Erinacenones A–L: Twelve New Isoindolinone Alkaloids from the Edible and Medicinal Mushroom Hericium erinaceus
Abstract
1. Introduction
2. Results and Discussion
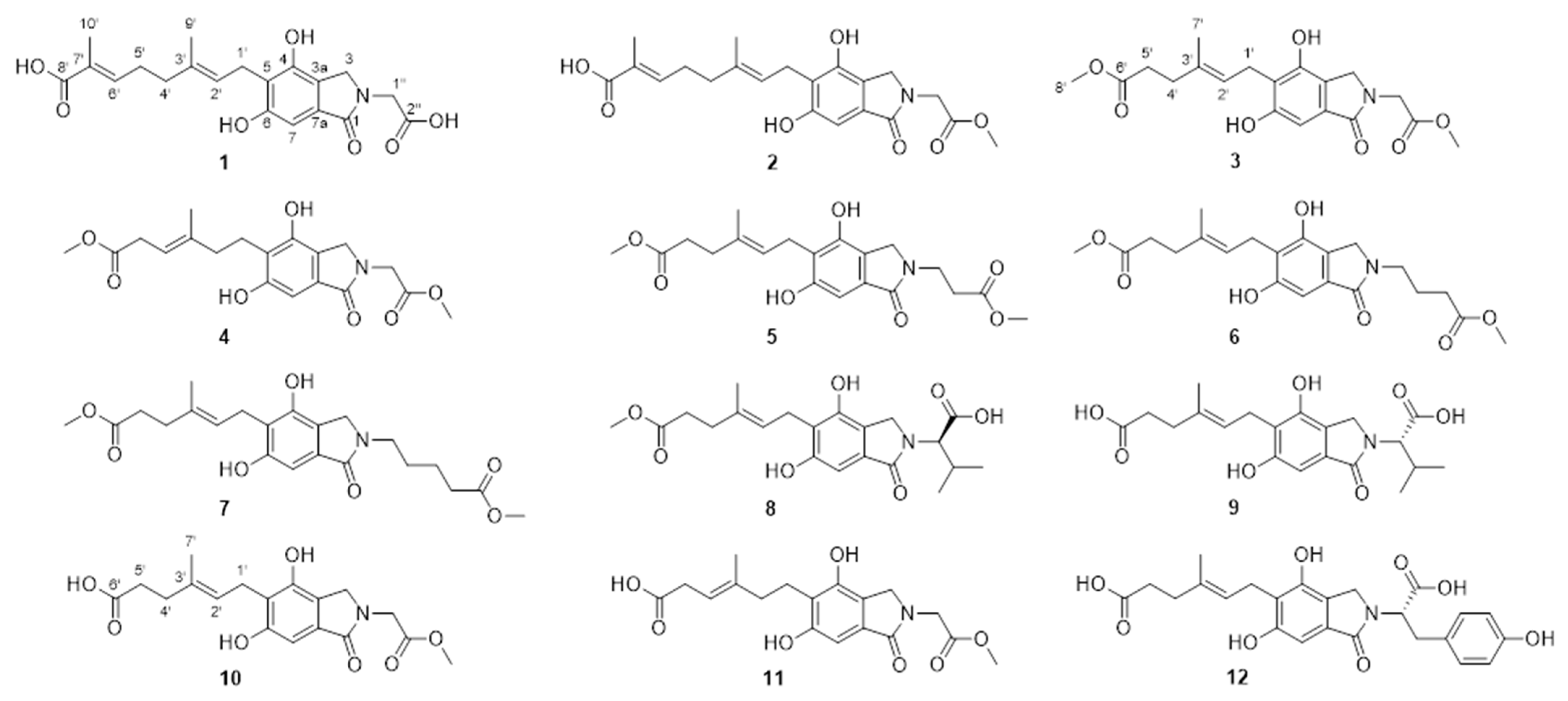
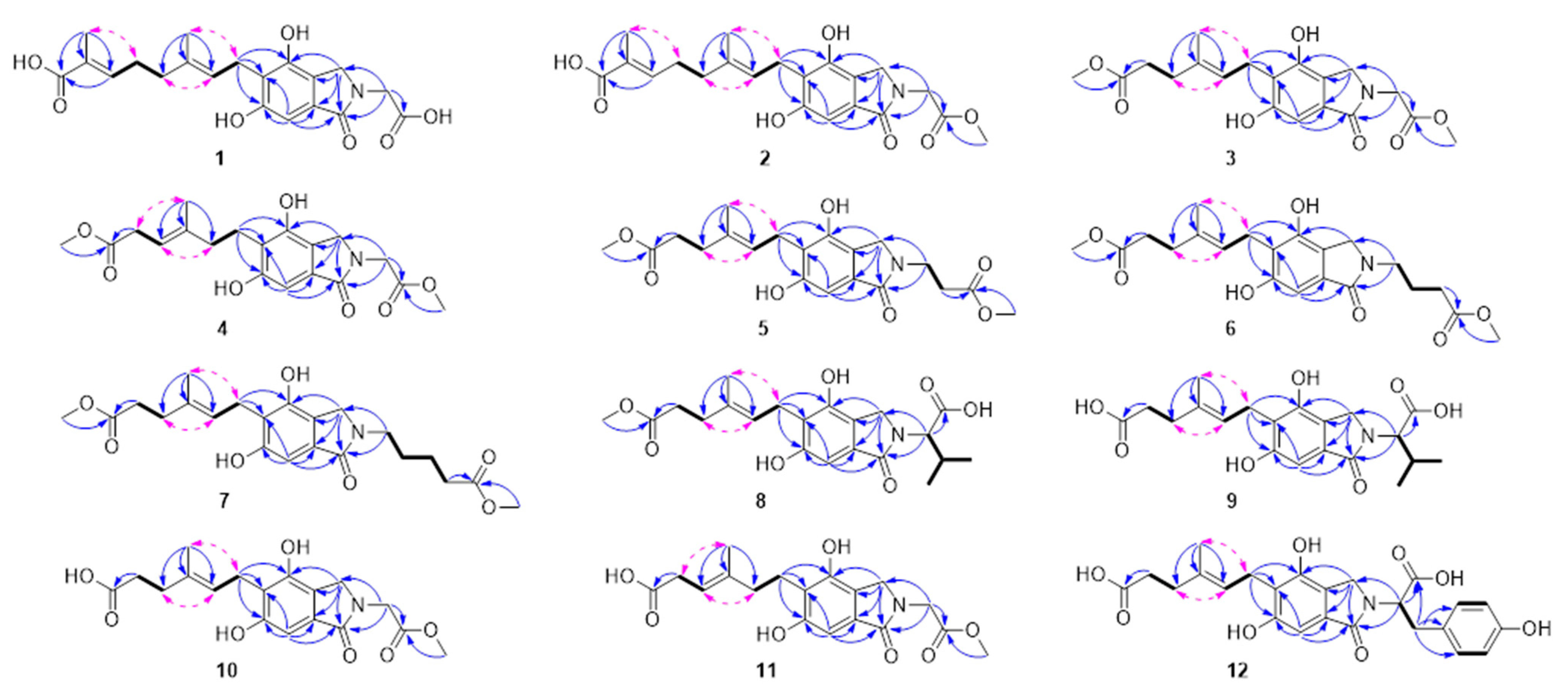
Structural Elucidation of the Previously Undescribed Compounds
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Extraction and Isolation
3.4. Characterization Data
3.4.1. Erinacenone A (1)
3.4.2. Erinacenone B (2)
3.4.3. Erinacenone C (3)
3.4.4. Erinacenone D (4)
3.4.5. Erinacenone E (5)
3.4.6. Erinacenone F (6)
3.4.7. Erinacenone G (7)
3.4.8. Erinacenone H (8)
3.4.9. Erinacenone I (9)
3.4.10. Erinacenone J (10)
3.4.11. Erinacenone K (11)
3.4.12. Erinacenone L (12)
3.5. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Szućko-Kociuba, I.; Trzeciak-Ryczek, A.; Kupnicka, P.; Chlubek, D. Neurotrophic and Neuroprotective Effects of Hericium erinaceus. Int. J. Mol. Sci. 2023, 24, 15960. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M. Chemistry, Nutrition, and Health-Promoting Properties of Hericium erinaceus (Lion’s Mane) Mushroom Fruiting Bodies and Mycelia and Their Bioactive Compounds. J. Agric. Food Chem. 2015, 63, 7108–7123. [Google Scholar] [CrossRef] [PubMed]
- Gravina, A.G.; Pellegrino, R.; Auletta, S.; Palladino, G.; Brandimarte, G.; D’Onofrio, R.; Arboretto, G.; Imperio, G.; Ventura, A.; Cipullo, M.; et al. Hericium erinaceus, a medicinal fungus with a centuries-old history: Evidence in gastrointestinal diseases. World J. Gastroenterol. 2023, 29, 3048–3065. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Hu, S.H.; Su, C.H.; Lee, T.M. Antitumor and immunoenhancing activities of polysaccharide from culture broth of Hericium spp. Kaohsiung J. Med. Sci. 2001, 17, 461–467. [Google Scholar] [PubMed]
