Abstract
Acute liver injury (ALI) is a significant global public health issue that can rapidly develop into acute liver failure, seriously endangering the safety of patients. Eugenol has various pharmacological effects such as antioxidant, anti-inflammatory, antibacterial, and neuroprotective properties. Through pathological section observation, eugenol can alleviate the degree of liver damage caused by thioacetamide. Based on metabolomics, a total of 87 metabolites were found to have differences in content between the control group and the thioacetamide group. Compared with the control group, the contents of 42 metabolites had increased and 45 metabolites had decreased in the thioacetamide group. These differential expressed metabolites mainly indicate inflammatory damage, oxidative damage, and abnormal energy metabolism. There were 269 metabolites with differences in content between the eugenol intervention group and the thioacetamide group. Compared with the thioacetamide group, there were 101 metabolites with increased content and 168 metabolites with decreased content in the eugenol intervention group. These differential expressed metabolites suggest that eugenol intervention can correct inflammation damage, oxidative damage, and energy metabolism abnormalities caused by TAA. This study found through pathological section observation and metabolomics that eugenol has a protective effect on acute liver injury caused by thioacetamide, and the protective mechanism may be related to the antioxidant and anti-inflammatory effects of eugenol.
1. Introduction
Liver disease impacts millions globally and stands as a major contributor to morbidity and mortality worldwide [1]. In China, liver diseases, predominantly drug-induced liver injury, nonalcoholic fatty liver disease, and alcoholic liver disease, afflict approximately 300 million individuals [2]. Acute liver injury denotes the abrupt harm or necrosis of liver cells resulting from a variety of factors over a brief period. It is characterized by its swift onset and diverse etiologies. Without timely intervention, the condition can progressively worsen, potentially culminating in acute liver failure at a later stage [3].
Eugenol is a simple phenolic compound characterized by the chemical formula C10H12O2. From a pharmacological standpoint, eugenol exhibits a range of beneficial activities, including antioxidant, anti-inflammatory, antibacterial, anesthetic, neuroprotective, antidiabetic, and anticancer properties [4,5,6,7]. Studies have demonstrated that eugenol possesses the ability to combat oxidation and neutralize reactive oxygen species (ROS). Additionally, it exerts a dose-dependent inhibitory effect on both DPPH and hydroxyl radicals [5]. It plays a significant role in delaying aging, enhancing skin health, and providing neuroprotection. Furthermore, the antioxidant properties of eugenol can also neutralize nitric oxide [6], thereby preventing the formation of reactive oxygen species (ROS) both in vivo and in vitro. It safeguards against lipid peroxidation and DNA damage induced by ROS and augments the antioxidant activity of the cellular glutathione system [8,9,10].
According to the literature, eugenol exerts its anti-inflammatory effects by inhibiting prostaglandin E (PGE) synthesis and the chemotaxis of neutrophils or macrophages. Kim et al. found in animal experiments that, at a drug dose of 160 mg/kg, alveolar collapse and multinucleated cell infiltration in experimental animal models were significantly reduced, indicating that eugenol can reduce TNF-α during lung infection and prevent neutrophil infiltration, thereby weakening the inflammatory response [11]. Additionally, eugenol can also cause dysfunction of macrophages, which balances the anti-inflammatory mediators of macrophages in the peritoneum of mice [12]. In summary, eugenol possesses pharmacological activities of anti-inflammatory and antioxidant properties.
Metabolomics is a branch of biological sciences that has emerged subsequent to genomics and proteomics. It focuses on the study of small molecule metabolites with a molecular weight below 1000 within the body, aiming to mirror the body’s condition. This field has found extensive application in pharmacological and toxicological research [13,14,15].
This study aims to employ thioacetamide in the creation of an acute liver injury model and to conduct a preliminary investigation into the protective mechanism of eugenol against acute liver injury through metabolomics, thus laying the groundwork for future clinical treatments.
2. Results
2.1. HE Staining Results
Through HE staining (as shown in Figure 1), it can be found that TAA has successfully induced acute liver injury and, after intervention with eugenol, the symptoms of acute liver injury have been alleviated.
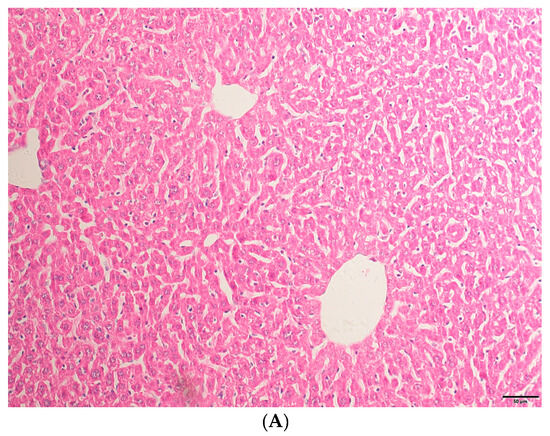
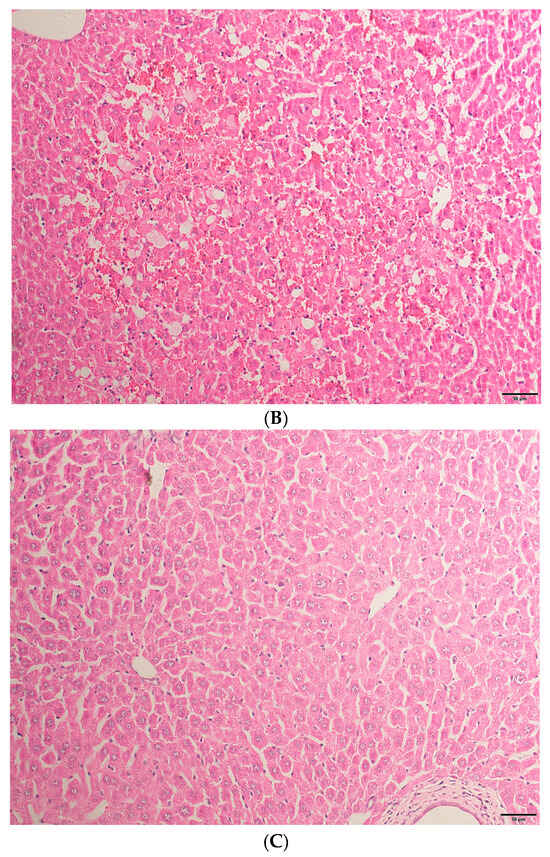
Figure 1.
