Metabolic Engineering of Komagataella phaffii for Xylose Utilization from Cellulosic Biomass
Abstract
1. Introduction
2. Results
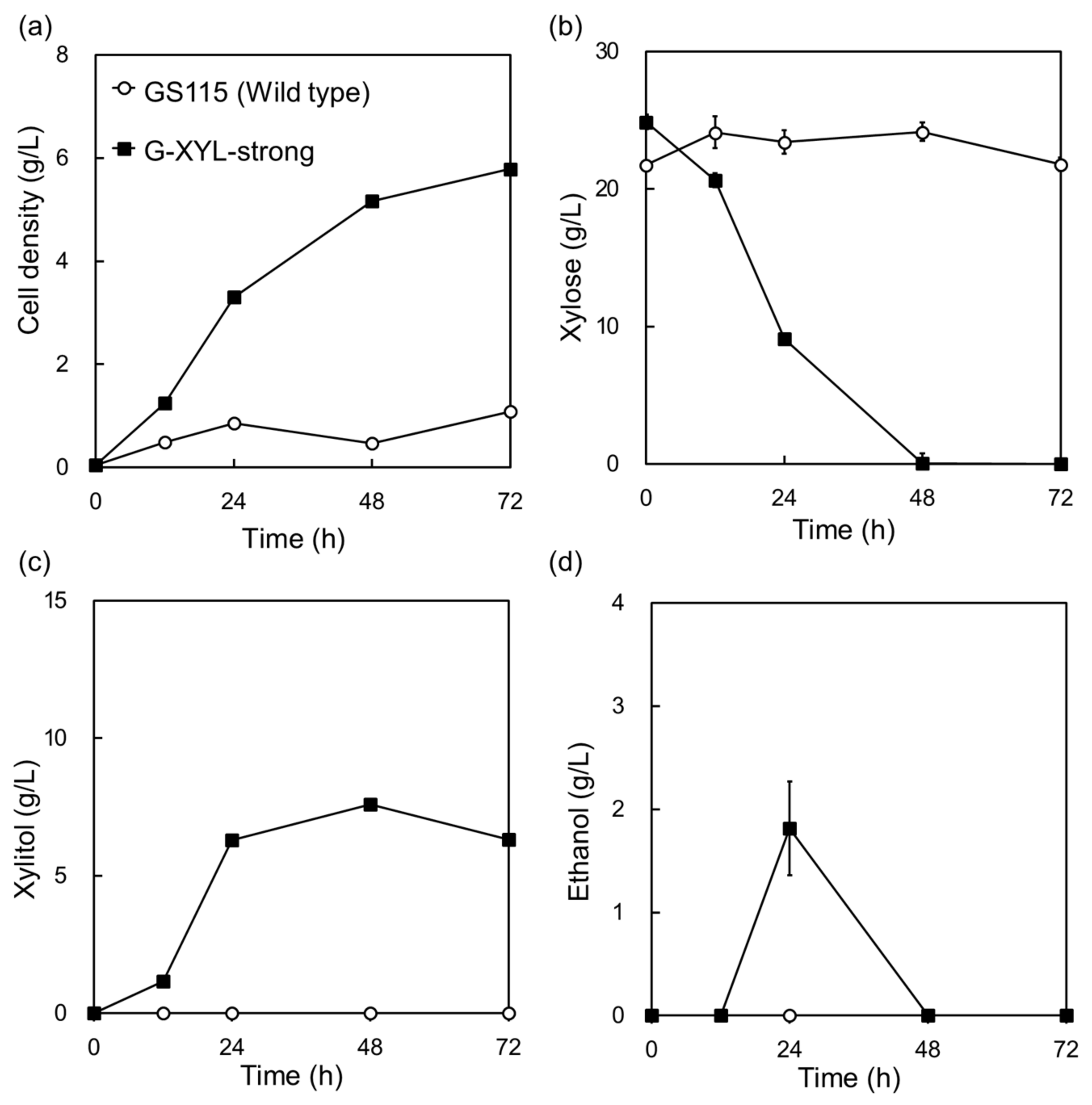
2.1. Expression of a Heterologous Xylose-Metabolic Pathway in Komagataella phaffii Under the Control of Strong Promoters
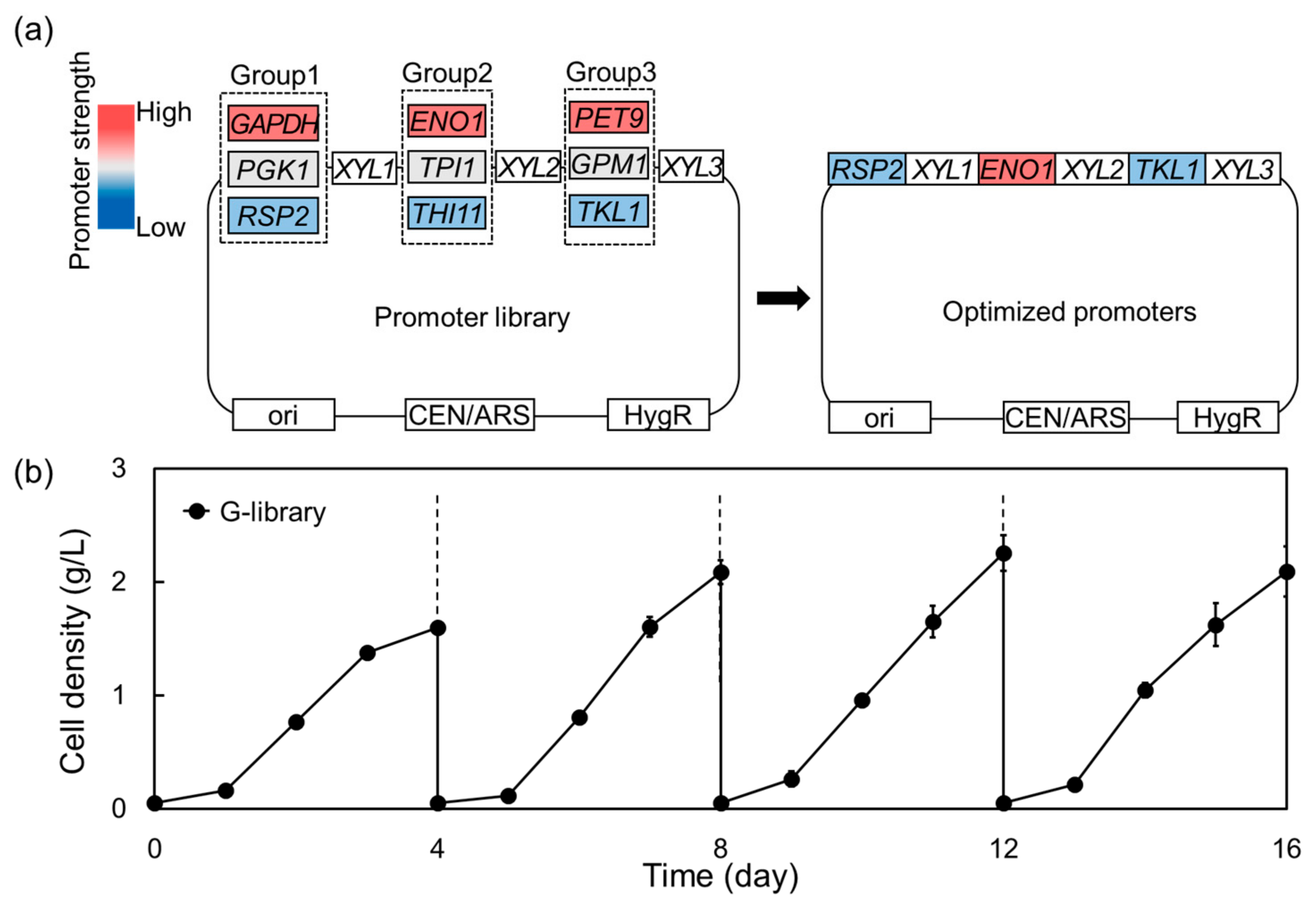
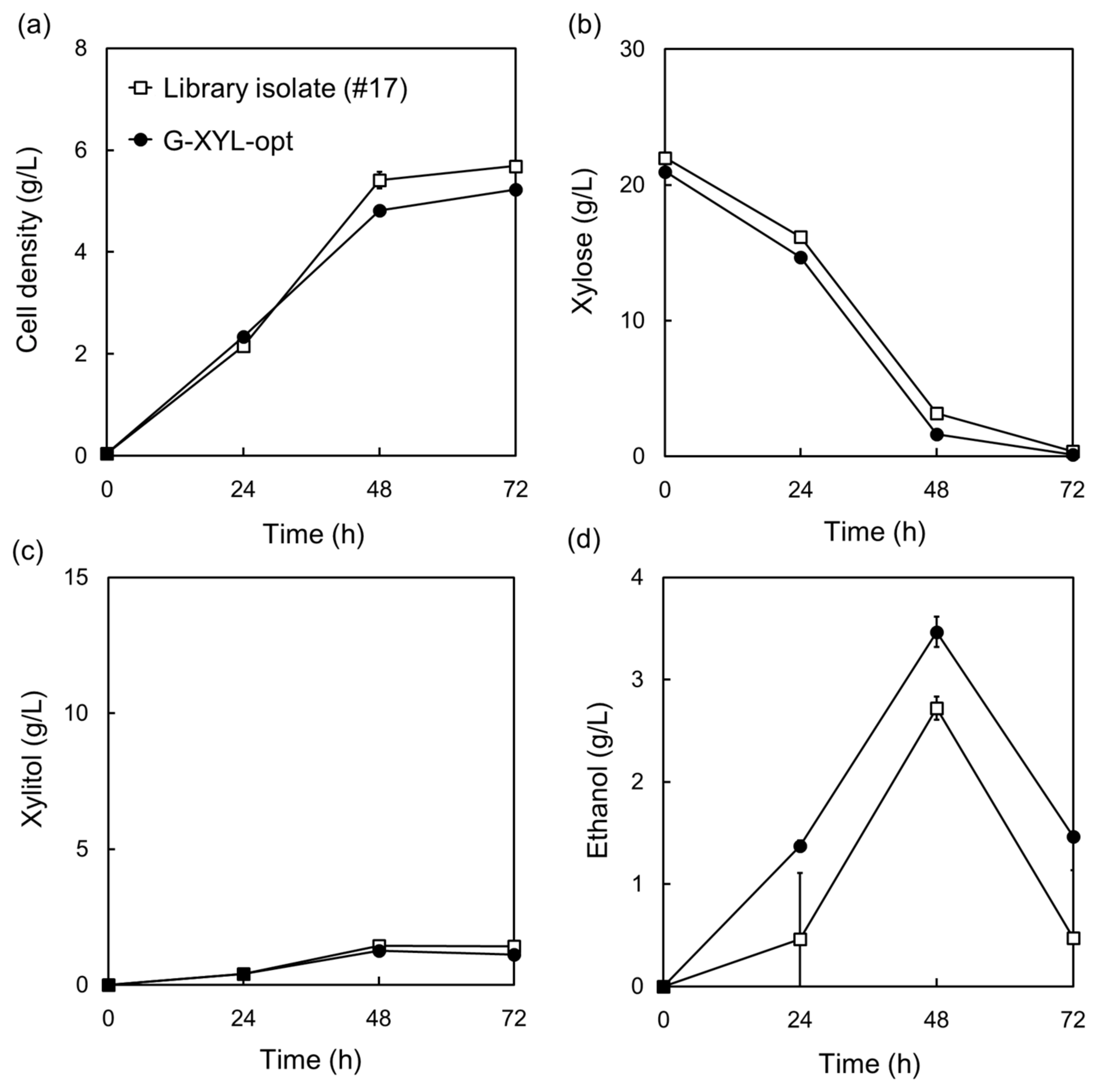
2.2. Optimization of a Heterologous Xylose-Metabolic Pathway in Komagataella phaffii Using a Promotor Library
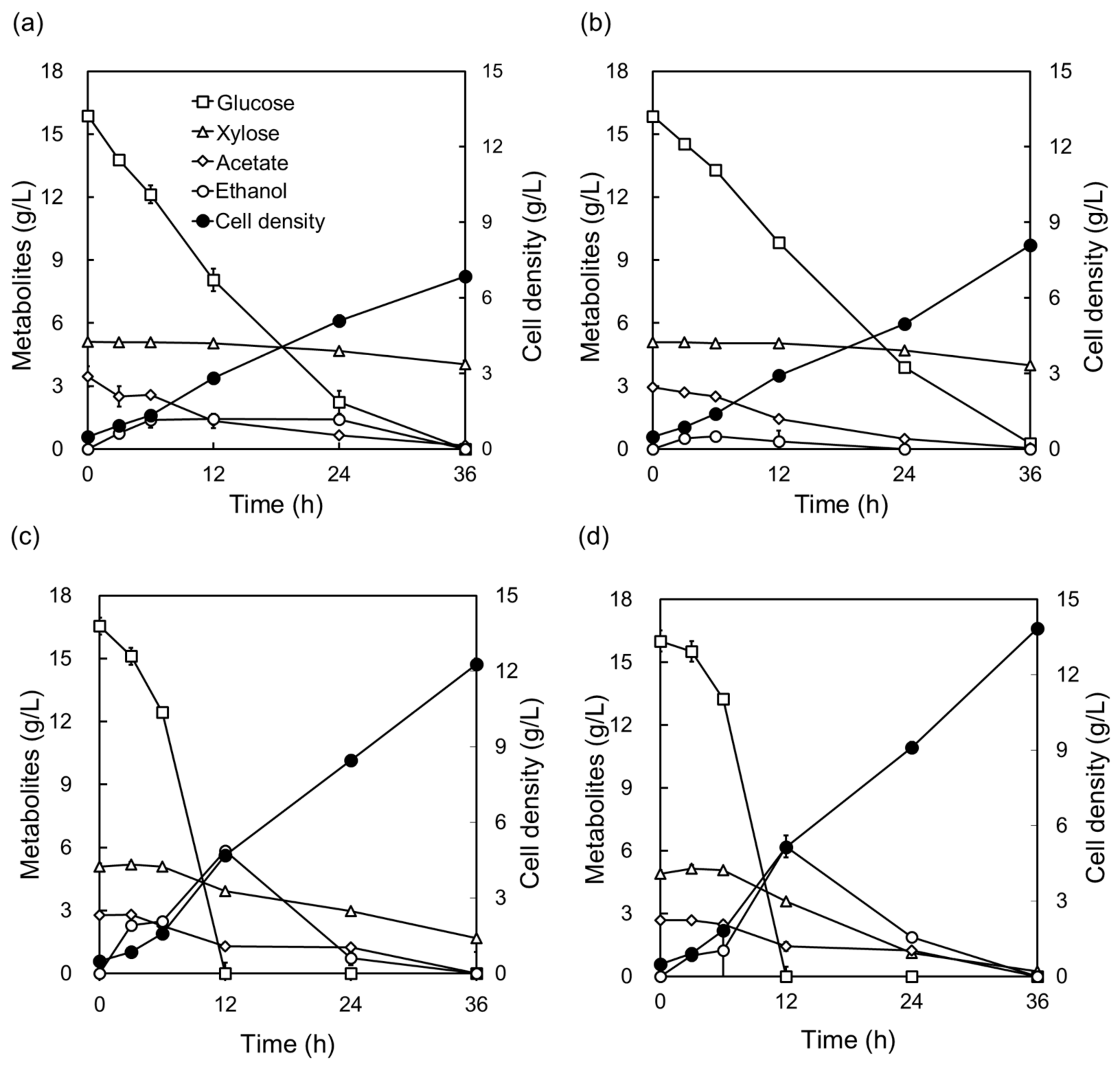
2.3. Cellulosic Hydrolysate Fermentation
3. Discussion
4. Materials and Methods
4.1. Plasmids and Strains
4.2. Culture Conditions
4.3. Genome Integration of a Heterologous Xylose-Metabolic Pathway
4.4. Construction of Promoter Library for Xylose-Metabolic Pathway
4.5. Cellulosic Biomass Hydrolysate Fermentation
4.6. High-Performance Liquid Chromatography
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ali, F.; Dawood, A.; Hussain, A.; Alnasir, M.H.; Khan, M.A.; Butt, T.M.; Janjua, N.K.; Hamid, A. Fueling the future: Biomass applications for green and sustainable energy. Discov. Sustain. 2024, 5, 156. [Google Scholar] [CrossRef]
- Ao, S.; Changmai, B.; Vanlalveni, C.; Chhandama, M.V.L.; Wheatley, A.E.; Rokhum, S.L. Biomass waste-derived catalysts for biodiesel production: Recent advances and key challenges. Renew. Energy 2024, 223, 120031. [Google Scholar] [CrossRef]
- Shen, F.; Xiong, X.; Fu, J.; Yang, J.; Qiu, M.; Qi, X.; Tsang, D.C. Recent advances in mechanochemical production of chemicals and carbon materials from sustainable biomass resources. Renew. Sustain. Energy Rev. 2020, 130, 109944. [Google Scholar] [CrossRef]
- Ma, T.-Y.; Lin, T.-H.; Hsu, T.-C.; Huang, C.-F.; Guo, G.-L.; Hwang, W.-S. An improved method of xylose utilization by recombinant Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2012, 39, 1477–1486. [Google Scholar] [CrossRef]
- Zoghlami, A.; Paës, G. Lignocellulosic biomass: Understanding recalcitrance and predicting hydrolysis. Front. Chem. 2019, 7, 874. [Google Scholar] [CrossRef]
- Sun, L.; Jin, Y.S. Xylose assimilation for the efficient production of biofuels and chemicals by engineered Saccharomyces cerevisiae. Biotechnol. J. 2021, 16, 2000142. [Google Scholar] [CrossRef]
