Abstract
The presented study depicts the synthesis of 11 carborane–thiazole conjugates with anticancer activity, as well as an evaluation of their biological activity as inhibitors of two enzymes: tyrosinase and 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1). The overexpression of tyrosinase results in the intracellular accumulation of melanin and can be observed in melanoma. The overexpression of 11β-HSD1 results in an elevation of glucocorticoid levels and has been associated with the aggravation of metabolic disorders such as type II diabetes mellitus and obesity. Recently, as the comorbidity of melanomas and metabolic disorders is being recognized as an important issue, the search for new therapeutic options has intensified. This study demonstrates that carborane–thiazole derivatives inhibit both enzymes, exerting beneficial effects. The antiproliferative action of all newly synthesized compounds was evaluated using three cancer cell lines, namely A172 (human brain glioblastoma), B16F10 (murine melanoma) and MDA-MB-231 (human breast adenocarcinoma), as well as a healthy control cell line of HUVEC (human umbilical vein endothelial cells). The results show that 9 out of 11 newly synthesized compounds demonstrated similar antiproliferative action against the B16F10 cell line to the reference drug, and three of these compounds surpassed it. To the best of our knowledge, this study is the first to demonstrate dual inhibitory action of carborane–thiazole derivatives against both tyrosinase and 11β-HSD1. Therefore, it represents the first step towards the simultaneous treatment of melanoma and comorbid diseases such as type II diabetes mellitus.
1. Introduction
Cancer is one of the main causes of morbidity and mortality worldwide and was responsible for approximately 9.7 million deaths in 2022 [1]. According to estimates by Global Cancer Data (GLOBOCAN), in 2022, there were about 20 million new cancer cases, and by 2050, a total of 35 million new cases will be diagnosed per year [2]. Currently, the most frequently diagnosed cancer is breast cancer in women, which accounts for 11.7% of new cases, followed by lung (11.4%), colorectal (10.0%), prostate (7.3%), and gastric (5.6%) cancer [2]. Compared to the abovementioned cancers, melanoma—an aggressive type of skin cancer—is less often diagnosed (estimated as 5% of all new cases in 2024 [3]), but due to its rapid growth and potential to spread to any organ, it is a serious health threat. The increased activity of tyrosinase in this type of cancer leads to the accumulation of melanin in the cells, which protects it from radiotherapy due to the high capacity of melanin to absorb radiation [4]. The antioxidant properties of melanin may reduce the effectiveness of chemotherapy, and the intermediates formed in the melanogenesis process, such as 3,4-dihydroxy-L-phenylalanine (L-DOPA) and quinones, may also have an immunosuppressive effect, which makes the applied immunotherapy ineffective. A possible solution to this problem is the use of tyrosinase inhibitors, which inhibit the conversion of L-tyrosine to L-DOPA and then to L-DOPA-quinone, which is one of the precursors of melanin [5].
Diabetes is now one of the major diseases worldwide, and the high prevalence of the main risk factors for developing diabetes, namely obesity, a poor diet, and physical inactivity, causes the further growth of the number of diagnosed cases. It is estimated that 463 million people had diabetes in 2019, and this number could rise to 578 million in 2030 and up to 700 million in 2045 [6]. Type II diabetes mellitus (T2DM), which accounts for about 90% of cases, is associated with the progressive impairment of insulin secretion by pancreatic β-cells and the development of peripheral tissue resistance to insulin (insulin resistance) and consequently elevated blood glucose levels [7]. Chronic hyperglycaemia and the associated dysfunction of protein, lipid, and carbohydrate metabolism lead to numerous microvascular and macrovascular disorders, such as neuropathies, retinopathies, nephropathies and atherosclerosis, which worsen patients’ quality of life and also increase the risk of other serious diseases [8].
11β-Hydroxysteroid dehydrogenases (11β-HSD) are prereceptor enzymes that regulate the intracellular ability of active glucocorticoids (GC) to bind and activate the glucocorticoid receptor (GR). There are two types (isoforms): 11β-HSD1 and 11β-HSD2. They are products of two different genes, are present in different tissues, and have different physiological roles. 11β-HSD2, the first to be discovered, converts active glucocorticoids, such as cortisol (in humans) or corticosterone (in mice/rats), to inactive forms, such as cortisone (in humans) or 11-dihydrocorticosterone (DHC in rodents). The second enzyme isoform, 11β-HSD1, occurs in key metabolic tissues such as the liver, adipose tissue, and muscles, and works bidirectionally—it is capable of acting as both a reductase (activating GC) and dehydrogenase (deactivating GC) [9]. However, in intact cells such as hepatocytes and adipocytes, 11β-HSD1 has a higher affinity for cortisone than for cortisol. Excess glucocorticoids lead to abdominal obesity, insulin resistance, dyslipidemia, and hypertension, which are the most commonly recognized risk factors for metabolic syndrome [10]. Lowering the level of glucocorticoids may be a potential therapy for insulin resistance and other disorders in patients with T2DM. The inhibition of 11β-HSD1 activity causes a decrease in glucocorticoid levels. In consequence, it may result in a decrease in body fat mass, a decrease in glucose levels in the blood of patients with T2DM, and a reduction in the total cholesterol level [11].