- Zhang, Z.; Liu, R.N.; Tang, Q.J.; Zhang, J.S.; Yang, Y.; Shang, X.D. A new diterpene from the fungal mycelia of Hericium erinaceus. Phytochem. Lett. 2015, 11, 151–156. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, S.L.; Wang, A.H.; Sun, Z.C.; Zhuo, Y.F.; Xu, Y.T.; He, Y.L. Protective effect of ethanol extracts of Hericium erinaceus on alloxan-induced diabetic neuropathic pain in rats. Evid.-Based Complement. Altern. Med. 2015, 2015, 595480. [Google Scholar] [CrossRef]
- Liang, B.; Guo, Z.D.; Xie, F.; Zhao, A.N. Antihyperglycemic and antihyperlipidemic activities of aqueous extract of Hericium erinaceus in experimental diabetic rats. BMC Complement. Altern. Med. 2013, 13, 253. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.K.; Park, J.B.; Song, C.H. Hypolipidemic effect of an exo-biopolymer produced from a submerged mycelial culture of Hericium erinaceus. Biosci. Biotechnol. Biochem. 2003, 67, 1292–1298. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Ouchi, K.; Hirasawa, N. The anti-inflammatory effects of lion’s mane culinary-medicinal mushroom, Hericium erinaceus (higher basidiomycetes) in a coculture system of 3T3-L1 adipocytes and RAW264 macrophages. Int. J. Med. Mushrooms 2015, 17, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Abdullah, N.; Aminudin, N. Inhibitory effect on in vitro LDL oxidation and HMG Co-A reductase activity of the liquid-liquid partitioned fractions of Hericium erinaceus (Bull.) Persoon (Lion’s mane mushroom). BioMed Res. Int. 2014, 828149. [Google Scholar] [CrossRef]
- Tzeng, T.T.; Chen, C.C.; Lee, L.Y.; Chen, W.P.; Lu, C.K.; Shen, C.C.; Huang, F.C.Y.; Chen, C.C.; Shiao, Y.J. Erinacine A-enriched Hericium erinaceus mycelium ameliorates Alzheimer’s disease-related pathologies in APPswe/PS1dE9 transgenic mice. J. Biomed. Sci. 2016, 23, 49. [Google Scholar] [CrossRef]
- Mori, K.; Inatomi, S.; Ouchi, K.; Azumi, Y.; Tuchida, T. Improving effects of the mushroom Yamabushitake (Hericium erinaceus) on mild cognitive impairment: A double-blind placebo-controlled clinical trial. Phytother. Res. 2009, 23, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.F.; Chen, J.H.; Teng, C.C.; Shen, C.H.; Hsieh, M.C.; Lu, C.C.; Lee, K.C.; Lee, L.Y.; Chen, W.P.; Chen, C.C.; et al. Protective effects of Hericium erinaceus mycelium and its isolated erinacine A against ischemia-injury-induced neuronal cell death via the inhibition of iNOS/p38 MAPK and nitrotyrosine. Int. J. Mol. Sci. 2014, 15, 15073–15089. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.H.; Chyau, C.C.; Chen, C.C.; Lee, L.Y.; Chen, W.P.; Liu, J.L.; Lin, W.H.; Mong, M.C. Erinacine A-enriched Hericium erinaceus mycelium produces antidepressant-like effects through modulating BDNF/PI3K/Akt/GSK-3β signaling in mice. Int. J. Mol. Sci. 2018, 19, 341. [Google Scholar] [CrossRef]
- Ryu, S.; Kim, H.G.; Kim, J.Y.; Kim, S.Y.; Cho, K.O. Hericium erinaceus extract reduces anxiety and depressive behaviors by promoting hippocampal neurogenesis in the adult mouse brain. J. Med. Food 2018, 21, 174–180. [Google Scholar] [CrossRef]
- Yao, W.; Zhang, J.C.; Dong, C.; Zhuang, C.; Hirota, S.; Inanaga, K.; Hashimoto, K. Effects of amycenone on serum levels of tumor necrosis factor-α, interleukin-10, and depression-like behavior in mice after lipopolysaccharide administration. Pharmacol. Biochem. Behav. 2015, 136, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Bao, L.; Qi, Q.X.; Zhao, F.; Ma, K.; Pei, Y.F.; Liu, H.W. Erinacerins C–L, Isoindolin-1-ones with α-Glucosidase Inhibitory Activity from Cultures of the Medicinal Mushroom Hericium erinaceus. J. Nat. Prod. 2015, 78, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A simple method of estimating fifty percent endpoint. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Lin, J.Y.; Chen, Y.P.; Lin, T.W.; Li, T.J.; Chen, Y.W.; Li, I.C.; Chen, C.C. Discovery of a New Compound, Erinacerin W, from the Mycelia of Hericium erinaceus, with Immunomodulatory and Neuroprotective Effects. Molecules 2024, 29, 812. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Z.H.; Yao, J.N.; Peng, Y.L.; Huang, R.; Feng, T.; Liu, J.K. Isoindolinone-containing meroterpenoids with α-Glucosidase Inhibitory activity from mushroom Hericium caput-medusae. Fitoterapia 2017, 122, 107–114. [Google Scholar] [CrossRef] [PubMed]