- Park, H.; Park, S.U.; Jang, B.K.; Lee, J.J.; Chung, Y.S. Germplasm evaluation of Kenaf (Hibiscus cannabinus) for alternative biomass for cellulosic ethanol production. GCB Bioenergy 2021, 13, 201–210. [Google Scholar] [CrossRef]
- Kim, S.; Jeong, D.; Jang, B.; Park, S.; Oh, E.J.; Kim, I.J.; Kim, S.R. Coupled engineering strategy of CYB2 deletion and ACS1 overexpression improves cellulosic lactic acid production by Saccharomyces cerevisiae. Biomass Bioenergy 2024, 185, 107249. [Google Scholar] [CrossRef]
- Rathour, R.K.; Behl, M.; Dhashmana, K.; Sakhuja, D.; Ghai, H.; Sharma, N.; Meena, K.R.; Bhatt, A.K.; Bhatia, R.K. Non-food crops derived lignocellulose biorefinery for sustainable production of biomaterials, biochemicals and bioenergy: A review on trends and techniques. Ind. Crops Prod. 2023, 204, 117220. [Google Scholar] [CrossRef]
- Kaur, G.; Kaur, P.; Kaur, J.; Singla, D.; Taggar, M.S. Xylanase, xylooligosaccharide and xylitol production from lignocellulosic biomass: Exploring biovalorization of xylan from a sustainable biorefinery perspective. Ind. Crops Prod. 2024, 215, 118610. [Google Scholar] [CrossRef]
- Ju, Y.; Kim, I.J.; Kim, S.; Olawuyi, I.F.; Kim, K.M.; Kim, S.R. Deacetylation kinetics of promising energy crops, hemp and kenaf, for cellulosic ethanol production. GCB Bioenergy 2022, 14, 1150–1161. [Google Scholar] [CrossRef]
- Ahmad, M.; Hirz, M.; Pichler, H.; Schwab, H. Protein expression in Pichia pastoris: Recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biotechnol. 2014, 98, 5301–5317. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Farajnia, S.; Ghasemi, Y.; Mortazavi, M.; Zarghami, N.; Samadi, N. New developments in Pichia pastoris expression system, review and update. Curr. Pharm. Biotechnol. 2018, 19, 451–467. [Google Scholar] [CrossRef] [PubMed]
- Jach, M.E.; Serefko, A.; Ziaja, M.; Kieliszek, M. Yeast protein as an easily accessible food source. Metabolites 2022, 12, 63. [Google Scholar] [CrossRef]
- Ma, J.; Sun, Y.; Meng, D.; Zhou, Z.; Zhang, Y.; Yang, R. Yeast proteins: The novel and sustainable alternative protein in food applications. Trends Food Sci. Technol. 2023, 135, 190–201. [Google Scholar] [CrossRef]