An increasing number of scientific studies indicate that cancer and diabetes mellitus are commonly coexisting illnesses. Postmenopausal breast cancer, endometrial cancer, and colorectal cancer closely correlate with diabetes [12]. Recent studies also suggest that type II diabetes mellitus is associated with increased cutaneous melanoma aggressiveness at diagnosis [13]. Also, evidence suggests that cancer and diabetes patients have a higher mortality rate, so a multi-pronged approach is important in treating patients.
Previous studies on the search for tyrosinase and selective 11β-HSD1 inhibitors, including our group’s research, were directed towards several groups of compounds, i.e., flavones, coumarins, chalcones, thiosemicarbazones, thiazole, kojic acid derivatives, cyreneTM, 2-(cyclopentylamino)thiazol-4(5H)-one, 2-amino-1,3-thiazol-4(5H)-ones, and thiazolo[3,2-a]pyrimidine-5-one derivatives [14,15,16,17,18,19,20]. However, to the best of our knowledge, carborane derivatives have never been tested as tyrosinase and 11β-HSD1 inhibitors.
Carboranes (dicarba-closo-dodecaboranes, C2H12B10) belong to the class of boron clusters in which BH vertices are substituted with neutral CH groups [21]. The interest in the use of carboranes in different areas of medicinal chemistry has grown over the years due to their properties including their abiotic origin, unique interaction properties with biomolecules, hydrophobicity or amphiphilicity, nearly spherical geometry, established chemistry, chemical stability and biological stability in vivo, resistance to catabolism, and low toxicity [22,23]. So far, many compounds bearing carborane clusters have revealed promising activity in several fields, such as in boron neutron capture therapy (BNCT) and the synthesis of nucleosides [24], amino acids [25], and potent antiviral [26], antimicrobial [27,28], and antiparasitic agents [29]. Biological enzymes have also been targeted using carborane derivatives [22,23]. Such conjugates show promising activity against cancer-associated carbonic anhydrase IX (CA IX) [30], hypoxia-inducible factor-1 (HIF-1) [31,32], nicotinamide phosphoribosyltransferase (NAMPT) [33], cyclooxygenase (COX) [34,35], lipoxygenase (LO) [36,37], and indoleamine-2,3-dioxygenase-1 (IDO1) [38].
Considering the abovementioned findings, we decided to design and synthesize hybrid structures incorporating both the carborane moiety and thiazole ring, the latter being a very good scaffold for obtaining antityrosinase activity, as confirmed by numerous studies by our group [39,40,41]. Additionally, the use of electron-donating, electron-withdrawing, and bulky character substituents enabled us to modify the hydrophilic and hydrophobic properties of the designed molecules. Next, the antiproliferative activity of these compounds against three human cancer cells lines, namely brain glioblastoma A172, murine melanoma B16F10, and human breast adenocarcinoma MDA-MB-231, as well as healthy human umbilical vein endothelial cells (HUVEC), was evaluated. To recognize the possible mechanism of action of the products, the tyrosinase inhibitory potential with an inhibitory mechanism was assessed, followed by molecular docking studies. Finally, we tested the inhibitory potential of our compounds against 11β-HSD1 and 11β-HSD2.
2. Results and Discussion
2.1. Chemistry
The synthesis of the title compounds was accomplished following the well-known Hantzsch cyclization reaction. In the first step, C-formyl-o-carborane (2) was obtained by means of the formylation reaction of o-carborane (1) through treatment with n-BuLi at −78 °C and the subsequent addition of methyl formate. Hydrazonecarbothioamide 3 was synthesized with 89% yield through condensation of C-formyl-o-carborane (2) with thiosemicarbazide in absolute ethyl alcohol containing a catalytic amount of glacial acetic acid. In the next step, cyclization of hydrazonecarbothioamide 3 with a variety of substituted bromoacetophenones in ethanolic solution and under reflux produced 2-(2-(1,2-dicarba-closo-dodecaboranylmethylene)hydrazinyl)thiazole 4a–4k with a high yield (60–99%). The reaction pathway is summarized in Scheme 1. All compounds were fully characterized spectroscopically using 1H, 13C, and 11B NMR and ESI-HRMS analysis. The 1H NMR spectrum of hydrazonecarbothioamide (3) showed three characteristic peaks derived from NH2 and NH groups at 8.08, 8.49, and 11.58 ppm, respectively. Carborane–thiazoles showed characteristic singlets derived from the CH group of the o-carborane moiety at 5.35–5.45 ppm and from the NH proton at about 12.17–12.60 ppm, whereas in the 13C NMR spectrum, we observe characteristic peaks at about 101–109 ppm and at about 170 ppm derived from C=N group.
