Heme Spin Distribution in the Substrate-Free and Inhibited Novel CYP116B5hd: A Multifrequency Hyperfine Sublevel Correlation (HYSCORE) Study
Abstract
1. Introduction
2. Results and Analysis
2.1. Effect of Nuclear Spin Labeling on the CW-EPR Spectrum
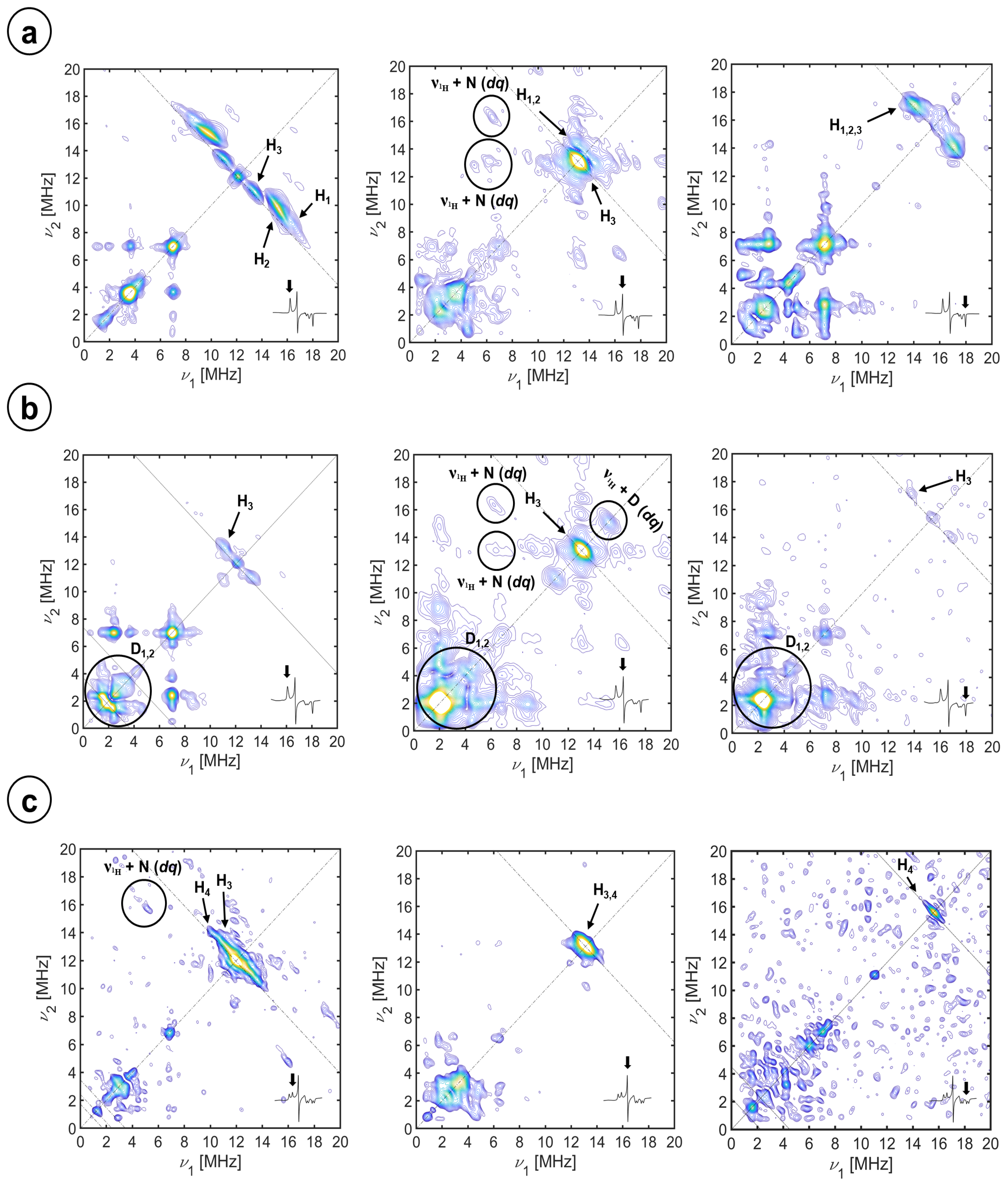
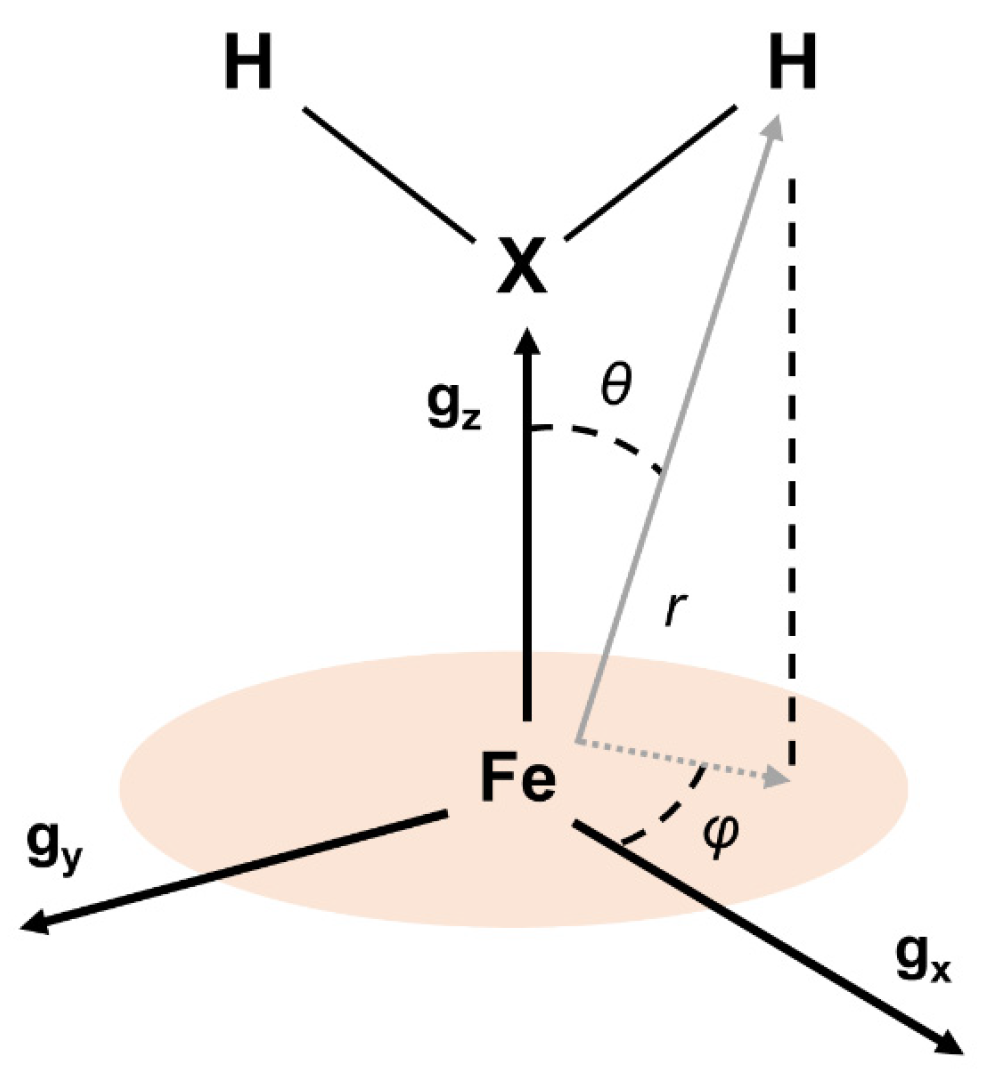
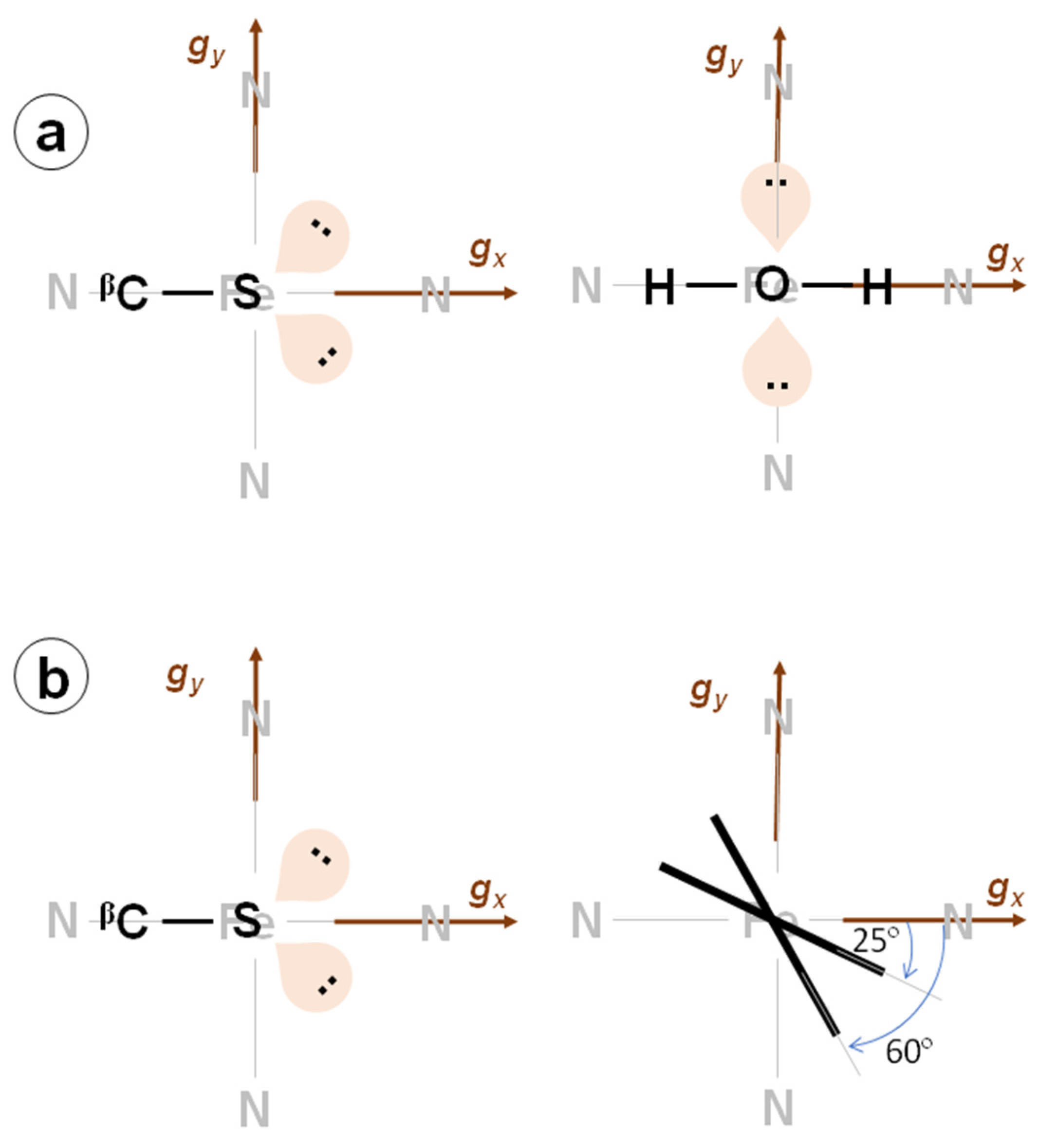
2.2. Hyperfine Interactions with Hydrogen Nuclei
| Species | Label | aiso [MHz] | T [MHz] | EPR α, β [°] | EPR r (Fe-H) [Å] | Crystal str. r (Fe-H) [Å] | Crystal str. θ [°] | Crystal str. φ [°] |
|---|---|---|---|---|---|---|---|---|
| H2O | H1 | −0.09 ± 0.06 | 5.60 ± 0.02 | 0 ± 5, 22 ± 5 | 2.42 a | 2.9 b | 23 b | 5 b |
| H2O | H2 | −1.095 ± 0.080 | 5.20 ± 0.02 | 0 ± 5, 16 ± 5 | 2.48 a | 2.7 b | 19 b | 0 b |
| D2O | D1 | −0.014 ± 0.01 | 0.860 ± 0.003 | 0 ± 5, 22 ± 5 | 2.42 a | |||
| D2O | D2 | −0.17 ± 0.03 | 0.800 ± 0.003 | 0 ± 5, 16 ± 5 | 2.48 a | |||
| Cysteine | H3 | 0.79 ± 0.22 | 2.60 ± 0.04 | 0 ± 5, 47 ± 5 | 3.12 a | 3.078, 4.256 [49] | 45, 66 c | 4, 13 c |
| Imidazole (2) | H4 | 1.76 ± 0.11 | 2.66 ± 0.08 | −25 ± 5, 40 ± 5 | 3.10 a | 3.142, 3.487 [60] | 41, 38 e | N.A. |
| Imidazole (1) | 1.76 ± 0.11 | 2.66 ± 0.08 | −60 d, 40 ± 5 |
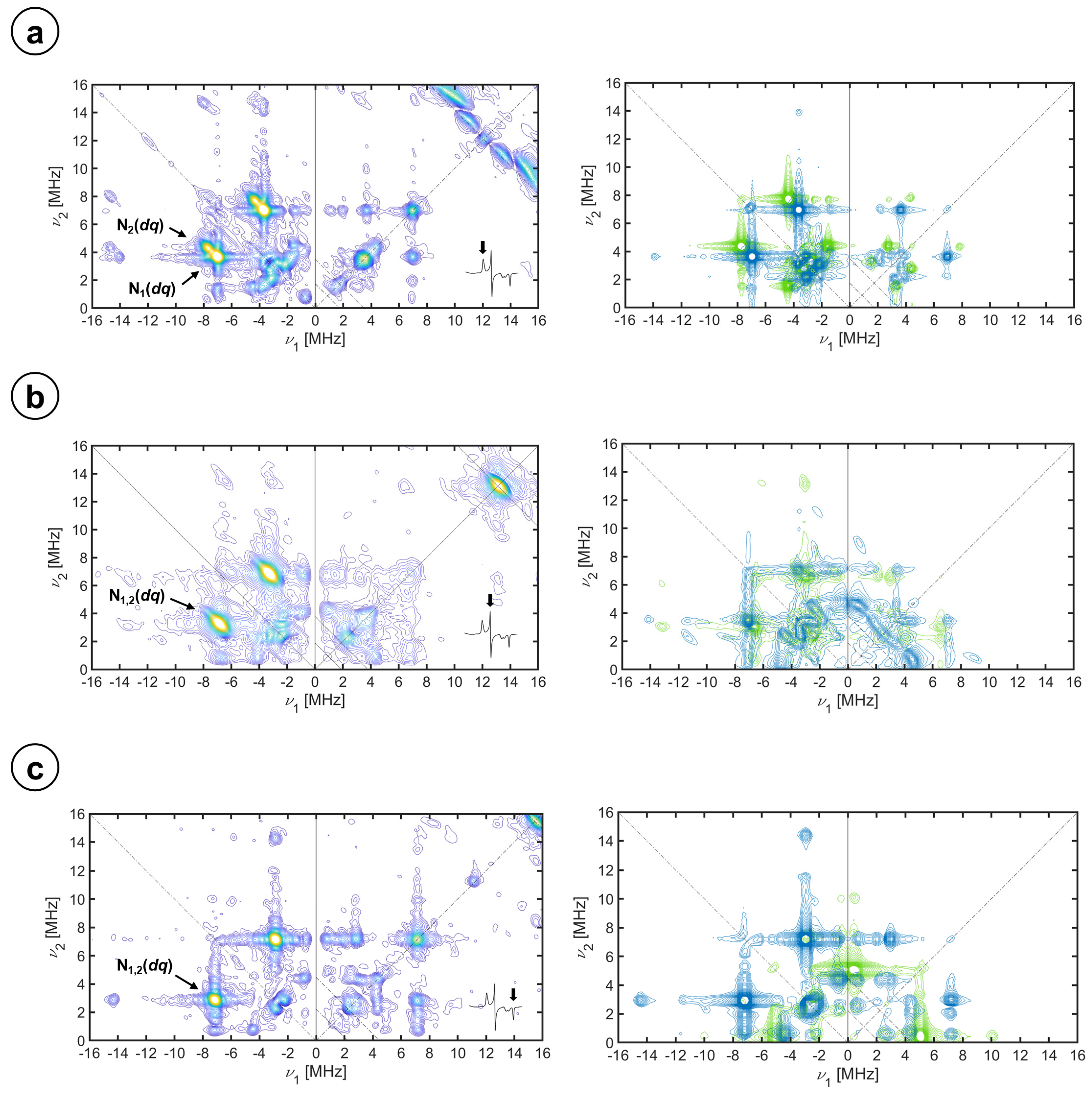
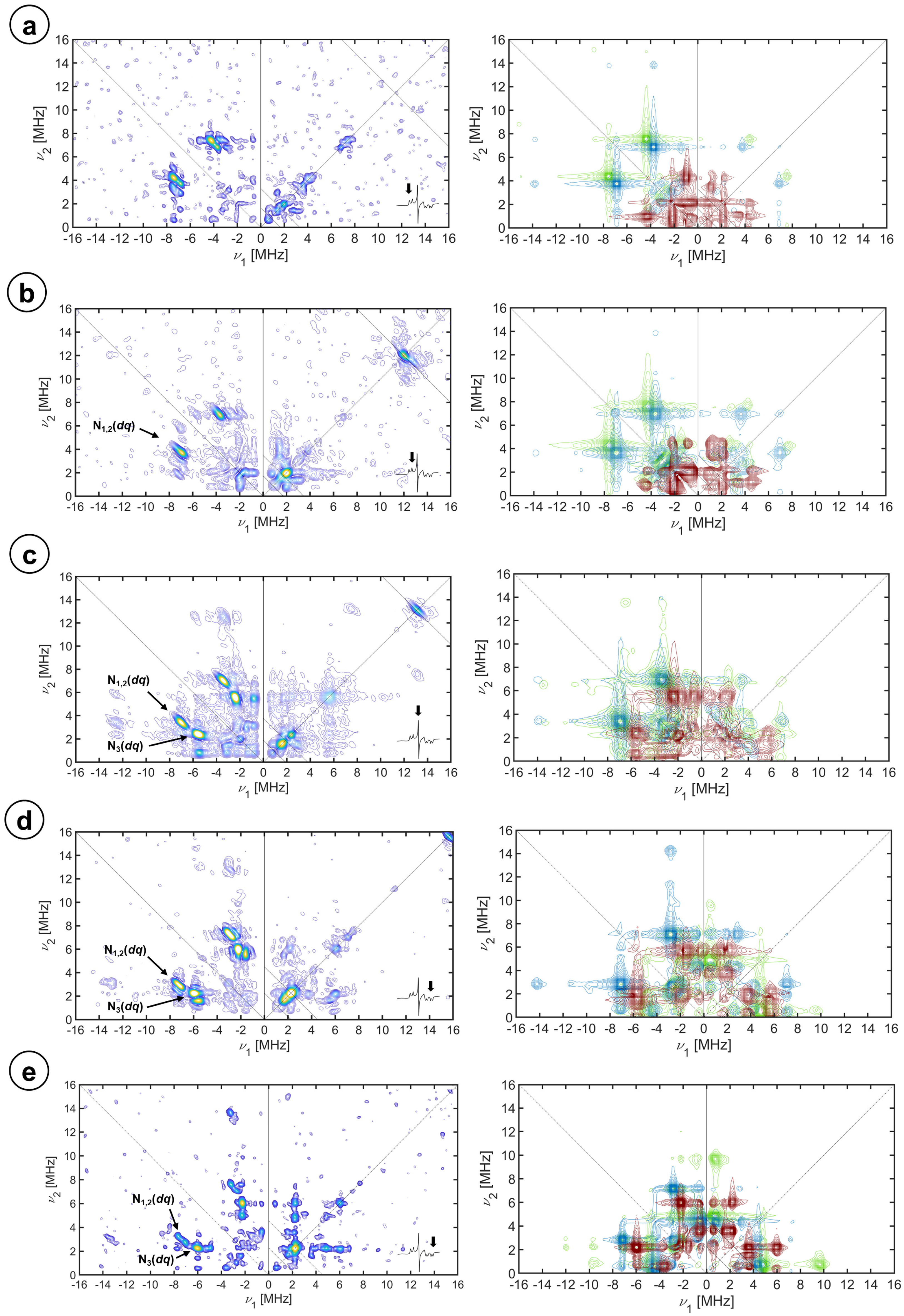
2.3. Hyperfine Interactions with Nitrogen Nuclei
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ortiz de Montellano, P.R. Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th ed.; Ortiz de Montellano, P.R., Ed.; Springer International Publishing: Cham, Switzerland, 2015; ISBN 978-3-319-12107-9. [Google Scholar]
- Lamb, D.C.; Lei, L.; Warrilow, A.G.S.; Lepesheva, G.I.; Mullins, J.G.L.; Waterman, M.R.; Kelly, S.L. The First Virally Encoded Cytochrome P450. J. Virol. 2009, 83, 8266–8269. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R. Cytochrome P450 Diversity in the Tree of Life. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Hammer, S.C.; Kubik, G.; Watkins, E.; Huang, S.; Minges, H.; Arnold, F.H. Anti-Markovnikov Alkene Oxidation by Metal-Oxo–Mediated Enzyme Catalysis. Science (1979) 2017, 358, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Tavanti, M.; Porter, J.L.; Sabatini, S.; Turner, N.J.; Flitsch, S.L. Panel of New Thermostable CYP116B Self-Sufficient Cytochrome P450 Monooxygenases That Catalyze C−H Activation with a Diverse Substrate Scope. ChemCatChem 2018, 10, 1042–1051. [Google Scholar] [CrossRef]
- Correddu, D.; Di Nardo, G.; Gilardi, G. Self-Sufficient Class VII Cytochromes P450: From Full-Length Structure to Synthetic Biology Applications. Trends Biotechnol. 2021, 39, 1184–1207. [Google Scholar] [CrossRef]