- Looser, V.; Bruhlmann, B.; Bumbak, F.; Stenger, C.; Costa, M.; Camattari, A.; Fotiadis, D.; Kovar, K. Cultivation strategies to enhance productivity of Pichia pastoris: A review. Biotechnol. Adv. 2015, 33, 1177–1193. [Google Scholar] [CrossRef]
- Gasser, B.; Mattanovich, D. A yeast for all seasons—Is Pichia pastoris a suitable chassis organism for future bioproduction? FEMS Microbiol. Lett. 2018, 365, fny181. [Google Scholar] [CrossRef]
- Cereghino, J.L.; Cregg, J.M. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 2000, 24, 45–66. [Google Scholar] [CrossRef]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef]
- Xiao, A.; Zhou, X.; Zhou, L.; Zhang, Y. Improvement of cell viability and hirudin production by ascorbic acid in Pichia pastoris fermentation. Appl. Microbiol. Biotechnol. 2006, 72, 837–844. [Google Scholar] [CrossRef]
- Jordà, J.; Suarez, C.; Carnicer, M.; ten Pierick, A.; Heijnen, J.J.; van Gulik, W.; Ferrer, P.; Albiol, J.; Wahl, A. Glucose-methanol co-utilization in Pichia pastoris studied by metabolomics and instationary 13 C flux analysis. BMC Syst. Biol. 2013, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Peña, D.A.; Gasser, B.; Zanghellini, J.; Steiger, M.G.; Mattanovich, D. Metabolic engineering of Pichia pastoris. Metab. Eng. 2018, 50, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Heyland, J.; Fu, J.; Blank, L.M.; Schmid, A. Quantitative physiology of Pichia pastoris during glucose-limited high-cell density fed-batch cultivation for recombinant protein production. Biotechnol. Bioeng. 2010, 107, 357–368. [Google Scholar] [CrossRef]
- Zhao, Z.; Xian, M.; Liu, M.; Zhao, G. Biochemical routes for uptake and conversion of xylose by microorganisms. Biotechnol. Biofuels 2020, 13, 21. [Google Scholar] [CrossRef]
- Inan, M.; Meagher, M.M. Non-repressing carbon sources for alcohol oxidase (AOX1) promoter of Pichia pastoris. J. Biosci. Bioeng. 2001, 92, 585–589. [Google Scholar] [CrossRef]
- Li, P.; Sun, H.; Chen, Z.; Li, Y.; Zhu, T. Construction of efficient xylose utilizing Pichia pastoris for industrial enzyme production. Microb. Cell Factories 2015, 14, 22. [Google Scholar] [CrossRef]
- Kawaguchi, H.; Vertès, A.A.; Okino, S.; Inui, M.; Yukawa, H. Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2006, 72, 3418–3428. [Google Scholar] [CrossRef]