| No. | 1 | 2 | ||
|---|---|---|---|---|
| 1 | 171.8 | 171.9 | ||
| 3 | 50.0 | 4.39, s | 44.7 | 4.39, s |
| 3a | 131.3 | 131.1 | ||
| 4 | 151.6 | 151.7 | ||
| 5 | 121.7 | 121.9 | ||
| 6 | 158.0 | 158.1 | ||
| 7 | 102.0 | 6.76, s | 102.0 | 6.76, s |
| 7a | 121.0 | 121.0 | ||
| 1′ | 23.6 | 3.42, dd (7.1) | 23.6 | 3.43, dd (7.1) |
| 2′ | 124.5 | 5.30, t (7.1) | 124.3 | 5.30, t (7.1) |
| 3′ | 134.8 | 135.0 | ||
| 4′ | 39.4 | 2.09, t (7.3) | 39.5 | 2.09, t (7.4) |
| 5′ | 28.3 | 2.28, dd (14.9, 7.3) | 28.4 | 2.28, dd (14.9, 7.4) |
| 6′ | 143.8 | 6.72, t (7.3) | 143.3 | 6.71, t (7.4) |
| 7′ | 128.8 | 129.2 | ||
| 8′ | 171.7 | 172.4 | ||
| 9′ | 16.3 | 1.82, s | 16.3 | 1.82, s |
| 10′ | 12.4 | 1.76, s | 12.5 | 1.76, s |
| 1″ | 45.0 | 4.32, s | 50.0 | 4.39, overlapped |
| 2″ | 173.0 | 171.2 | ||
| 3″ | 52.8 | 3.76, s |
| No. | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|
| 3 | 4.38, s | 4.39, s | 4.34, s | 4.30, s | 4.30, s |
| 7 | 6.76, s | 6.76, s | 6.72, s | 6.73, s | 6.73, s |
| 1′ | 3.40, d (7.1) | 2.83, m | 3.39, d (7.1) | 3.40, d (7.1) | 3.40, d (7.1) |
| 2′ | 5.28, t (7.1) | 2.24, t (7.8) | 5.27, t (7.1) | 5.27, t (7.1) | 5.28, t (7.1) |
| 4′ | 2.26, t (7.6) | 5.26, t (7.1) | 2.25, t (7.6) | 2.25, t (7.6) | 2.25, t (7.5) |
| 5′ | 2.39, m | 3.04, d (7.1) | 2.39, t (7.6) | 2.39, overlapped | 2.39, overlapped |
| 7′ | 1.80, s | 1.74, s | 1.79, s | 1.79, s | 1.80, s |
| 8′ | 3.55, s | 3.64, s | 3.55, s | 3.55, s | 3.55, s |
| 1″ | 4.38, s | 4.39, s | 3.85, t (6.7) | 3.62, t (6.8) | 3.60, s |
| 2″ | 2.73, t (6.7) | 1.98, m | 1.71, m | ||
| 3″ | 3.76, s | 2.37, overlapped | 1.63, m | ||
| 4″ | 3.75, s | 3.67, s | 2.40, overlapped | ||
| 5″ | 3.58, s | ||||
| 6″ | 3.64, s |
| No. | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|