(A) The liver tissue structure of the NC group was basically normal. (B) The TAA group had disordered liver lobular structure and hepatic sinusoids congestion, accompanied by inflammatory infiltration and lipid droplet vacuolar formation. (C) The liver tissue structure in the eugenol treatment group was significantly improved compared to the TAA group (200× magnifications, bar = 50 µm).
2.2. ALT, AST, and SOD Results
As depicted in Figure 2, in comparison to the control group, the TAA group exhibited a marked elevation in ALT and AST levels. However, subsequent to the intervention with eugenol, there was a significant reduction in both ALT and AST levels. Additionally, the SOD level in the TAA group was notably diminished relative to the control group but, after the eugenol intervention, the SOD level was significantly enhanced compared to the TAA group.
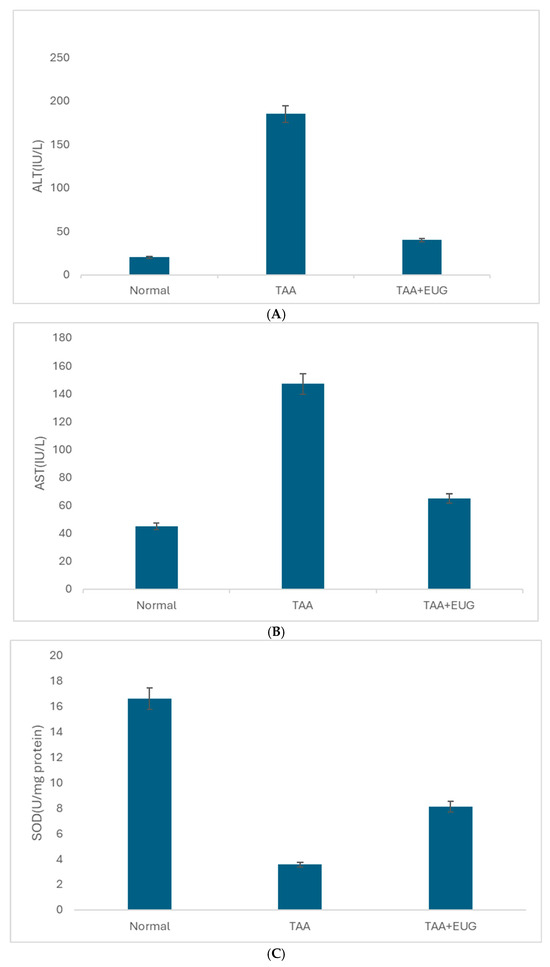
Figure 2.
(A) The expression levels of ALT in the control, TAA, and EUG intervention groups; (B) AST in the control, TAA, and EUG intervention groups; (C) SOD in the control, TAA, and EUG intervention groups.
2.3. Metabolomics Quality Control Results
The correlation analysis of samples can assess the biological replicability within and between groups. A higher correlation coefficient between samples within the same group and those from different groups indicates a more reliable set of differential metabolites. Spearman Rank Correlation r is used as the metric to evaluate the correlation of biological replicates. The closer r is to 1, the stronger the correlation between the two repeated samples as shown in Figure 3.
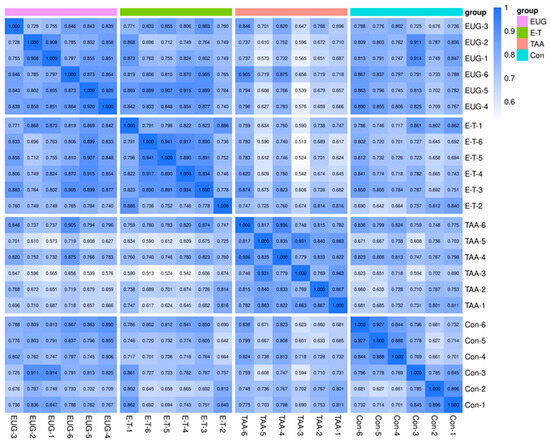
Figure 3.
Sperman Rank Correlation across all samples. Note: the horizontal and vertical co-ordinates represent the sample name, the color depth represents the magnitude of the correlation coefficient r, and Group represents the grouping.