- Zhang, M.; Eddy, C.; Deanda, K.; Finkelstein, M.; Picataggio, S. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 1995, 267, 240–243. [Google Scholar] [CrossRef]
- Ledesma-Amaro, R.; Lazar, Z.; Rakicka, M.; Guo, Z.; Fouchard, F.; Crutz-Le Coq, A.-M.; Nicaud, J.-M. Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab. Eng. 2016, 38, 115–124. [Google Scholar] [CrossRef]
- Kwak, S.; Jin, Y.-S. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: A review and perspective. Microb. Cell Factories 2017, 16, 82. [Google Scholar] [CrossRef]
- Subudhi, S.; Mudgil, D.; Saha, K.; kumar Sarangi, P.; Pal, P. Yeast as a cell factory for fermentative production of ethanol from xylose. J. Taiwan Inst. Chem. Eng. 2024, 105616. [Google Scholar] [CrossRef]
- Bankefa, O.E.; Samuel-Osamoka, F.C.; Oladeji, S.J. Improved enzyme production on corncob hydrolysate by a xylose-evolved Pichia pastoris cell factory. J. Food Sci. Technol. 2022, 59, 1280–1287. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.; Oh, E.J.; Ko, J.K.; Nam, J.-O.; Park, H.-S.; Jin, Y.-S.; Lee, E.J.; Kim, S.R. Metabolic engineering considerations for the heterologous expression of xylose-catabolic pathways in Saccharomyces cerevisiae. PLoS ONE 2020, 15, e0236294. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Park, Y.-C.; Jin, Y.-S.; Seo, J.-H. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol. Adv. 2013, 31, 851–861. [Google Scholar] [CrossRef]
- Gao, S.; Zhou, H.; Zhou, J.; Chen, J. Promoter-library-based pathway optimization for efficient (2S)-naringenin production from p-coumaric acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 2020, 68, 6884–6891. [Google Scholar] [CrossRef]
- Meinander, N.Q.; Hahn-Hägerdal, B. Fed-batch xylitol production with two recombinant Saccharomyces cerevisiae strains expressing XYL1 at different levels, using glucose as a cosubstrate: A comparison of production parameters and strain stability. Biotechnol. Bioeng. 1997, 54, 391–399. [Google Scholar] [CrossRef]
- Snoep, J.L.; Yomano, L.P.; Westerhoff, H.V.; Ingram, L.O. Protein burden in Zymomonas mobilis: Negative flux and growth control due to overproduction of glycolytic enzymes. Microbiology 1995, 141, 2329–2337. [Google Scholar] [CrossRef]