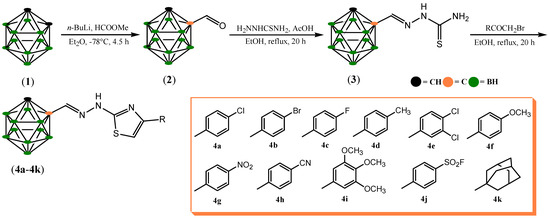
Scheme 1.
Synthesis of the carborane–thiazoles 4a–4k.
2.2. Antiproliferative Activity
The antiproliferative activity of all newly synthesized carborane–thiazole derivates was evaluated on three cancer cell lines, namely A172 (human brain glioblastoma), B16F10 (murine melanoma) and MDA-MB-231 (human breast adenocarcinoma), as well as a healthy control cell line, HUVEC (human umbilical vein endothelial cells). As presented in Table 1, particularly good results were observed for the carborane–thiazole derivatives numbered 4a–4e, 4g, and 4j. For all of these compounds, the selectivity index (SI) based on the ratios of recorded IC50 values was favorable. That is, the concentrations of inhibitors required to prevent proliferation of cancer cell lines were notably lower than the concentrations required to prevent the growth of healthy cell line (above 1.0). The following was also observed, albeit not in all cases for compounds 4f, 4h, and 4i. In these cases, for compound 4f, favorable SI values were observed only for the B16F10 and MDA-MB-231 cell lines, while for compounds 4h and 4i, they were only observed for the B16F10 cell line. On the contrary, unfavorable results were observed for compound 4k, which was not effective against any of the cancer cell lines.

Table 1.
Effect of newly synthetized carborane–thiazole derivatives on the proliferation of human brain glioblastoma (A172), murine melanoma (B16F10), human breast adenocarcinoma (MDA-MB-231), and human umbilical vein endothelial cells (HUVEC) (shown as the IC50 values).
Considering the effects of investigated compounds on particular cell lines, the most favorable SI values were observed for the murine melanoma cell line. Therefore, the evaluated compounds are most suitable for the treatment of this cancer type. Moreover, the activity of all newly synthesized compounds (except for 4d and 4k) is comparable to the activity of the reference inhibitor of chlorambucil. Especially favorable were some of the recorded IC50 values, which surpassed the results observed for the reference inhibitor. Such was the case with compounds 4a (5.03 ± 0.69 µM), 4b (5.37 ± 0.11 µM), and 4g (5.11 µM). Finally, the results of this study showed that the activity of the newly synthesized compounds was similar against all of the evaluated cancer cell lines, with all recorded IC50 values in similar ranges: for A172, from 5.77 ± 0.96 to 7.88 ± 0.98 µM; for B16F10, from 5.03 ± 0.69 to 5.88 µM; and for MDA-MB-231, from 6.02 ± 0.08 to 7.32 ± 1.26 µM. This is an especially beneficial trait, as it suggests universal applicability of the evaluated compounds.
The SAR analysis showed that the differences in activity between individual substituents were small, at around 2 µM. However, it can be easily seen that (-3,4,5-tri-OCH3) and adamantyl substituents drastically reduce the antiproliferative activity, most likely due to their bulky structure and high lipophilicity, especially for the compound 4k containing an adamantyl substituent.
2.3. Mushroom Tyrosinase Inhibitory Effect and Kinetic Analysis of Compounds
The studies of inhibition activity against the mushroom tyrosinase and enzyme inhibition kinetics were performed for all synthesized compounds. The recorded IC50 values (shown in Table 2) ranged from 11.27 ± 5.90 µM for compound 4h to 360.51 ± 118.59 µM for compound 4j. After compound 4h, the second most effective compound was 4f with an IC50 value of 49.36 ± 8.01 µM; however, it appears to be a tyrosinase activator.

Table 2.
Inhibition activity towards the mushroom tyrosinase and enzyme inhibition kinetics of the evaluated compounds.
Based on the Lineweaver–Burk double reciprocal plots presented in Figure 1, the inhibition mode for these two compounds was established as mixed type inhibition. Furthermore, out of all of the investigated compounds, 4h and 4f were the only ones determined to be more active against mushroom tyrosinase than the reference inhibitor of kojic acid. However, it should be stressed that the gold standard of mushroom tyrosinase inhibition is displayed by ascorbic acid rather than kojic acid. Remarkably, all of the synthesized compounds surpassed this widely established reference as enzyme inhibitors.
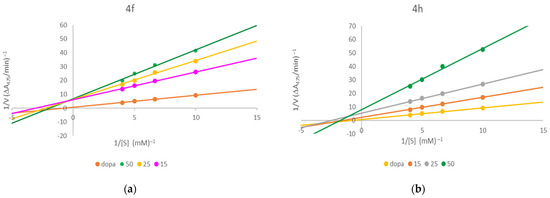
Figure 1.