2.4. PCA and OPLS-DA Results
The PCA distribution results among the various groups are depicted in Figure 4. In the graph, the horizontal axis corresponds to PC1, while the vertical axis corresponds to PC2, representing the scores of the first and second principal components, respectively. The scatter points of varying colors signify samples from distinct experimental groups, and the ellipses denote the 95% confidence intervals. The outcomes indicate that the data separation among the different groups is ideal.
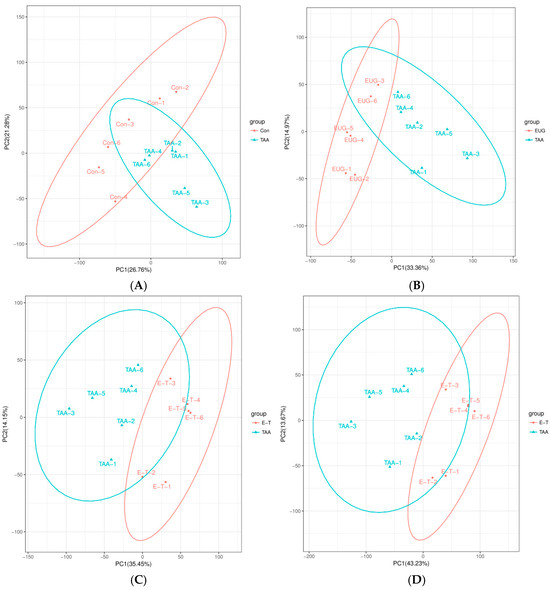
Figure 4.
(A,B) are the PCA plots of TAA compared to the control group, where (A) represents in negative ion monitoring mode and (B) represents in positive ion monitoring mode; (C,D) is the PCA plot of the eugenol intervention group compared to the TAA group, where (C) represents in negative ion monitoring mode and (D) represents in positive ion monitoring mode.
Orthogonal Partial Least Squares (OPLS) distinguishes between modeling and response-related and orthogonal predictor variables based on PLS. The evaluation of Q2Y indicators and permutation testing is crucial for avoiding overfitting and assessing the statistical significance of the model. In general, a Q2Y value exceeding 0.5 indicates an effective model as shown in Figure 5.
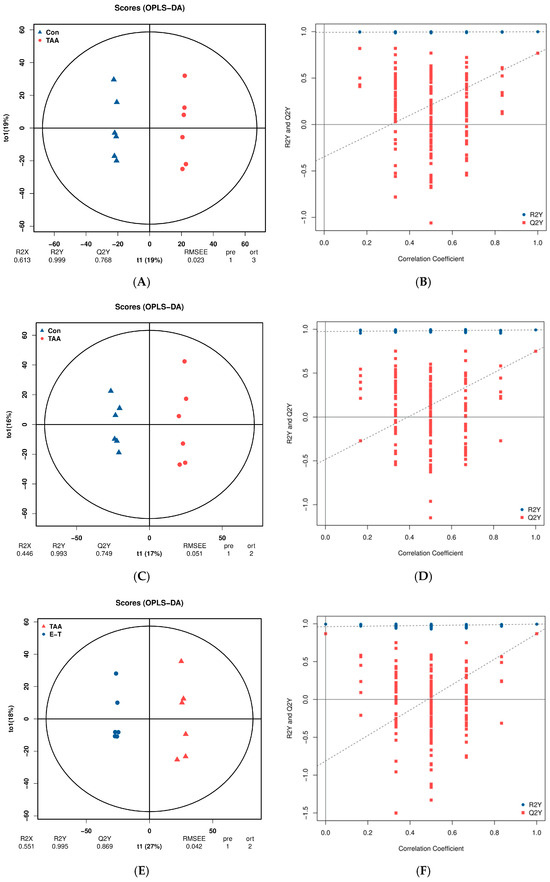
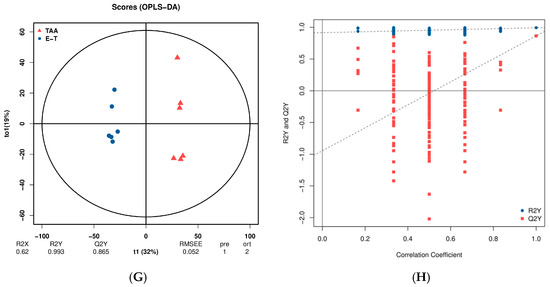
Figure 5.
(A,C) represents OPLS-DA plot between the TAA group and the control groups; (B,D) represents OPLS-DA permutation between the TAA group and the control groups; (E,G) represents OPLS-DA plot between the TAA group and the eugenol treatment group; (F,H) represent OPLS-DA permutation between the TAA group and the eugenol intervention group; (A,B,E,F) represents in negative ion monitoring mode; (C,D,F,H) represents in positive ion monitoring mode.
2.5. Differential Expressed Metabolites
The cluster heatmap analysis is depicted in Figure 6. It is evident that there exists a distinct differentiation in metabolic patterns between the TAA group and the control group. Additionally, the metabolic patterns of the eugenol intervention group are also distinctly differentiated from those of the TAA group.