- Eliasson, A.; Christensson, C.; Wahlbom, C.F.; Hahn-Hägerdal, B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 2000, 66, 3381–3386. [Google Scholar] [CrossRef]
- Kötter, P.; Ciriacy, M. Xylose fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1993, 38, 776–783. [Google Scholar] [CrossRef]
- Kim, S.R.; Kwee, N.R.; Kim, H.; Jin, Y.-S. Feasibility of xylose fermentation by engineered Saccharomyces cerevisiae overexpressing endogenous aldose reductase (GRE3), xylitol dehydrogenase (XYL2), and xylulokinase (XYL3) from Scheffersomyces stipitis. FEMS Yeast Res. 2013, 13, 312–321. [Google Scholar] [CrossRef]
- Vellanki, R.N.; Potumarthi, R.; Doddapaneni, K.K.; Anubrolu, N.; Mangamoori, L.N. Constitutive Optimized Production of Streptokinase in Saccharomyces cerevisiae Utilizing Glyceraldehyde 3-Phosphate Dehydrogenase Promoter of Pichia pastoris. BioMed Res. Int. 2013, 2013, 268249. [Google Scholar] [CrossRef] [PubMed]
- Çalık, P.; Ata, Ö.; Güneş, H.; Massahi, A.; Boy, E.; Keskin, A.; Öztürk, S.; Zerze, G.H.; Özdamar, T.H. Recombinant protein production in Pichia pastoris under glyceraldehyde-3-phosphate dehydrogenase promoter: From carbon source metabolism to bioreactor operation parameters. Biochem. Eng. J. 2015, 95, 20–36. [Google Scholar] [CrossRef]
- Gupta, A.; Rangarajan, P.N. Histidine is essential for growth of Komagataella phaffii cultured in YPA medium. FEBS Open Bio 2022, 12, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.S.M.; Luthfi, A.A.I.; Low, K.O.; Harun, S.; Manaf, S.F.A.; Illias, R.M.; Jahim, J.M. Preparation of kenaf stem hemicellulosic hydrolysate and its fermentability in microbial production of xylitol by Escherichia coli BL21. Sci. Rep. 2019, 9, 4080. [Google Scholar] [CrossRef]
- Lee, Y.-G.; Jin, Y.-S.; Cha, Y.-L.; Seo, J.-H. Bioethanol production from cellulosic hydrolysates by engineered industrial Saccharomyces cerevisiae. Bioresour. Technol. 2017, 228, 355–361. [Google Scholar] [CrossRef]
- Ünver, H.; Polat, E.; Altınbaş, M. Screening the Lipid Production Potential of Oleaginous Yeast Yarrowia lipolytica under Wood Hydrolysates. Separations 2023, 10, 371. [Google Scholar] [CrossRef]
- Bettiga, M.; Hahn-Hägerdal, B.; Gorwa-Grauslund, M.F. Comparing the xylose reductase/xylitol dehydrogenase and xylose isomerase pathways in arabinose and xylose fermenting Saccharomyces cerevisiae strains. Biotechnol. Biofuels 2008, 1, 16. [Google Scholar] [CrossRef]