- Carta, M.; Malpass-Evans, R.; Croad, M.; Rogan, Y.; Jansen, J.C.; Bernardo, P.; Bazzarelli, F.; McKeown, N.B. An Efficient Polymer Molecular Sieve for Membrane Gas Separations. Science (1979) 2013, 339, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Farwell, C.C.; McIntosh, J.A.; Hyster, T.K.; Wang, Z.J.; Arnold, F.H. Enantioselective Imidation of Sulfides via Enzyme-Catalyzed Intermolecular Nitrogen-Atom Transfer. J. Am. Chem. Soc. 2014, 136, 8766–8771. [Google Scholar] [CrossRef]
- Farwell, C.C.; Zhang, R.K.; McIntosh, J.A.; Hyster, T.K.; Arnold, F.H. Enantioselective Enzyme-Catalyzed Aziridination Enabled by Active-Site Evolution of a Cytochrome P450. ACS Cent. Sci. 2015, 1, 89–93. [Google Scholar] [CrossRef]
- Zhang, X.; Li, S. Expansion of Chemical Space for Natural Products by Uncommon P450 Reactions. Nat. Prod. Rep. 2017, 34, 1061–1089. [Google Scholar] [CrossRef]
- Prier, C.K.; Zhang, R.K.; Buller, A.R.; Brinkmann-Chen, S.; Arnold, F.H. Enantioselective, Intermolecular Benzylic C–H Amination Catalysed by an Engineered Iron-Haem Enzyme. Nat. Chem. 2017, 9, 629–634. [Google Scholar] [CrossRef]
- Correddu, D.; Helmy Aly, S.; Di Nardo, G.; Catucci, G.; Prandi, C.; Blangetti, M.; Bellomo, C.; Bonometti, E.; Viscardi, G.; Gilardi, G. Enhanced and Specific Epoxidation Activity of P450 BM3 Mutants for the Production of High Value Terpene Derivatives. RSC Adv. 2022, 12, 33964–33969. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Antona, C.; Ingelman-Sundberg, M. Cytochrome P450 Pharmacogenetics and Cancer. Oncogene 2006, 25, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.; Dahal, U.P.; Rock, D.; Peng, C.-C.; Schenk, J.O.; Joswig-Jones, C.; Jones, J.P. The Kinetic Mechanism for Cytochrome P450 Metabolism of Type II Binding Compounds: Evidence Supporting Direct Reduction. Arch. Biochem. Biophys. 2011, 511, 69–79. [Google Scholar] [CrossRef]
- Deodhar, M.; Al Rihani, S.B.; Arwood, M.J.; Darakjian, L.; Dow, P.; Turgeon, J.; Michaud, V. Mechanisms of CYP450 Inhibition: Understanding Drug-Drug Interactions Due to Mechanism-Based Inhibition in Clinical Practice. Pharmaceutics 2020, 12, 846. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Inhibition of Cytochrome P450 Enzymes by Drugs-Molecular Basis and Practical Applications. Biomol. Ther. (Seoul) 2022, 30, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Minerdi, D.; Sadeghi, S.J.; Di Nardo, G.; Rua, F.; Castrignanò, S.; Allegra, P.; Gilardi, G. CYP116B5: A New Class VII Catalytically Self-sufficient Cytochrome P450 from Acinetobacter radioresistens That Enables Growth on Alkanes. Mol. Microbiol. 2015, 95, 539–554. [Google Scholar] [CrossRef] [PubMed]
- Eser, B.E.; Zhang, Y.; Zong, L.; Guo, Z. Self-Sufficient Cytochrome P450s and Their Potential Applications in Biotechnology. Chin. J. Chem. Eng. 2021, 30, 121–135. [Google Scholar] [CrossRef]
- Ciaramella, A.; Catucci, G.; Di Nardo, G.; Sadeghi, S.J.; Gilardi, G. Peroxide-Driven Catalysis of the Heme Domain of A. radioresistens Cytochrome P450 116B5 for Sustainable Aromatic Rings Oxidation and Drug Metabolites Production. New Biotechnol. 2020, 54, 71–79. [Google Scholar] [CrossRef]