Lineweaver–Burk plots for tyrosinase inhibition for compounds 4f (a) and 4h (b).
Summarizing the structure–activity relationship (SAR) findings, it can be seen that the type of halogen substituent, i.e., electron-withdrawing substituents (-Cl, -Br, -F), did not cause significant differences in the tyrosinase inhibition capacity of compounds 4a–4c (IC50 = 130–144 mM). Also, the addition of a second chlorine atom did not lead to increased activity (compound 4e (IC50 = 142 mM)). However, compound 4h, containing an electron-withdrawing group –CN, showed the highest inhibitory activity (IC50 = 11 mM). This can be explained by the linear shape of this group, the strong electronegativity of the nitrogen atom, and the most favorable binding mode with the Cu ions present in the protein structure. The molecular docking confirms that this group has the lowest binding energy with tyrosinase (−7.9 kcal/mol). Compounds with electron-donating (-CH3) and bulky (adamantyl) groups showed similar activities (4d (IC50 = 121 mM) and 4k (IC50 = 134 mM), respectively). Superior activity is shown by compound 4f (IC50 = 49 mM) containing an electron-donating (-OCH3) group. At the same time, compounds containing the (-OCH3) or (-CN) ligands display the highest values of binding energy (−7.3–−7.9 kcal/mol).
2.4. Inhibition of 11β-HSD1
The seven derivatives were tested for their inhibition of 11β-hydroxysteroid dehydrogenase type 1. The inhibition of the dehydrogenase activity was determined as a percentage of the inhibition at the concentration of potential inhibitor of 10 μM. All tested derivatives showed inhibitory activity in the range of 55.54 to 73.51% at a concentration of 10 μM (see Table 3). The IC50 for all analyzed compounds was lower than 10 µM (in the range 3.90–9.35 µM). The known 11β-HSD1 inhibitor, carbenoxolone, is characterized by a higher percent of inhibition of this enzyme activity and a lower IC50 value. Furthermore, the analyzed compounds are not selective. They also inhibit, albeit to a lesser extent (in the range from 19.34 to 32.55% at a concentration of 10 μM), the activity of isoform 2 of the enzyme (11β-HSD2). However, carborane derivatives are a group of compounds that have not been previously tested in the literature for 11β-HSD1 inhibition. Therefore, the fact that all analyzed compounds showed significant inhibitory activity in relation to this enzyme isoform, and less activity in relation to isoform 2, demonstrates the possibility of searching for a selective 11β-HSD1 inhibitor in this group of compounds.

Table 3.
Inhibitory activity against the 11β-hydroxysteroid dehydrogenases type 1 and type 2 of the evaluated compounds.
Summarizing the structure–activity relationship (SAR), it can be concluded that the obtained values of inhibition of 11β-HSD1 are not very dependent on the type of substituent and are very similar to each other. However, the best activity is observed for the compounds with electron-withdrawing substituents (-Cl, -Br, -NO2) for compounds 4a, 4b and 4g (IC50 = 6.90–7.05 mM), while the second group, containing substituents such as -F and -diCl, including compounds 4c and 4e (IC50 = 7.90–8.10), is characterized by slightly lower activity, probably due to the high electronegativity of these substituents. However, the lowest activity was shown by compounds 4d and 4f containing substituents with electron-donating groups (-CH3, -OCH3), with IC50 = 9.05 mM and 9.35 mM, respectively.
2.5. Molecular Docking Study
The binding energies found during docking simulations are given in Table 4 and graphically illustrated in Figure 2. In the case of tyrosinase, apart from the two ligands that do not belong to the o-carborane family (i.e., ascorbic acid and kojic acid), the relatively limited magnitude of the obtained binding energies can be noticed (−6.2–−7.9 kcal/mol). The strongest binding is exhibited by 4h, whereas the weakest is displayed by 4i and 4j, which correctly reflects the limiting behavior found in the experimental studies. More quantitative analysis reveals that the binding energies calculated for o-carborane derivatives are strongly correlated with the experimentally determined values of ln(IC50) (see Figure 2). The ascorbic acid and kojic acid do not follow this trend, probably due to divergent molecular topologies when compared to the o-carborane ligands, which enhances the possible docking-related errors. Nevertheless, the docking results still correctly predict the weakest potency of ascorbate ions in relation to all remaining ligands. In the case of 11β-HSD1, the scatter of the obtained binding energy values is even narrower (−11.0–−10.6 kcal/mol), which partially results from the smaller pool of considered compounds. As in the previous case, the theoretically determined binding energies correlate reasonably well with the experimental ln(IC50) values, including the correct predictions of the most (4a, 4b and 4g) and least potent (4f) compounds. However, one can note that this apparently good correlation is a consequence of slightly distinct binding energies obtained for 4f and 4d, whereas the remaining compounds display extremely similar affinities (see Table 4). Carbenoxolone, exhibiting a unique molecular structure in comparison to remaining ligands, differs also with respect to the magnitude of determined binding energy (Table 4) and does not follow the above-described correlation.