- Li, X.; Park, A.; Estrela, R.; Kim, S.-R.; Jin, Y.-S.; Cate, J.H. Comparison of xylose fermentation by two high-performance engineered strains of Saccharomyces cerevisiae. Biotechnol. Rep. 2016, 9, 53–56. [Google Scholar] [CrossRef]
- Eliasson, A.; Hofmeyr, J.-H.S.; Pedler, S.; Hahn-Hägerdal, B. The xylose reductase/xylitol dehydrogenase/xylulokinase ratio affects product formation in recombinant xylose-utilising Saccharomyces cerevisiae. Enzym. Microb. Technol. 2001, 29, 288–297. [Google Scholar] [CrossRef]
- Jin, Y.-S.; Alper, H.; Yang, Y.-T.; Stephanopoulos, G. Improvement of xylose uptake and ethanol production in recombinant Saccharomyces cerevisiae through an inverse metabolic engineering approach. Appl. Environ. Microbiol. 2005, 71, 8249–8256. [Google Scholar] [CrossRef]
- Nambu-Nishida, Y.; Sakihama, Y.; Ishii, J.; Hasunuma, T.; Kondo, A. Selection of yeast Saccharomyces cerevisiae promoters available for xylose cultivation and fermentation. J. Biosci. Bioeng. 2018, 125, 76–86. [Google Scholar] [CrossRef]
- Lou, J.; Wang, J.; Yang, Y.; Yang, Q.; Li, R.; Hu, M.; He, Q.; Du, J.; Wang, X.; Li, M.; et al. Development and characterization of efficient xylose utilization strains of Zymomonas mobilis. Biotechnol. Biofuels 2021, 14, 231. [Google Scholar] [CrossRef] [PubMed]
- Ata, Ö.; Rebnegger, C.; Tatto, N.E.; Valli, M.; Mairinger, T.; Hann, S.; Steiger, M.G.; Çalık, P.; Mattanovich, D. A single Gal4-like transcription factor activates the Crabtree effect in Komagataella phaffii. Nat. Commun. 2018, 9, 4911. [Google Scholar] [CrossRef] [PubMed]
- Karaoğlan, M.; Erden-Karaoğlan, F.; Yılmaz, S.; İnan, M. Identification of major ADH genes in ethanol metabolism of Pichia pastoris. Yeast 2020, 37, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Siripong, W.; Wolf, P.; Kusumoputri, T.P.; Downes, J.J.; Kocharin, K.; Tanapongpipat, S.; Runguphan, W. Metabolic engineering of Pichia pastoris for production of isobutanol and isobutyl acetate. Biotechnol. Biofuels 2018, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Inan, M.; Meagher, M.M. The effect of ethanol and acetate on protein expression in Pichia pastoris. J. Biosci. Bioeng. 2001, 92, 337–341. [Google Scholar] [CrossRef]