- Groves, J.T. Key Elements of the Chemistry of Cytochrome P-450: The Oxygen Rebound Mechanism. J. Chem. Educ. 1985, 62, 928. [Google Scholar] [CrossRef]
- Sarkar, M.R.; Houston, S.D.; Savage, G.P.; Williams, C.M.; Krenske, E.H.; Bell, S.G.; De Voss, J.J. Rearrangement-Free Hydroxylation of Methylcubanes by a Cytochrome P450: The Case for Dynamical Coupling of C-H Abstraction and Rebound. J. Am. Chem. Soc. 2019, 141, 19688–19699. [Google Scholar] [CrossRef]
- Correddu, D.; Catucci, G.; Giuriato, D.; Di Nardo, G.; Ciaramella, A.; Gilardi, G. Catalytically Self-sufficient CYP116B5: Domain Switch for Improved Peroxygenase Activity. Biotechnol. J. 2023, 18, 2200622. [Google Scholar] [CrossRef]
- Munro, A.W.; Leys, D.G.; McLean, K.J.; Marshall, K.R.; Ost, T.W.B.; Daff, S.; Miles, C.S.; Chapman, S.K.; Lysek, D.A.; Moser, C.C.; et al. P450 BM3: The Very Model of a Modern Flavocytochrome. Trends Biochem. Sci. 2002, 27, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Peisach, J.; Blumberg, W.E. Electron Paramagnetic Resonance Study of the High- and Low-Spin Forms of Cytochrome P-450 in Liver and in Liver Microsomes from a Methylcholanthrene-Treated Rabbit. Proc. Natl. Acad. Sci. USA 1970, 67, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Lipscomb, J.D. Electron Paramagnetic Resonance Detectable States of Cytochrome P-450cam. Biochemistry 1980, 19, 3590–3599. [Google Scholar] [CrossRef] [PubMed]
- Davydov, R.; Matsui, T.; Fujii, H.; Ikeda-Saito, M.; Hoffman, B.M. Kinetic Isotope Effects on the Rate-Limiting Step of Heme Oxygenase Catalysis Indicate Concerted Proton Transfer/Heme Hydroxylation. J. Am. Chem. Soc. 2003, 125, 16208–16209. [Google Scholar] [CrossRef]
- Aldag, C.; Gromov, I.A.; García-Rubio, I.; von Koenig, K.; Schlichting, I.; Jaun, B.; Hilvert, D. Probing the Role of the Proximal Heme Ligand in Cytochrome P450cam by Recombinant Incorporation of Selenocysteine. Proc. Natl. Acad. Sci. USA 2009, 106, 5481–5486. [Google Scholar] [CrossRef]
- Rittle, J.; Green, M.T. Cytochrome P450 Compound I: Capture, Characterization, and C-H Bond Activation Kinetics. Science (1979) 2010, 330, 933–937. [Google Scholar] [CrossRef]
- Maurelli, S.; Chiesa, M.; Giamello, E.; Di Nardo, G.; Ferrero, V.E.V.; Gilardi, G.; Van Doorslaer, S. Direct Spectroscopic Evidence for Binding of Anastrozole to the Iron Heme of Human Aromatase. Peering into the Mechanism of Aromatase Inhibition. Chem. Commun. 2011, 47, 10737. [Google Scholar] [CrossRef]
- Lockart, M.M.; Rodriguez, C.A.; Atkins, W.M.; Bowman, M.K. CW EPR Parameters Reveal Cytochrome P450 Ligand Binding Modes. J. Inorg. Biochem. 2018, 183, 157–164. [Google Scholar] [CrossRef]