Table 4.
The binding energies recovered during docking with respect to the studied ligands. The calculation results were averaged over 4 or 5 structures available for the given type of protein; the corresponding standard deviations are given.
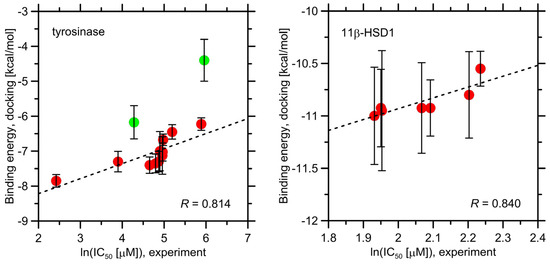
Figure 2.
The correlation between the binding energies calculated during docking studies for a series of ligands to the two studied proteins and the experimental values of ln(IC50). The vertical bars denote the standard deviation values calculated with respect to various crystal structures of the same protein. The ligands containing the o-carborane moiety are represented by red dots whereas kojic acid and ascorbic acid by green dots. The corresponding results for carbenoxolone are not shown due to magnitude mismatch.
In summary, one can conclude that a reasonably good agreement between the theoretical and experimental, affinity-related results exists.
The results of the docking studies were analyzed with respect to the mechanistic interaction pattern that may be significant in the context of interpretation of the obtained binding energies. The summary given below relies on analyzing the ligand–protein contacts that take place if the distance between any corresponding atom pair is smaller than the arbitrarily accepted value of 0.38 nm. The description provided below concerns (1) in the case of tyrosinase, the most potent compound (4h), and (2) in the case of 11β-HSD1, compound 4a. A similar interaction pattern is seen for the given protein and most of the studied carborane–thiazole derivatives; the exceptions are compounds 4i and 4j interacting with tyrosinase. The graphical illustration of the docking results (on example of 4h and 4a) is given in Figure 3.
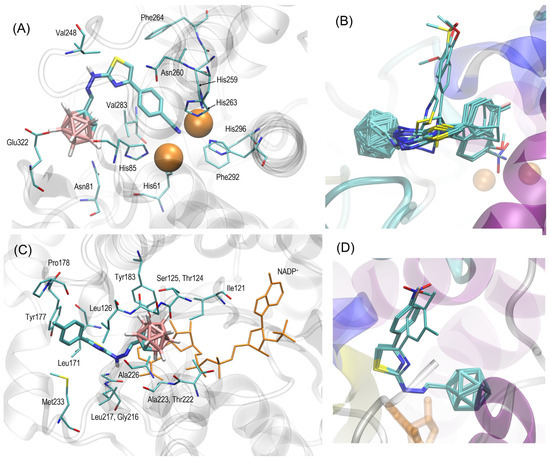
Figure 3.
(A) The location of the 4h ligand molecule bound to the tyrosinase (PDB:2y9x) structure and found in the docking procedure. The ligand molecule is shown as thick sticks, whereas all the closest amino acid residues (of a distance no larger than 0.38 nm) are represented by thin sticks. Copper ions are shown as orange balls. The description of the interaction types is given in the text. (B) The superposition of the most favorable poses of all ligands in the tyrosinase-containing complexes. Compounds 4i and 4j exhibit the orientation in the binding cavity, which is notably different from those of remaining ligands; compounds 4f, 4g, and 4h are located in the closest vicinity of Cu ions present in the protein structure. (C) The location of the 4a ligand molecule bound to the 11β-HSD1 (PDB:3czr) structure and found in the docking procedure. The NADP+ molecule is highlighted in orange. Other details as in (A). (D) The superposition of the most favorable poses of ligands 4a–4g in the 11β-HSD1-containing complexes.
The tyrosinase-containing complexes are considered in the first order. The most favorable binding mode is correlated with the intensive interactions of the nucleophilic substituent of the phenyl group (e.g., the –CN moiety) with the Cu ions present in the protein structure. The corresponding interatomic distances are close to the van der Waals radii; thus, a strong contribution of the electrostatic interactions or, possibly, coordination bonding can be expected to play a role in the ligand–protein interactions. Additionally, the presence of cluster composed of positively charged sidechains of histidines (His259, His263, His61, His85, and His296) favorably influences the interactions of nucleophilic substituents attached to the phenyl ring. The phenyl group interacts with Val283 and Asn260 via the π-stacking interactions involving either the aliphatic hydrogen atoms of Val or the lone electron pairs of Asn. The thiazole ring, being the central part of the ligand molecule, is in contact with Asn260 and Phe264. The latter interactions, driven by the CH–π forces, is characteristic also of other ligands containing a thiazole moiety [39]. Surprisingly, the o-carborane group is not involved in any type of non-polar interactions. Instead, it maintains close contact with the hydrophilic sidechains of Glu322, Asn81, and the backbone fragment of His85. Moreover, this part of the ligand molecule adopts a fixed position across the whole set of docked molecules, indicating the conserved character of the involved interactions.