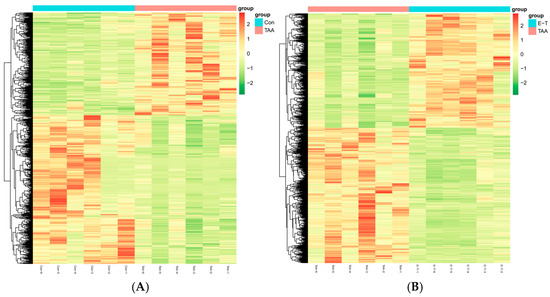
Figure 6.
Cluster heatmap (A) represents the TAA group and control group; (B) represents the clustering diagram between TAA and eugenol intervention groups.
Compared with the control group, the differentially expressed metabolites in the TAA group are detailed in Supplementary Table S1. Utilizing metabolomics, a total of 87 metabolites were identified with significant differences in content between the control group and the thioacetamide group. Specifically, 42 metabolites exhibited increased content, while 45 metabolites showed decreased content in the thioacetamide group. Our focus is on the differentially expressed metabolites enriched by KEGG, as presented in Table 1.

Table 1.
Differential expressed metabolites between the TAA group and control group.
After intervention with eugenol, the differential metabolites between the eugenol treatment group and the thioacetamide group are presented in Supplementary Table S2. A total of 269 metabolites exhibited differences in content between the eugenol treatment group and the thioacetamide group. Compared to the thioacetamide group, the eugenol treatment group had 101 metabolites with increased content and 168 metabolites with decreased content. The significant differential metabolites of interest in this study are detailed in Table 2.

Table 2.
Differential expressed metabolites between the E + T group and TAA group.
2.6. Enrichment of KEGG Pathway
We enriched the KEGG metabolic pathway with significantly different metabolites and discovered that, as illustrated in Figure 7, the differential metabolites in the TAA groups, compared to the control group, were predominantly concentrated in energy metabolism pathways. These included pyrimidine metabolism, purine metabolism, lipid metabolism, steroid hormone biosynthesis, glycerophospholipid metabolism, ferroptosis in terms of oxidation–reduction, biosynthesis of unsaturated fatty acids, glutathione metabolism, arachidonic acid metabolism, and primary bile acid biosynthesis, which are related to the liver and gallbladder.
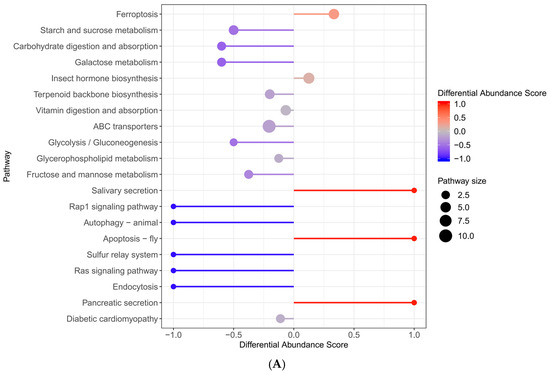
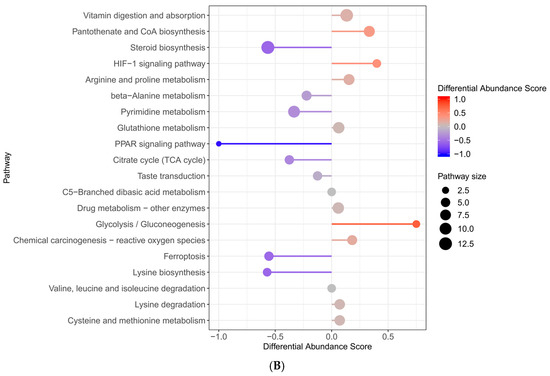
Figure 7.
KEGG enrichment plot ((A) represents TAA vs. control; (B) represents E + T vs. TAA).
The distinct metabolites identified between the E + T and TAA groups were primarily associated with vitamin absorption and degradation, as well as steroid hormone biosynthesis. Regarding energy metabolism, the focus was on pyrimidine and purine metabolism. For oxidation–reduction processes, the metabolites included glutathione metabolism and unsaturated fatty acids. In terms of inflammation, arachidonic acid was a key metabolite of interest.
To assist readers in better comprehending the therapeutic effect of eugenol, Table 3 illustrates that, following eugenol intervention, the metabolite levels which underwent alterations in the TAA group exhibited a contrasting pattern of change compared to the control group.

Table 3.
The differential expressed metabolites between TAA vs. control and E + T vs. T.
3. Discussion
Acute liver injury is a common liver disease in clinical practice, often accompanied by elevated serum transaminase and bilirubin levels [16]. The liver, as the most important metabolic organ in the human body, has functions such as detoxification, coagulation, immunity, and bile secretion. It is also susceptible to stimulation by toxic chemicals, drugs, or ethanol, which can cause acute or chronic liver damage, leading to a series of metabolic diseases and even death. The severity of the condition can range from asymptomatic drug-induced elevation of liver enzymes to liver fibrosis, cirrhosis, liver cancer, and even liver failure, seriously endangering human health [17]. Currently, in clinical practice, symptomatic treatment is carried out using chemically synthesized nucleoside analogues [18], vitamin-based prophylaxis, and hepatoprotective drugs [19]. These methods can control or improve symptoms to a certain extent, but there are limitations such as insignificant therapeutic effects and side effects such as myopathy and lactic acidosis. Therefore, it is of great significance to search for novel, safe, and effective liver injury protective agents. Thioacetamide is a commonly used modeling drug for acute liver injury [20,21]. Studies have found that administering TAA to rats can cause liver damage, including inflammation, oxidative stress, and the production of profibrotic markers [22,23,24].