- Greule, A.; Izoré, T.; Machell, D.; Hansen, M.H.; Schoppet, M.; De Voss, J.J.; Charkoudian, L.K.; Schittenhelm, R.B.; Harmer, J.R.; Cryle, M.J. The Cytochrome P450 OxyA from the Kistamicin Biosynthesis Cyclization Cascade Is Highly Sensitive to Oxidative Damage. Front. Chem. 2022, 10, 868240. [Google Scholar] [CrossRef]
- Famulari, A.; Correddu, D.; Di Nardo, G.; Gilardi, G.; Chiesa, M.; García-Rubio, I. EPR Characterization of the Heme Domain of a Self-Sufficient Cytochrome P450 (CYP116B5). J. Inorg. Biochem. 2022, 231, 111785. [Google Scholar] [CrossRef] [PubMed]
- Famulari, A.; Correddu, D.; Di Nardo, G.; Gilardi, G.; Chiesa, M.; García-Rubio, I. CYP116B5hd, a Self-Sufficient P450 Cytochrome: A Dataset of Its Electronic and Geometrical Properties. Data Brief. 2022, 42, 108195. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.P.S. The EPR of Low Spin Heme Complexes Relation of the Τ2g Hole Model to the Directional Properties of the g Tensor, and a New Method for Calculating the Ligand Field Parameters. Biochim. Biophys. Acta (BBA)—Protein Struct. 1977, 491, 137–148. [Google Scholar] [CrossRef]
- Alonso, P.; Martinez, J.; García-Rubio, I. The Study of the Ground State Kramers Doublet of Low-Spin Heminic System Revisited. A Comprehensive Description of the EPR and Mössbauer Spectra. Coord. Chem. Rev. 2007, 251, 12–24. [Google Scholar] [CrossRef]
- García-Rubio, I.; Martínez, J.I.; Picorel, R.; Yruela, I.; Alonso, P.J. HYSCORE Spectroscopy in the Cytochrome b559 of the Photosystem II Reaction Center. J. Am. Chem. Soc. 2003, 125, 15846–15854. [Google Scholar] [CrossRef] [PubMed]
- Vinck, E.; Van Doorslaer, S.; Dewilde, S.; Mitrikas, G.; Schweiger, A.; Moens, L. Analyzing Heme Proteins Using EPR Techniques: The Heme-Pocket Structure of Ferric Mouse Neuroglobin. JBIC J. Biol. Inorg. Chem. 2006, 11, 467–475. [Google Scholar] [CrossRef]
- García-Rubio, I.; Alonso, P.J.; Medina, M.; Martínez, J.I. Hyperfine Correlation Spectroscopy and Electron Spin Echo Envelope Modulation Spectroscopy Study of the Two Coexisting Forms of the Hemeprotein Cytochrome c6 from Anabaena PCC 7119. Biophys. J. 2009, 96, 141–152. [Google Scholar] [CrossRef]
- García-Rubio, I.; Mitrikas, G. Structure and Spin Density of Ferric Low-Spin Heme Complexes Determined with High-Resolution ESEEM Experiments at 35 GHz. JBIC J. Biol. Inorg. Chem. 2010, 15, 929–941. [Google Scholar] [CrossRef]
- Conner, K.P.; Vennam, P.; Woods, C.M.; Krzyaniak, M.D.; Bowman, M.K.; Atkins, W.M. 1,2,3-Triazole–Heme Interactions in Cytochrome P450: Functionally Competent Triazole–Water–Heme Complexes. Biochemistry 2012, 51, 6441–6457. [Google Scholar] [CrossRef]
- Podgorski, M.N.; Harbort, J.S.; Coleman, T.; Stok, J.E.; Yorke, J.A.; Wong, L.-L.; Bruning, J.B.; Bernhardt, P.V.; De Voss, J.J.; Harmer, J.R.; et al. Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases. Biochemistry 2020, 59, 1038–1050. [Google Scholar] [CrossRef]