When considering the complete series of ligands, the same pattern of interactions with tyrosinase as that discussed above can be observed for 4g and 4f, i.e., ligands that, together with 4h, display the highest values of binding energy (−7.3–−7.9 kcal/mol). All of these ligands bear nucleophilic substituents of relatively small dimensions attached to the phenyl ring. The role of the –CN group is played by either the –NO2 or -OMe moieties instead. The ligands with moderate binding energies exhibit a slight reorientation of the phenyl (or aliphatic, in the case of 4k) group, which is moved further from the Cu ions and closer to Phe264. Finally, ligands with the smallest binding energies (4i and 4j) are reoriented in such a way that the phenyl moiety and its substituent are located closer to Phe264 and Val248, whereas the interactions with Cu ions are less intensive due to a much larger (>0.4 nm) ligand–ion distance. We speculate that such drastic reorientation is the result of larger dimensions of the corresponding substituents which cannot be accommodated within the relatively narrow subcavity created by Cu ions and neighboring histidines.
In the case of ligands interacting with 11β-HSD1, the most intensive contacts are observed in the region of the o-carborane group. This group maintains contacts with the aromatic ring and furanose residue of the NADP+ molecule, also present in the 11β-HSD1 structure, as well as with sidechains of Ala223, Ala226, Thr222, Ile121, and Tyr183. Moreover, this interaction also involves backbone fragments of residues 124–126. The chemical character of the abovementioned residues is different, however; the non-polar contacts seem to be preferred, in contrast with the results found for tyrosinase. The amine group of ligands is located close to the backbone carbonyl atoms of Ala226 and Gly216, which indicates the possibility of hydrogen bonding. The thiazole ring interacts via CH–π or π–π forces with Leu171, Leu126, Leu217, and Tyr177. Some parts of these residues (namely Leu126, Tyr177, and Leu171) are also involved in the aromatic ring of the ligand molecule. Moreover, additional amino acids display contacts with this group: Met233 and Pro178. For sterical reasons, it can also be expected that this region of the ligand molecule can interact with water, which is not explicitly present in our system. Overall, the intensity of the interactions in which the ring substituents differing between compounds are involved is rather moderated and mainly includes Tyr177 and Pro178 (and possibly a solvent). This may explain the narrow range within which the binding energies vary. Finally, it is worth noting that the scatter of the ligand position in the binding cavity is much smaller than in the case of tyrosinase; this may result from both the smaller pool of considered compounds but also from the location of the crucial substituent which interacts with the protein less intensively when compared to the remaining parts of the molecule.
3. Materials and Methods
3.1. Chemistry
All reactions were performed under an argon atmosphere. All reagents and starting materials were purchased from commercial suppliers and used without further purification. The dichloromethane was dried over calcium hydride. 1H NMR (400 MHz) and 13C NMR (100 MHz) spectra were recorded on a Bruker Avance III multinuclear instrument. High-resolution mass spectrometry (HRMS) measurements were performed using a Synapt G2-Si mass spectrometer (Waters) equipped with an ESI source and quadrupole-Time-of-Flight mass analyzer. The mass spectrometer was operated in the positive- or negative-ion detection modes. To ensure accurate mass measurements, data were collected in centroid mode, and the mass was corrected during acquisition using a leucine encephalin solution as an external reference, Lock-SprayTM, (Waters Corp., Milford, MA, USA), which generated reference ions at m/z 554.2615 ([M − H]−) in negative-ESI mode and at m/z 556.2771 Da ([M + H]+) in positive-ESI mode. The results of the measurements were processed using the MassLynx 4.1 software (Waters). Melting points were determined in open glass capillaries and were uncorrected. Analytical TLC was performed using Macherey-Nagel Polygram Sil G/UV254 0.2 mm plates. The malonyl dichloride and appropriate thioureas were commercial materials (Merck, Darmstadt, Germany).
3.1.1. C-Formyl-o-Carborane (2)
o-Carborane (144 mg, 1 mmol) was dissolved in dry diethyl ether (3 mL) at room temperature and cooled to −78 °C under an argon atmosphere. To the stirred solution of o-carborane was slowly added n-BuLi (0.69 mL, 1.1 mmol, 1.6 M in hexane) over 10 min and stirring was continued for another 2.5 h at the same temperature. Methyl formate (0.22 mL, 3.6 mmol) was added to the reaction mixture, and stirring was continued for an additional 2 h. The reaction was quenched with dilute HCl (4%, 1.8 mL) at −78 °C and warmed to room temperature. Excess ether was removed under reduced pressure, the aqueous layer was extracted with hexane (3 × 15 mL), and the combined organic layers were dried over MgSO4. Subsequently, the solvent was evaporated to dryness under vacuum, and the product was purified by means of column chromatography on silica gel (230–400 mesh) using 100% hexane and a gradient of CHCl3 in hexane (0–50%) to afford a pure product, yield 0.127 g, 74%, eluent (25% CH2Cl2/hexane − 3×), Rf = 0.52 [42].