- Zepeda, A.B.; Pessoa, A., Jr.; Farías, J.G. Carbon metabolism influenced for promoters and temperature used in the heterologous protein production using Pichia pastoris yeast. Braz. J. Microbiol. 2018, 49, 119–127. [Google Scholar] [CrossRef]
- Cavka, A.; Jo, L.J. Comparison of the growth of filamentous fungi and yeasts in lignocellulose-derived media. Biocatal. Agric. Biotechnol. 2014, 3, 197–204. [Google Scholar] [CrossRef]
- Rebnegger, C.; Graf, A.B.; Valli, M.; Steiger, M.G.; Gasser, B.; Maurer, M.; Mattanovich, D. In Pichia pastoris, growth rate regulates protein synthesis and secretion, mating and stress response. Biotechnol. J. 2014, 9, 511–525. [Google Scholar] [CrossRef]
- Gassler, T.; Heistinger, L.; Mattanovich, D.; Gasser, B.; Prielhofer, R. CRISPR/Cas9-mediated homology-directed genome editing in Pichia pastoris. In Recombinant Protein Production in Yeast; Humana Press: New York, NY, USA, 2019; pp. 211–225. [Google Scholar]
- Liu, Q.; Shi, X.; Song, L.; Liu, H.; Zhou, X.; Wang, Q.; Zhang, Y.; Cai, M. CRISPR–Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb. Cell Factories 2019, 18, 144. [Google Scholar] [CrossRef]
- Gasser, B.; Prielhofer, R.; Marx, H.; Maurer, M.; Nocon, J.; Steiger, M.; Puxbaum, V.; Sauer, M.; Mattanovich, D. Pichia pastoris: Protein production host and model organism for biomedical research. Future Microbiol. 2013, 8, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wen, A.; Shen, B.; Lu, J.; Huang, Y.; Chang, Y. FastCloning: A highly simplified, purification-free, sequence-and ligation-independent PCR cloning method. BMC Biotechnol. 2011, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Park, B.-R.; Jeong, D.; Park, J.; Ko, J.K.; Kim, S.-J.; Kim, J.-S.; Jin, Y.-S.; Kim, S.R. Functional expression of RuBisCO reduces CO2 emission during fermentation by engineered Saccharomyces cerevisiae. Process Biochem. 2023, 134, 286–293. [Google Scholar] [CrossRef]
- Stadlmayr, G.; Mecklenbräuker, A.; Rothmüller, M.; Maurer, M.; Sauer, M.; Mattanovich, D.; Gasser, B. Identification and characterisation of novel Pichia pastoris promoters for heterologous protein production. J. Biotechnol. 2010, 150, 519–529. [Google Scholar] [CrossRef]