- Loftfield, R.B.; Eigner, E.A.; Pastuszyn, A.; Lövgren, T.N.; Jakubowski, H. Conformational Changes during Enzyme Catalysis: Role of Water in the Transition State. Proc. Natl. Acad. Sci. USA 1980, 77, 3374–3378. [Google Scholar] [CrossRef] [PubMed]
- Meyer, E. Internal Water Molecules and H-bonding in Biological Macromolecules: A Review of Structural Features with Functional Implications. Protein Sci. 1992, 1, 1543–1562. [Google Scholar] [CrossRef] [PubMed]
- Thellamurege, N.; Hirao, H. Water Complexes of Cytochrome P450: Insights from Energy Decomposition Analysis. Molecules 2013, 18, 6782–6791. [Google Scholar] [CrossRef] [PubMed]
- Shaik, S.; Dubey, K.D. The Catalytic Cycle of Cytochrome P450: A Fascinating Choreography. Trends Chem. 2021, 3, 1027–1044. [Google Scholar] [CrossRef]
- Yadav, S.; Kardam, V.; Tripathi, A.; T G, S.; Dubey, K.D. The Performance of Different Water Models on the Structure and Function of Cytochrome P450 Enzymes. J. Chem. Inf. Model. 2022, 62, 6679–6690. [Google Scholar] [CrossRef] [PubMed]
- Thomann, H.; Bernardo, M.; Goldfarb, D.; Kroneck, P.M.H.; Ullrich, V. Evidence for Water Binding to the Fe Center in Cytochrome P450cam Obtained by 17O Electron Spin-Echo Envelope Modulation Spectroscopy. J. Am. Chem. Soc. 1995, 117, 8243–8251. [Google Scholar] [CrossRef]
- Schweiger, A.; Jeschke, G. Principles of Pulse Electron Paramagnetic Resonance; Oxford University Press: Oxford, UK, 2001; ISBN 0198506341. [Google Scholar]
- Ciaramella, A.; Catucci, G.; Gilardi, G.; Di Nardo, G. Crystal Structure of Bacterial CYP116B5 Heme Domain: New Insights on Class VII P450s Structural Flexibility and Peroxygenase Activity. Int. J. Biol. Macromol. 2019, 140, 577–587. [Google Scholar] [CrossRef]
- LoBrutto, R.; Scholes, C.P.; Wagner, G.C.; Gunsalus, I.C.; Debrunner, P.G. Electron Nuclear Double Resonance of Ferric Cytochrome P450CAM. J. Am. Chem. Soc. 1980, 102, 1167–1170. [Google Scholar] [CrossRef]
- Scholes, C.P.; Falkowski, K.M.; Chen, S.; Bank, J. Electron Nuclear Double Resonance (ENDOR) of Bis(Imidazole) Ligated Low-Spin Ferric Heme Systems. J. Am. Chem. Soc. 1986, 108, 1660–1671. [Google Scholar] [CrossRef]
- Goldfarb, D.; Bernardo, M.; Thomann, H.; Kroneck, P.M.H.; Ullrich, V. Study of Water Binding to Low-Spin Fe(III) in Cytochrome P450 by Pulsed ENDOR and Four-Pulse ESEEM Spectroscopies. J. Am. Chem. Soc. 1996, 118, 2686–2693. [Google Scholar] [CrossRef]
- García-Rubio, I.; Medina, M.; Cammack, R.; Alonso, P.J.; Martínez, J.I. CW-EPR and ENDOR Study of Cytochrome c6 from Anabaena PCC 7119. Biophys. J. 2006, 91, 2250–2263. [Google Scholar] [CrossRef] [PubMed]
