.26 (m, 1H) | 5 | 15.6 | 0.86 (d, J = 6.7 Hz, 3H) |
| 6 | 128.4 | 7.32 (t, J = 7.5 Hz, 1H) | 6 | 10.0 | 0.78 (t, J = 7.5 Hz, 3H) |
| 7 | 126.7 | 7.22 (m, 1H) | NH | 8.32 (brs, 1H) | |
| 8 | 128.4 | 7.32 (t, J = 7.5 Hz, 1H) | |||
| 9 | 129.2 | 7.26 (m, 1H) | |||
| NH | 8.41 (d, J = 8.5 Hz, 1H) | ||||
| IC50 Values (μM) of 1–4 and 7 a | ||||||
|---|---|---|---|---|---|---|
| Cell lines | 1 | 2 | 3 | 4 | 7 | adriamycin |
| A549 | 38.60 ± 0.40 | 45.20 ± 0.42 | 13.69 ± 0.27 | 6.52 ± 0.25 | 26.36 ± 0.32 | 4.26 ± 0.17 |
| NCI-H1944 | 28.60 ± 0.12 | 37.66 ± 0.15 | 23.66 ± 0.37 | 24.60 ± 0.20 | 39.50 ± 0.30 | 4.35 ± 0.27 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Liu, B.; Wang, Y.; Xu, Y.; Ma, H.; Sun, Y. Cytotoxic and Antibacterial Cyclodepsipeptides from an Endophytic Fungus Fusarium avenaceum W8. Molecules 2024, 29, 5746. https://doi.org/10.3390/molecules29235746
Wang Z, Liu B, Wang Y, Xu Y, Ma H, Sun Y. Cytotoxic and Antibacterial Cyclodepsipeptides from an Endophytic Fungus Fusarium avenaceum W8. Molecules. 2024; 29(23):5746. https://doi.org/10.3390/molecules29235746
Chicago/Turabian StyleWang, Zimo, Bo Liu, Yanlei Wang, Yicen Xu, Hai Ma, and Yi Sun. 2024. "Cytotoxic and Antibacterial Cyclodepsipeptides from an Endophytic Fungus Fusarium avenaceum W8" Molecules 29, no. 23: 5746. https://doi.org/10.3390/molecules29235746
APA StyleWang, Z., Liu, B., Wang, Y., Xu, Y., Ma, H., & Sun, Y. (2024). Cytotoxic and Antibacterial Cyclodepsipeptides from an Endophytic Fungus Fusarium avenaceum W8. Molecules, 29(23), 5746. https://doi.org/10.3390/molecules29235746