| 1 | 171.8 | 171.1 | 171.4 | 171.5 | 171.4 |
| 3 | 49.9 | 44.7 | 49.7 | 48.8 | 48.8 |
| 3a | 131.1 | 131.1 | 131.7 | 131.8 | 132.0 |
| 4 | 151.7 | 151.8 | 151.6 | 151.6 | 151.6 |
| 5 | 121.8 | 122.4 | 121.4 | 121.3 | 121.3 |
| 6 | 158.1 | 158.3 | 158.1 | 158.1 | 158.1 |
| 7 | 102.0 | 101.9 | 101.7 | 101.8 | 101.8 |
| 7a | 121.0 | 121.0 | 120.6 | 120.4 | 120.4 |
| 1′ | 23.6 | 23.4 | 23.6 | 23.6 | 23.6 |
| 2′ | 124.5 | 39.5 | 124.6 | 124.6 | 124.6 |
| 3′ | 134.3 | 140.7 | 134.2 | 134.2 | 134.2 |
| 4′ | 35.9 | 116.8 | 36.0 | 36.0 | 36.0 |
| 5′ | 33.9 | 34.3 | 33.9 | 33.9 | 33.9 |
| 6′ | 175.8 | 174.8 | 175.8 | 175.8 | 175.8 |
| 7′ | 16.1 | 16.4 | 16.1 | 16.1 | 16.1 |
| 8′ | 51.9 | 52.3 | 51.9 | 51.9 | 51.9 |
| 1″ | 44.7 | 50.0 | 39.8 | 42.9 | 43.0 |
| 2″ | 171.1 | 171.8 | 33.9 | 24.7 | 28.7 |
| 3″ | 52.8 | 52.8 | 173.7 | 32.0 | 23.1 |
| 4″ | 52.3 | 175.2 | 34.1 | ||
| 5″ | 52.1 | 175.6 | |||
| 6″ | 52.0 |
| No. | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|
| 3 | 4.60, d (17.0); 4.32, d (17.0) | 4.59, overlapped; 4.32, d (17.0) | 4.38, s | 4.39, s | 4.37, d (16.6); 4.27, d (16.6) |
| 7 | 6.75, s | 6.75, s | 6.76, s | 6.75, s | 6.67, s |
| 1′ | 3.41, d (7.1) | 3.42, d (7.2) | 3.42, d (7.1) | 2.83, t (7.6) | 3.39, m |
| 2′ | 5.28, t (7.1) | 5.31, t (7.2) | 5.30, t (7.1) | 2.21, t (7.6) | 5.29, t (7.4) |
| 4′ | 2.26, t (7.6) | 2.26, t (7.2) | 2.26, m | 5.39, m | 2.25, overlapped |
| 5′ | 2.39, m | 2.36, overlapped | 2.32, m | 2.97, m | 2.35, m |
| 7′ | 1.80, d | 1.81, s | 1.81, s | 1.74, s | 1.80, s |
| 8′ | 3.55, s | ||||
| 1″ | 4.56, m | 4.58, overlapped | 4.39, s | 4.39, s | 5.10, dd (11.2, 4.7) |
| 2″ | 2.34, m | 2.35, overlapped | 3.11, dd (14.6, 11.4); 2.25, overlapped | ||
| 3″ | 1.09, d (6.6) | 1.09, d (6.6) | 3.78, s | 3.76, s | |
| 4″ | 0.89, d (6.6) | 0.9, d (6.6) | 7.04, d (8.5) | ||
| 5″ | 6.65, d (8.5) | ||||
| 7″ | 6.65, d (8.5) | ||||
| 8″ | 7.04, d (8.5) |
| No. | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|