3.1.2. 2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinecarbothioamide (3)
Thiosemicarbazide (0.292 g, 3.20 mmol) was added to a stirred solution of C-formyl-o-carborane (2) (0.547 g, 3.20 mmol) in absolute ethyl alcohol (20 mL) under an argon atmosphere and then acetic acid (0.2 mL) was added. The reaction mixture was stirred under reflux for 20 h. The product was filtered off to yield 0.69 g, 89%; mp 195–197 °C; eluent: dichloromethane/methanol (95:15), Rf = 0.58. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.42–2.90 (bs, 10H, 10B-H); 5.83 (bs, 1H, CH-carborane); 7.45 (s, 1H, CH = N); 8.08 (s, 1H, NH); 8.49 (s, 1H, NH); 11.58 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 59.15 (C); 72.32 (C); 133.97 (C); 179.07 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −2.52; −4.00; −9.07; −12.16.
3.1.3. 4-(4-Chlorophenyl)-2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)Thiazole (4a)—Typical Procedure
2-(1,2-Dicarba-closo-dodecaboranylmethylene)hydrazinecarbothioamide (3) (0.086 g, 0.35 mmol) was added under an argon atmosphere to a stirred solution of 2-bromo-1-(4-chlorophenyl)ethanone (0.082 g, 0.35 mmol) in absolute ethyl alcohol (10 mL). The reaction mixture was stirred under reflux for 20 h. Next, the reaction mixture was added to water (50 mL) and neutralized with NaHCO3 solution. The separate precipitate was collected by means of filtration and dried in vacuum to afford the desired product: 0.120 g, 90%; mp 218–220 °C, eluent: dichloromethane/methanol (95:5); Rf = 0.93. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.37–3.09 (bs, 10H, 10B-H); 5.44 (bs, 1H, CH-carborane); 7.42–7.49 (m, 4H, 4CH); 7.84 (d, 2H, 2CH, J = 8.4 Hz); 12.57 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 61.60 (C); 73.16 (C); 106.15 (C); 127.70 (2C); 129.13 (2C); 132.58 (C); 132.94 (C); 133.60 (C); 149.63 (C); 167.99 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −3.13; −9.71; −11.96. ESI-HRMS (m/z) average mass for C12H19B10ClN3S: 382.1951 [M + H]+. Found: 382.1946 [M + H]+.
3.1.4. 4-(4-Bromophenyl)-2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)Thiazole (4b)
Yield: 0.14 g, 93%, (dichloromethane/methanol (95:5), Rf = 0.75); mp 223–225 °C. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.45–3.10 (bs, 10H, 10B-H); 5.44 (bs, 1H, CH-carborane); 7.43 (s, 1H, CH); 7.48 (s, 1H, CH); 7.59 (d, 2H, 2CH, J = 8.8 Hz); 7.78 (d, 2H, 2CH, J = 8.8 Hz); 12.44 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 61.60 (C); 73.15 (C); 106.24 (C); 121.17 (C); 128.02 (2C); 132.05 (2C); 132.97 (C); 133.95 (C); 149.71 (C); 168.01 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −2.52; −4.04; −9.32; −11.05; −12.29. ESI-HRMS (m/z) average mass for C12H19B10BrN3S: 426.1460 [M + H]+. Found: 426.1454 [M + H]+.
3.1.5. 2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)-4-(4-Fluorphenyl)Thiazole (4c)
Yield: 0.12 g, 94%, (dichloromethane/methanol (95:5), Rf = 0.87); mp 237–238 °C. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.43–2.94 (bs, 10H, 10B-H); 5.44 (bs, 1H, CH-carborane); 7.23 (t, 2H, 2CH, J = 9.2 Hz); 7.38 (s, 1H, CH); 7.42 (s, 1H, CH); 7.86 (q, 2H, 2CH, J = 9.2 Hz); 12.42 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 61.63 (C); 73.18 (C); 105.09 (C); 115.94 (d, 2C, JC-F = 21.6 Hz); 128.00 (d, 2C, JC-F = 7.6 Hz); 131.34 (d, C, JC-F = 2.9 Hz); 132.90 (C); 149.79 (C); 162.13 (d, C, JC-F = 243.8 Hz); 167.96 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −3.19; −9.83; −12.51. ESI-HRMS (m/z) average mass for C12H19B10FN3S: 366.2235 [M + H]+. Found: 366.2230 [M + H]+.