- Davydov, R.; Makris, T.M.; Kofman, V.; Werst, D.E.; Sligar, S.G.; Hoffman, B.M. Hydroxylation of Camphor by Reduced Oxy-Cytochrome P450cam: Mechanistic Implications of EPR and ENDOR Studies of Catalytic Intermediates in Native and Mutant Enzymes. J. Am. Chem. Soc. 2001, 123, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Davydov, R.; Hoffman, B.M. Active Intermediates in Heme Monooxygenase Reactions as Revealed by Cryoreduction/Annealing, EPR/ENDOR Studies. Arch. Biochem. Biophys. 2011, 507, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Davydov, R.; Dawson, J.H.; Perera, R.; Hoffman, B.M. The Use of Deuterated Camphor as a Substrate in 1 H ENDOR Studies of Hydroxylation by Cryoreduced Oxy P450cam Provides New Evidence of the Involvement of Compound I. Biochemistry 2013, 52, 667–671. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Davydov, R.; Im, S.; Shanmugam, M.; Gunderson, W.A.; Pearl, N.M.; Hoffman, B.M.; Waskell, L. Role of the Proximal Cysteine Hydrogen Bonding Interaction in Cytochrome P450 2B4 Studied by Cryoreduction, Electron Paramagnetic Resonance, and Electron–Nuclear Double Resonance Spectroscopy. Biochemistry 2016, 55, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Dikanov, S.A.; Bowman, M.K. Cross-Peak Lineshape of Two-Dimensional ESEEM Spectra in Disordered S = 1/2, I = 1/2 Spin Systems. J. Magn. Reson. A 1995, 116, 125–128. [Google Scholar] [CrossRef]
- Dikanov, S.A.; Tyryshkin, A.M.; Bowman, M.K. Intensity of Cross-Peaks in Hyscore Spectra of S = 1/2, I = 1/2 Spin Systems. J. Magn. Reson. 2000, 144, 228–242. [Google Scholar] [CrossRef]
- Verras, A.; Alian, A.; Montellano, P.R.O.D. Cytochrome P450 Active Site Plasticity: Attenuation of Imidazole Binding in Cytochrome P450cam by an L244A Mutation. Protein Eng. Des. Sel. 2006, 19, 491–496. [Google Scholar] [CrossRef]
- Podgorski, M.N.; Harbort, J.S.; Lee, J.H.Z.; Nguyen, G.T.H.; Bruning, J.B.; Donald, W.A.; Bernhardt, P.V.; Harmer, J.R.; Bell, S.G. An Altered Heme Environment in an Engineered Cytochrome P450 Enzyme Enables the Switch from Monooxygenase to Peroxygenase Activity. ACS Catal. 2022, 12, 1614–1625. [Google Scholar] [CrossRef]
- Vinck, E.; Van Doorslaer, S. Analysing Low-Spin Ferric Complexes Using Pulse EPR Techniques: A Structure Determination of Bis (4-Methylimidazole)(Tetraphenylporphyrinato)Iron(Iii). Phys. Chem. Chem. Phys. 2004, 6, 5324. [Google Scholar] [CrossRef]
- Ashby, C.I.H.; Cheng, C.P.; Brown, T.L. Nitrogen-14 Nuclear Quadrupole Resonance Spectra of Coordinated Imidazole. J. Am. Chem. Soc. 1978, 100, 6057–6063. [Google Scholar] [CrossRef]
- Griffith, J.S. Theory of E.P.R. in Low-Spin Ferric Haemoproteins. Mol. Phys. 1971, 21, 135–139. [Google Scholar] [CrossRef]
- Zoppellaro, G.; Bren, K.L.; Ensign, A.A.; Harbitz, E.; Kaur, R.; Hersleth, H.; Ryde, U.; Hederstedt, L.; Andersson, K.K. Review: Studies of Ferric Heme Proteins with Highly Anisotropic/Highly Axial Low Spin (S = 1/2) Electron Paramagnetic Resonance Signals with Bis-Histidine and Histidine-methionine Axial Iron Coordination. Biopolymers 2009, 91, 1064–1082. [Google Scholar] [CrossRef] [PubMed]