| 1 | 171.8 | 171.9 | 171.8 | 171.9 | 171.8 |
| 3 | 46.8 | 46.8 | 50.0 | 44.7 | 47.0 |
| 3a | 131.2 | 131.1 | 131.1 | 131.0 | 131.4 |
| 4 | 151.6 | 151.6 | 151.7 | 151.8 | 151.5 |
| 5 | 121.6 | 121.7 | 121.9 | 122.8 | 121.5 |
| 6 | 158.1 | 158.1 | 158.2 | 158.3 | 158.0 |
| 7 | 101.9 | 102.0 | 102.0 | 6.75 | 101.9 |
| 7a | 120.9 | 120.9 | 121.0 | 121.2 | 120.9 |
| 1′ | 23.6 | 23.6 | 23.6 | 23.6 | 23.6 |
| 2′ | 124.6 | 124.2 | 123.9 | 39.8 | 124.2 |
| 3′ | 134.2 | 134.5 | 135.1 | 138.6 | 134.5 |
| 4′ | 36.0 | 36.0 | 36.6 | 119.5 | 35.9 |
| 5′ | 33.9 | 34.1 | 35.4 | 37.1 | 34.0 |
| 6′ | 175.8 | 177.6 | 179.6 | nd a | 177.6 |
| 7′ | 16.1 | 16.2 | 16.3 | 16.4 | 16.2 |
| 8′ | 51.9 | ||||
| 1″ | 62.8 | 62.5 | 44.7 | 50.0 | 57.5 |
| 2″ | 30.1 | 30.1 | 171.1 | 171.2 | 35.9 |
| 3″ | 20.1 | 20.0 | 52.8 | 52.8 | 129.3 |
| 4″ | 19.6 | 19.2 | 130.6 | ||
| 5″ | nd a | 174.5 | 116.4 | ||
| 6″ | 157.2 | ||||
| 7″ | 116.4 | ||||
| 8″ | 130.6 | ||||
| 9″ | 174.5 |
| Compounds | MCF-7 |
|---|---|
| 4 | 24.7 |
| 5 | 18.4 |
| Cisplatin a | 9.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, L.-L.; Liu, J.-K. Erinacenones A–L: Twelve New Isoindolinone Alkaloids from the Edible and Medicinal Mushroom Hericium erinaceus. Molecules 2024, 29, 4901. https://doi.org/10.3390/molecules29204901
Yuan L-L, Liu J-K. Erinacenones A–L: Twelve New Isoindolinone Alkaloids from the Edible and Medicinal Mushroom Hericium erinaceus. Molecules. 2024; 29(20):4901. https://doi.org/10.3390/molecules29204901
Chicago/Turabian StyleYuan, Lin-Lin, and Ji-Kai Liu. 2024. "Erinacenones A–L: Twelve New Isoindolinone Alkaloids from the Edible and Medicinal Mushroom Hericium erinaceus" Molecules 29, no. 20: 4901. https://doi.org/10.3390/molecules29204901
APA StyleYuan, L.-L., & Liu, J.-K. (2024). Erinacenones A–L: Twelve New Isoindolinone Alkaloids from the Edible and Medicinal Mushroom Hericium erinaceus. Molecules, 29(20), 4901. https://doi.org/10.3390/molecules29204901