Metabolomics can effectively identify and quantify the levels of metabolites, often used to elucidate the pathogenesis of diseases and the mechanisms of action of drugs [25]. Through comprehensive analysis of biological samples (such as blood, urine, liver, etc.), the changes in metabolites within the body are evaluated to reflect the physiological and pathological states of the organism. This technology has been widely applied in disease diagnosis, mechanism research, and drug development [26,27]. Utilizing metabolomics, it is observed that TAA induces inflammation and oxidative stress, evidenced by a significant elevation in metabolites associated with the arachidonic acid pathway compared to the control group; metabolites linked to oxidative stress, such as an increase in oxidized glutathione, are also noted, while retinol levels decline. Additionally, there is an increase in metabolic products from the primary bile acid biosynthesis pathway and steroid hormone biosynthesis. These findings align with previously reported liver toxicity of TAA [28,29].
In the TAA group, the levels of certain substances associated with oxidation–reduction processes were observed to decrease, including alpha tocotrienol [30], retinol [31], ascorbic acid [31], and D-Octopine [32]. Conversely, the content of oxidized glutathione, 4-hydroxy-L-trhreonine, and L-4-hydroxyglutamate semialdehyde exhibited an increase, indicating oxidative damage within the TAA group. Additionally, the concentrations of inflammation-related substances such as prostaglandin D2, (15S)-15-Hydroxy-5,8,11-cis-13-trans-eicosatenoate, 20-COOH-Leukotriene B4, and 12-Keto-leukotriene B4 were found to be elevated.
Substances associated with primary bile acid biosynthesis, including 3alpha, 7alpha, 12alpha, 26-Tetrahydroxy-5beta-cholestane and 3alpha, 7alpha, 12alpha-Trihydroxy-5beta-cholestane, also experienced an increase. Conversely, the levels of unsaturated acids such as Docosenoyl CoA and Montanoyl CoA diminished. Additionally, substances linked to steroid hormone biosynthesis, such as 7alpha-Hydroxydehydroepiandrosterone and Cortolone, exhibited an increase in the TAA group.
Certain small-molecule substances associated with energy metabolism, including deoxyadenosine monophosphate, GTP, stachyose, and L-rhamnulose, were also found to be significantly different compared to the control group. Furthermore, metabolomic analysis revealed that TAA-induced liver injury is associated with ferroptosis.
Some studies have reported the anti-inflammatory properties of eugenol [6,7,33]. Through the intervention of eugenol, it was found that the content of metabolites related to arachidonic acid decreased, which is consistent with the results observed in pathological sections. Compared with the TAA group, eugenol can elevate the SOD level. Some substances that can resist oxidative stress increase, such as alpha tocotrienol, retinol, ascorbic acid, and D-octopine, which is also consistent with the antioxidant properties of eugenol [4,5,34]. The intervention of eugenol also corrected the primary bile acid biosynthesis, steroid hormone biosynthesis, and abnormal metabolism of fatty acids and energy metabolism caused by TAA. And it suggests that the protective mechanism of eugenol against acute liver injury may be mediated through the ferroptosis pathway.
4. Materials and Methods
4.1. Chemicals and Reagents
Eugenol (analytical standard, HPLC ≥ 98%) and thioacetamide reagent were purchased from Shanghai Yuanye Co., Ltd. (Shanghai, China), while formic acid, methanol, and acetonitrile were purchased from Thermo Fisher (Waltham, MA, USA). DMSO, PEG300, and Tween 80 were purchased from China National Pharmaceutical Group Chemical Reagent Co., Ltd. in Shenyang, China. The SOD assay kit, ALT, and AST assay kits were purchased from Beyotime Biotechnology Co., Ltd. (Wuhan, China).
4.2. Animals Grouping
(a) Preparation method of eugenol suspension: an adequate amount of eugenol was dissolved in DMSO to formulate a solution with a concentration of 150 mg/mL; subsequently, It was diluted to a concentration of 15 mg/mL using PEG300, Tween 80, and normal saline in a volume ratio of 8:1:9. A total of 0.1 mL of eugenol solution (15 mg/mL) was administered to mice at a dose of 150 mg/kg per 10 g body weight by gavage. The dosage of eugenol was selected based on literature reports [20,21]. The eugenol was administered by gavage two weeks before the administration of thioacetamide for a total of two weeks.
(b) Thioacetamide injection method: 0.1 mL of thioacetamide solution (10 mg/mL) was given by intraperitoneal injection to every 10 g body weight mice, which is a dose of 100 mg/kg per mouse [22,23]. Thioacetamide was administered intraperitoneally on the 14th day of eugenol gavage, with a total of one intraperitoneal injection. The experimental animals used in this study were all SPF-grade wild-type C57BL/6J mice (male, 10–12 weeks old, weighing 25 ± 2 g, at a total of 24, purchased from Beijing Huafukang Biotechnology Co., Ltd., Beijing, China). All experimental animals were housed in the Barrier Laboratory of the Experimental Animal Department of China Medical University, with 4–5 animals per cage. The ambient temperature was maintained at 22 ± 1 °C, and the circadian rhythm was maintained for 12 h. They were free to drink and eat. After one week of adaptive feeding, mice were randomly divided into four groups, with six mice in each group. According to the experimental protocol, they were divided into a control group, a thioacetamide model group (TAA group), and a eugenol intervention group (E + T). All experimental procedures comply with the “Regulations on the Management of Experimental Animals” issued by the National Science and Technology Commission of the People’s Republic of China and follow the “Guidelines for the Operation of Experimental Animals at China Medical University” issued by the Experimental Animal Center of China Medical University. All experiments have been approved by the Animal Ethics Committee of China Medical University (No. KT2022353). After intraperitoneal injection of thioacetamide for 24 h, all mice were humanely euthanized using compressed gas within their home cages by trained personnel. Subsequently, liver tissue and serum were collected for further testing.