- Conner, K.P.; Cruce, A.A.; Krzyaniak, M.D.; Schimpf, A.M.; Frank, D.J.; Ortiz de Montellano, P.; Atkins, W.M.; Bowman, M.K. Drug Modulation of Water–Heme Interactions in Low-Spin P450 Complexes of CYP2C9d and CYP125A1. Biochemistry 2015, 54, 1198–1207. [Google Scholar] [CrossRef] [PubMed]
- Le Breton, N.; Wright, J.J.; Jones, A.J.Y.; Salvadori, E.; Bridges, H.R.; Hirst, J.; Roessler, M.M. Using Hyperfine Electron Paramagnetic Resonance Spectroscopy to Define the Proton-Coupled Electron Transfer Reaction at Fe–S Cluster N2 in Respiratory Complex I. J. Am. Chem. Soc. 2017, 139, 16319–16326. [Google Scholar] [CrossRef]
- Kondo, H.X.; Kanematsu, Y.; Masumoto, G.; Takano, Y. PyDISH: Database and Analysis Tools for Heme Porphyrin Distortion in Heme Proteins. Database 2023, 2023, baaa066. [Google Scholar] [CrossRef] [PubMed]
- Peisach, J.; Mims, W.B.; Davis, J.L. Studies of the Electron-Nuclear Coupling between Fe(III) and 14N in Cytochrome P-450 and in a Series of Low Spin Heme Compounds. J. Biol. Chem. 1979, 254, 12379–12389. [Google Scholar] [CrossRef]
- Magliozzo, R.S.; Peisach, J. Evaluation of Nitrogen Nuclear Hyperfine and Quadrupole Coupling Parameters for the Proximal Imidazole in Myoglobin-Azide, -Cyanide, and -Mercaptoethanol Complexes by Electron Spin Echo Envelope Modulation Spectroscopy. Biochemistry 1993, 32, 8446–8456. [Google Scholar] [CrossRef]
- Garcia, M.L.S.; Smith, J.A.S.; Bavin, P.M.G.; Ganellin, C.R. 14N and 2H Quadrupole Double Resonance in Substituted Imidazoles. J. Chem. Soc. Perkin Trans. 1983, 2, 1391–1399. [Google Scholar] [CrossRef]
- Hsieh, Y.-N.; Rubenacker, G.V.; Cheng, C.P.; Brown, T.L. Nitrogen-14 Nuclear Quadrupole Resonance Spectra of Coordinated Pyridine. J. Am. Chem. Soc. 1977, 99, 1384–1389. [Google Scholar] [CrossRef]
- Omura, T.; Sato, R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. II. Solubilization, Purification, and Properties. J. Biol. Chem. 1964, 239, 2379–2385. [Google Scholar] [CrossRef] [PubMed]
- Höfer, P.; Grupp, A.; Nebenführ, H.; Mehring, M. Hyperfine Sublevel Correlation (Hyscore) Spectroscopy: A 2D ESR Investigation of the Squaric Acid Radical. Chem. Phys. Lett. 1986, 132, 279–282. [Google Scholar] [CrossRef]
- Bodenhausen, G.; Kogler, H.; Ernst, R.R. Selection of Coherence-Transfer Pathways in NMR Pulse Experiments. J. Magn. Reson. (1969) 1984, 58, 370–388. [Google Scholar] [CrossRef][Green Version]
- Mims, W.B. Pulsed Endor Experiments. Proc. R. Soc. Lond. A Math. Phys. Sci. 1965, 283, 452–457. [Google Scholar] [CrossRef]
- Stoll, S.; Schweiger, A. EasySpin, a Comprehensive Software Package for Spectral Simulation and Analysis in EPR. J. Magn. Reson. 2006, 178, 42–55. [Google Scholar] [CrossRef]

| Sample | gz | gy | gz | V/ξ | ∆/ξ | |