- Kim, S.R.; Ha, S.-J.; Kong, I.I.; Jin, Y.-S. High expression of XYL2 coding for xylitol dehydrogenase is necessary for efficient xylose fermentation by engineered Saccharomyces cerevisiae. Metab. Eng. 2012, 14, 336–343. [Google Scholar] [CrossRef]
| Strains and Plasmids | Features 1 | Reference |
|---|---|---|
| GS115 | Komagataella phaffii his4 | InvitrogenTM |
| G-XYL-strong | GS115 GAPDHp_XYL1_ICL1t_ENO1p_XYL2_DAS1t_PET9p_XYL3_CYC1t | This study |
| G-control | GS115 pBB3cH | This study |
| G-XYL-opt | GS115 pBB3cH-XYL-opt | This study |
| X-33 | Komagataella phaffii, prototrophic | InvitrogenTM |
| X-control | X-33 pBB3cH | This study |
| X-XYL-opt | X-33 pBB3cH-XYL-opt | This study |
| pBB3cH | Low-copy plasmid with CEN6/ARS4 and hph (constructed by removing Cas9 from pBB3cH-Cas9) | This study |
| pBB3cH-Cas9 | pBB3cH harboring Cas9 and sgRNA | [49] |
| pBB3cH-Cas9-FLD1UP | pBB3cH-Cas9 with sgRNA targeting the upstream of the FLD1 gene | This study |
| pBB3cH-Library | pBB3cH harboring a xylose pathway with promoter library; promoter1-XYL1-ICL1t-promoter2-XYL2-DAS1t-promoter3-XYL3-CYC1t | This study |
| pBB3cH-XYL-opt | pBB3cH-RSP2p-XYL1-ICL1t-ENO1p-XYL2-DAS1t-TKL1p-XYL3-CYC1t | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.; Park, S.; Evelina, G.; Kim, S.; Jin, Y.-S.; Chi, W.-J.; Kim, I.J.; Kim, S.R. Metabolic Engineering of Komagataella phaffii for Xylose Utilization from Cellulosic Biomass. Molecules 2024, 29, 5695. https://doi.org/10.3390/molecules29235695
Park J, Park S, Evelina G, Kim S, Jin Y-S, Chi W-J, Kim IJ, Kim SR. Metabolic Engineering of Komagataella phaffii for Xylose Utilization from Cellulosic Biomass. Molecules. 2024; 29(23):5695. https://doi.org/10.3390/molecules29235695
Chicago/Turabian StylePark, Jongbeom, Sujeong Park, Grace Evelina, Sunghee Kim, Yong-Su Jin, Won-Jae Chi, In Jung Kim, and Soo Rin Kim. 2024. "Metabolic Engineering of Komagataella phaffii for Xylose Utilization from Cellulosic Biomass" Molecules 29, no. 23: 5695. https://doi.org/10.3390/molecules29235695
APA StylePark, J., Park, S., Evelina, G., Kim, S., Jin, Y.-S., Chi, W.-J., Kim, I. J., & Kim, S. R. (2024). Metabolic Engineering of Komagataella phaffii for Xylose Utilization from Cellulosic Biomass. Molecules, 29(23), 5695. https://doi.org/10.3390/molecules29235695