4.3. Metabolomics
4.3.1. Metabolites Extraction
A total of 50 mg of liver tissue sample was weighed, 1000 μL of extraction solution containing internal standard (2-chlorophenylalanine) (methanol/acetonitrile/water volume ratio = 2:2:1, internal standard concentration 20 mg/L) was added, vortexed, and mixed for 30 s; steel balls were added and it was treated with a 45 Hz grinder for 10 min and sonicated for 10 min (in an ice water bath); it was allowed to stand at minus 20 °C for 1 h; the sample was centrifuged at 4 °C and 12,000 rpm for 15 min; 500 μL of supernatant was carefully removed into an EP tube; the extract was dried in a vacuum concentrator; 160 μL of extraction solution (acetonitrile/water volume ratio = 1:1) was added to the dried metabolites for reconstitution; it was vortexed for 30 s and placed in an ice water bath ultrasound for 10 min; the sample was centrifuged at 4 °C and 12,000 rpm for 15 min; 120 μL of supernatant was carefully taken out and injected into a 2 mL vial. A total of 10 μL of each sample were taken and mixed to form a QC sample for machine testing. The injection volume was 1 μL.
4.3.2. LC-MS/MS Analysis, Data Preprocessing
The LC/MS system employed for metabolomics is identical to that in previous studies [35]. The raw data obtained through MassLynx V4.2 are analyzed using Progenesis QI software (Version 2.0) for tasks such as peak extraction, alignment, and other processing activities. This analysis relies on the METLIN database available online via Progenesis QI for identification purposes. Additionally, both theoretical fragment identification and mass deviation are maintained within a threshold of 100 ppm.
4.3.3. Data Analysis
Following the normalization of the original peak area data against the total peak area, subsequent analyses were conducted, including principal component analysis and Spearman correlation analysis. The identified compounds were examined for classification and pathway information using the KEGG, HMDB, and LipidMaps databases. Based on the classification data, we calculated and compared the difference multiples. A t-test was employed to assess the statistical significance (p-value) of each compound’s differences. Additionally, the R language package (Version 3.5.3) ropls was utilized to conduct OPLS-DA analysis, and 200 times permutation tests were performed to verify the reliability of the model. The VIP value of the model was calculated using multiple cross-validation. The method of combining the difference multiple, the p value, and the VIP value of the OPLS-DA model was adopted to screen the differential metabolites. The screening criteria are FC > 1, p value < 0.05, and VIP > 1. The different metabolites of KEGG pathway enrichment significance were calculated using a hypergeometric distribution test.
4.4. Hematoxylin and Eosin (H&E) Staining
Livers from the different treatment groups were fixed in 4% paraformaldehyde for 24 h; the detailed procedure was identical to that reported in our previous study [35].
4.5. SOD, ALT and AST
According to the kit instruction, the levels of SOD, ALT, and AST were determined in each group.
4.6. Statistical Analysis
Statistical analysis was conducted using SPSS 20.0 software, with normally distributed quantitative data represented as x ± s. Analysis of variance was used for overall comparison, and Student’s t-test was used for intergroup comparison.
5. Conclusions
Through this experiment, it was discovered that the mechanism of acute liver injury induced by thioacetamide is intricate, encompassing oxidative damage, inflammatory damage, and, potentially, liver fibrosis. The protective mechanism of eugenol against acute liver injury is hypothesized, based on metabolomics, to involve the reduction in oxidative and inflammatory damage, as well as the correction of aberrant energy metabolism. Nevertheless, the study’s limitation lies in its reliance on metabolomics to primarily speculate on the protective mechanism of eugenol against liver injury, while its strength is the initial exploration of the body’s holistic response. The subsequent phase involves conducting a detailed exploration at the protein and DNA levels for a specific pathway.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29225288/s1. Table S1. Differential expressed metabolites between the TAA group and control group. Table S2: Differential expressed metabolites between the Eugenol treatment group and TAA group.
Author Contributions
H.C. was responsible for data curation and formal analysis. M.L. was responsible for the methodology and prepared Figure 2, Figure 3, Figure 4 and Figure 5. H.C. and M.L. are co-first-authors. H.Y. was responsible for the methodology and software. H.Y. prepared Figure 1. J.Y. prepared Figure 6 and Figure 7. G.W. was responsible for revising the manuscript. L.G. was responsible for supervision and wrote the original draft. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Liaoning province natural science foundation (2024-MS-027) and Liaoning province innovative training programs (S202410159005).