|---|---|---|---|---|---|---|
| CYP116B5hd in D2O | 2.443 ± 0.005 | 2.253 ± 0.002 | 1.923 ± 0.002 | 4.74 ± 0.06 | 5.44 ± 0.18 | |
| CYP116B5hd in H2O [32] | 2.440 ± 0.005 | 2.250 ± 0.002 | 1.920 ± 0.002 | 4.74 ± 0.06 | 5.44 ± 0.18 | |
| CYP116B5hd in H2O + 15N2-imidazole | (2) | 2.466 ± 0.005 | 2.258 ± 0.002 | 1.902 ± 0.002 | 3.47 ± 0.03 | 5.11 ± 0.17 |
| (1) | 2.589 ± 0.005 | 2.258 ± 0.002 | 1.857 ± 0.002 | 4.40 ± 0.05 | 5.13 ± 0.17 | |
| CYP116B5hd in H2O + Imidazole [32] | (2) | 2.468 ± 0.005 | 2.258 ± 0.002 | 1.902 ± 0.002 | 3.50 ± 0.03 | 5.13 ± 0.17 |
| (1) | 2.585 ± 0.005 | 2.258 ± 0.002 | 1.860 ± 0.002 | 4.39 ± 0.05 | 5.14 ± 0.17 |
| Species | Label | Ax [MHz] | Ay [MHz] | Az [MHz] | α, β, γ [°] | Qx [MHz] | Qy [MHz] | Qz [MHz] | α′, β′, γ′ [°] |
|---|---|---|---|---|---|---|---|---|---|
| Heme | N1 | −4.90 ± 0.1 | −4.80 ± 0.1 | −5.10 ± 0.1 | 90 ± 5, 0 ± 5, 0 ± 5 | 0.90 ± 0.10 | −0.60 ± 0.10 | −0.30 ± 0.10 | 90 ± 5, 0 ± 5, 0 ± 5 |
| N2 | −4.80 ± 0.1 | −4.70 ± 0.1 | −5.80 ± 0.1 | 0 ± 5, 0 ± 5, 0 ± 5 | 1.00 ± 0.10 | −0.60 ± 0.10 | −0.40 ± 0.10 | 0 ± 5, 0 ± 5, 0 ± 5 | |
| Imidazole (2) | N3 | −3.57 0.1 | −3.20 ± 0.1 | −2.54 ± 0.1 | 65 ± 5, 0 ± 5, 0 ± 5 | 0.30 ± 0.10 | 0.80 ± 0.10 | −1.10 ± 0.10 | 65 ± 5, 0 ± 5, 0 ± 5 |
| Imidazole (1) | N3 | −3.57 0.1 | −3.20 ± 0.1 | −2.54 ± 0.1 | 30 ± 5, 0 ± 5, 0 ± 5 | 0.30 ± 0.10 | 0.80 ± 0.10 | −1.10 ± 0.10 | 30 ± 5, 0 ± 5, 0 ± 5 |
| Imidazole-15N2 | N4 | 5.00 ± 0.1 | 4.48 ± 0.1 | 3.56 ± 0.1 | 65 ± 5, 0 ± 5, 0 ± 5 | N/A | N/A | N/A | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Famulari, A.; Correddu, D.; Di Nardo, G.; Gilardi, G.; Mitrikas, G.; Chiesa, M.; García-Rubio, I. Heme Spin Distribution in the Substrate-Free and Inhibited Novel CYP116B5hd: A Multifrequency Hyperfine Sublevel Correlation (HYSCORE) Study. Molecules 2024, 29, 518. https://doi.org/10.3390/molecules29020518
Famulari A, Correddu D, Di Nardo G, Gilardi G, Mitrikas G, Chiesa M, García-Rubio I. Heme Spin Distribution in the Substrate-Free and Inhibited Novel CYP116B5hd: A Multifrequency Hyperfine Sublevel Correlation (HYSCORE) Study. Molecules. 2024; 29(2):518. https://doi.org/10.3390/molecules29020518
Chicago/Turabian StyleFamulari, Antonino, Danilo Correddu, Giovanna Di Nardo, Gianfranco Gilardi, George Mitrikas, Mario Chiesa, and Inés García-Rubio. 2024. "Heme Spin Distribution in the Substrate-Free and Inhibited Novel CYP116B5hd: A Multifrequency Hyperfine Sublevel Correlation (HYSCORE) Study" Molecules 29, no. 2: 518. https://doi.org/10.3390/molecules29020518
APA StyleFamulari, A., Correddu, D., Di Nardo, G., Gilardi, G., Mitrikas, G., Chiesa, M., & García-Rubio, I. (2024). Heme Spin Distribution in the Substrate-Free and Inhibited Novel CYP116B5hd: A Multifrequency Hyperfine Sublevel Correlation (HYSCORE) Study. Molecules, 29(2), 518. https://doi.org/10.3390/molecules29020518