Institutional Review Board Statement
The animal study protocol was approved by the Ethics Committee of China Medical University (protocol code: KT2022353).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xiao, J.; Wang, F.; Wong, N.K.; He, J.; Zhang, R.; Sun, R.; Xu, Y.; Liu, Y.; Li, W.; Koike, K.; et al. Global liver disease burdens and research trends: Analysis from a Chinese perspective. J. Hepatol. 2019, 71, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.S.; Fan, J.G.; Zhang, Z.; Gao, B.; Wang, H.Y. The global burden of liver disease: The major impact of China. Hepatology 2014, 60, 2099–2108. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jaeschke, H.; Akakpo, J.Y.; Umbaugh, D.S.; Ramachandran, A. Novel Therapeutic Approaches Against Acetaminophen-induced Liver Injury and Acute Liver Failure. Toxicol. Sci. 2020, 174, 159–167. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Batiha, G.E.; Alkazmi, L.M.; Wasef, L.G.; Beshbishy, A.M.; Nadwa, E.H.; Rashwan, E.K. Syzygium aromaticum L. (Myrtaceae): Traditional Uses, Bioactive Chemical Constituents, Pharmacological and Toxicological Activities. Biomolecules 2020, 10, 202. [Google Scholar] [CrossRef]
- Ulanowska, M.; Olas, B. Biological Properties and Prospects for the Application of Eugenol-A Review. Int. J. Mol. Sci. 2021, 22, 3671. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Barboza, J.N.; da Silva Maia Bezerra Filho, C.; Silva, R.O.; Medeiros, J.V.R.; de Sousa, D.P. An Overview on the Anti-inflammatory Potential and Antioxidant Profile of Eugenol. Oxid. Med. Cell Longev. 2018, 2018, 3957262. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Marchese, A.; Barbieri, R.; Coppo, E.; Orhan, I.E.; Daglia, M.; Nabavi, S.F.; Izadi, M.; Abdollahi, M.; Nabavi, S.M.; Ajami, M. Antimicrobial activity of eugenol and essential oils containing eugenol: A mechanistic viewpoint. Crit. Rev. Microbiol. 2017, 43, 668–689. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Prasad, R.; Mahmood, A.; Routray, I.; Shinkafi, T.S.; Sahin, K.; Kucuk, O. Eugenol-rich Fraction of Syzygium aromaticum (Clove) Reverses Biochemical and Histopathological Changes in Liver Cirrhosis and Inhibits Hepatic Cell Proliferation. J. Cancer Prev. 2014, 19, 288–300. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Porto Mde, P.; da Silva, G.N.; Luperini, B.C.; Bachiega, T.F.; de Castro Marcondes, J.P.; Sforcin, J.M.; Salvadori, D.M. Citral and eugenol modulate DNA damage and pro-inflammatory mediator genes in murine peritoneal macrophages. Mol. Biol. Rep. 2014, 41, 7043–7051. [Google Scholar] [CrossRef] [PubMed]
- Kar Mahapatra, S.; Chakraborty, S.P.; Majumdar, S.; Bag, B.G.; Roy, S. Eugenol protects nicotine-induced superoxide mediated oxidative damage in murine peritoneal macrophages in vitro. Eur. J. Pharmacol. 2009, 623, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Oh, O.J.; Min, H.Y.; Park, E.J.; Kim, Y.; Park, H.J.; Nam Han, Y.; Lee, S.K. Eugenol suppresses cyclooxygenase-2 expression in lipopolysaccharide-stimulated mouse macrophage RAW264.7 cells. Life Sci. 2003, 73, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Raghavenra, H.; Diwakr, B.T.; Lokesh, B.R.; Naidu, K.A. Eugenol--the active principle from cloves inhibits 5-lipoxygenase activity and leukotriene-C4 in human PMNL cells. Prostagland. Leukot. Essent. Fat. Acids. 2006, 74, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J. Am. Soc. Mass. Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, R.; Li, B.; Lam, S.M.; Shui, G. Integration of lipidomics and metabolomics for in-depth understanding of cellular mechanism and disease progression. J. Genet. Genom. 2020, 47, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Shastry, A.; Dunham-Snary, K. Metabolomics and mitochondrial dysfunction in cardiometabolic disease. Life Sci. 2023, 333, 122137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Mao, Y.; Qiao, H.; Jiang, H.; Zhao, H.; Chen, X.; Tong, L.; Sun, X. Protective effects of taurine against endotoxin-induced acute liver injury after hepatic ischemia reperfusion. Amino Acids 2010, 38, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.C.; Cheng, L.Y.; Lu, J.; Huang, Y.H.; Chiou, S.H.; Tsai, P.H.; Huo, T.I.; Lin, H.C.; Lee, F.Y. The role of interferon-γ inducible protein-10 in a mouse model of acute liver injury post induced pluripotent stem cells transplantation. PLoS ONE 2012, 7, e50577. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Singer, G.A.; Zielsdorf, S.; Fleetwood, V.A.; Alvey, N.; Cohen, E.; Eswaran, S.; Shah, N.; Chan, E.Y.; Hertl, M.; Fayek, S.A. Limited hepatitis B immunoglobulin with potent nucleos(t)ide analogue is a cost effective prophylaxis against hepatitis B virus after liver transplantation. Transpl. Proc. 2015, 47, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Duan, J.; Feng, Y.; Huang, X.; Fan, W.; Wang, K.; Ouyang, P.; Deng, Y.; Du, Z.; Chen, D. Hepatoprotective Activity of Vitamin E and Metallothionein in Cadmium-Induced Liver Injury in Ctenopharyngodon idellus. Oxid Med Cell Longev. 2018, 2018, 9506543. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, M.; Wang, X.; Bu, Q.; Wang, Q.; Su, W.; Li, L.; Zhou, H.; Lu, L. XBP1 deficiency promotes hepatocyte pyroptosis by impairing mitophagy to activate mtDNA-cGAS-STING signaling in macrophages during acute liver injury. Redox Biol. 2022, 52, 102305. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Elnfarawy, A.A.; Nashy, A.E.; Abozaid, A.M.; Komber, I.F.; Elweshahy, R.H.; Abdelrahman, R.S. Vinpocetine attenuates thioacetamide-induced liver fibrosis in rats. Hum. Exp. Toxicol. 2021, 40, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Enciso, N.; Amiel, J.; Fabián-Domínguez, F.; Pando, J.; Rojas, N.; Cisneros-Huamaní, C.; Nava, E.; Enciso, J. Model of Liver Fibrosis Induction by Thioacetamide in Rats for Regenerative Therapy Studies. Anal. Cell Pathol. 2022, 2022, 2841894. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nishi, K.; Yagi, H.; Ohtomo, M.; Nagata, S.; Udagawa, D.; Tsuchida, T.; Morisaku, T.; Kitagawa, Y. A thioacetamide-induced liver fibrosis model for pre-clinical studies in microminipig. Sci. Rep. 2023, 13, 14996. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ezhilarasan, D. Molecular mechanisms in thioacetamide-induced acute and chronic liver injury models. Env. Toxicol. Pharmacol. 2023, 99, 104093. [Google Scholar] [CrossRef] [PubMed]
- Ta, N.; Erdunduleng, E.; Qi, R.; Mu, X.; Feng, L.; Ba, G.; Li, Y.; Zhang, J.; Bai, L.; Fu, M. Metabolomics analysis reveals amelioration effects of yellowhorn tea extract on hyperlipidemia, inflammation, and oxidative stress in high-fat diet-fed mice. Front. Nutr. 2023, 10, 1087256. [Google Scholar] [CrossRef]
- Xiao, G.; Xu, A.; Jiang, J.; Chen, Z.; Li, Y.; Li, S.; Chen, W.; Zhang, J.; Jia, C.; Zeng, Z.; et al. Metabolomics analysis delineates the therapeutic effects of Yinlan Tiaozhi capsule on triton WR-1339 -induced hyperlipidemia in mice. Front. Pharmacol. 2023, 14, 1252146. [Google Scholar] [CrossRef]
- Luo, D.; Chen, K.; Li, J.; Fang, Z.; Pang, H.; Yin, Y.; Rong, X.; Guo, J. Gut microbiota combined with metabolomics reveals the metabolic profile of the normal aging process and the anti-aging effect of FuFang Zhenshu TiaoZhi(FTZ) in mice. Biomed. Pharmacother. 2020, 121, 109550. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.R.; Lee, J.A.; Kim, M.; Lee, S.; Oh, M.; Moon, J.; Nam, J.W.; Choi, H.; Mun, Y.J.; Roh, S.S. Gardeniae Fructus Attenuates Thioacetamide-Induced Liver Fibrosis in Mice via Both AMPK/SIRT1/NF-κB Pathway and Nrf2 Signaling. Antioxidants 2021, 10, 1837. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- da Silva, B.S.; Paulino, A.M.B.; Taffarel, M.; Borba, I.G.; Telles, L.O.; Lima, V.V.; Aguiar, D.H.; Dias, M.C.; Nascimento, A.F.; Sinhorin, V.D.G.; et al. High sucrose diet attenuates oxidative stress, inflammation and liver injury in thioacetamide-induced liver cirrhosis. Life Sci. 2021, 267, 118944. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, A.; Tudawe, D.; Chakravarthi, S.; Chiew, G.S.; Haleagrahara, N. Effect of γ-tocotrienol in counteracting oxidative stress and joint damage in collagen-induced arthritis in rats. Exp. Ther. Med. 2014, 7, 1408–1414. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sies, H. Oxidative stress: Oxidants and antioxidants. Exp. Physiol. 1997, 82, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Liu, Z.; Chen, S. Study on Antioxidant Activity of Octopamine and Its Derivatives. Nat. Prod. Res. Dev. 2015, 27, 255–258. [Google Scholar]
- Chniguir, A.; Saguem, M.H.; Dang, P.M.; El-Benna, J.; Bachoual, R. Eugenol Inhibits Neutrophils Myeloperoxidase In Vitro and Attenuates LPS-Induced Lung Inflammation in Mice. Pharmaceuticals 2024, 17, 504. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bezerra, D.P.; Militão, G.C.G.; de Morais, M.C.; de Sousa, D.P. The Dual Antioxidant/Prooxidant Effect of Eugenol and Its Action in Cancer Development and Treatment. Nutrients 2017, 9, 1367. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guo, H.; Wang, G.; Huang, W.; Li, L.; Bai, Y.; Wang, H.; Gao, L. The Mechanism of Hepatic Encephalopathy Induced by Thioacetamide Based on Metabolomics and Proteomics: A Preliminary Study. Int. J. Mol. Sci. 2023, 25, 284. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).