3.1.6. 2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)-4-p-Tolylthiazole (4d)
Yield: 0.10 g, 79%, (dichloromethane/methanol (95:5), Rf = 0.85); mp 249–251 °C. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.42–3.06 (bs, 10H, 10B-H); 2.31 (s, 3H, CH3); 5.44 (bs, 1H, CH-carborane); 7.20 (d, 2H, 2CH, J = 8.4 Hz); 7.30 (s, 1H, CH); 7.43 (s, 1H, CH); 7.71 (d, 2H, 2CH, J = 8.4 Hz); 12.42 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 21.26 (C); 61.64 (C); 73.24 (C); 104.36 (C); 125.97 (2C); 129.67 (2C); 132.03 (C); 132.84 (C); 137.51 (C); 150.72 (C); 167.85 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −2.52; −4.00; −9.20; −11.19; −12.46. ESI-HRMS (m/z) average mass for C13H22B10N3S: 362.2487 [M + H]+. Found: 362.2484 [M + H]+.
3.1.7. 2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)-4-(3,4-Dichlorophenyl)Thiazole (4e)
Yield: 0.12 g, 83%, (dichloromethane/methanol (95:5), Rf = 0.74); mp 239–241 °C. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.49–2.99 (bs, 10H, 10B-H); 5.45 (bs, 1H, CH-carborane); 7.45 (s, 1H, CH); 7.62 (s, 1H, CH); 7.66 (d, 1H, CH, J = 8.4 Hz); 7.81 (dd, 1H, CH, J1 = 2.0 Hz J2 = 8.4 Hz); 8.05 (d, 1H, CH, J = 2.4 Hz); 12.52 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 61.57 (C); 73.10 (C); 107.60 (C); 126.03 (C); 127.62 (C); 130.36 (C); 131.35 (C); 131.94 (C); 133.16 (C); 135.31 (C); 148.35 (C); 168.11 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −2.48; −3.94; −9.16; −11.07; −12.45. ESI-HRMS (m/z) average mass for C12H18B10Cl2N3S: 416.1567 [M + H]+. Found: 416.1562 [M + H]+.
3.1.8. 2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)-4-(4-Methoxyphenyl)Thiazole (4f)
Yield: 0.13 g, 99%, (dichloromethane/methanol (95:5), Rf = 0.67); mp 228–230 °C. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.70–2.70 (bs, 10H, 10B-H); 3.75 (s, 3H, CH3); 5.41 (bs, 1H, CH-carborane); 6.93 (d, 2H, 2CH, J = 8.4 Hz); 7.18 (s, 1H, CH); 7.40 (s, 1H, CH); 7.72 (d, 2H, 2CH, J = 8.4 Hz); 12.17 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 55.63 (C); 61.63 (C); 73.24 (C); 103.12 (C); 114.52 (2C); 127.38 (2C); 127.48 (C); 132.86 (C); 150.43 (C); 159.43 (C); 167.81 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −2.58; −4.09; −9.21; −11.15; −12.42. ESI-HRMS (m/z) average mass for C13H22B10N3OS: 378.2437 [M + H]+. Found: 378.2429 [M + H]+.
3.1.9. 2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)-4-(4-Nitrophenyl)Thiazole (4g)
Yield: 0.12 g, 88%, (dichloromethane/methanol (95:5), Rf = 0.85); mp 248–249 °C. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.70–2.71 (bs, 10H, 10B-H); 5.43 (bs, 1H, CH-carborane); 7.42 (s, 1H, CH); 7.77 (s, 1H, CH); 8.06 (d, 2H, 2CH, J = 9.1 Hz); 8.25 (d, 2H, 2CH, J = 9.1 Hz); 12.56 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 61.56 (C); 73.05 (C); 110.18 (C); 124.58 (2C); 126.84 (2C); 133.30 (C); 140.73 (C); 146.78 (C); 148.91 (C); 168.31 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −3.12; −9.96; −12.36. ESI-HRMS (m/z) average mass for C12H19B10N4O2S: 393.2181 [M + H]+. Found: 393.2172 [M + H]+.
3.1.10. 4-(2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)Thiazol-4-yl)Benzonitrile (4h)
Yield: 0.10 g, 77%, (dichloromethane/methanol (95:5), Rf = 0.70); mp 244–246 °C. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.69–2.80 (bs, 10H, 10B-H); 5.42 (bs, 1H, CH-carborane); 7.41 (s, 1H, CH); 7.69 (s, 1H, CH); 7.84 (d, 2H, 2CH, J = 8.4 Hz); 7.98 (d, 2H, 2CH, J = 8.4 Hz); 12.53 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 61.57 (C); 73.07 (C); 109.21 (C); 110.25 (C); 119.38 (C); 126.59 (2C); 133.08 (C); 133.19 (2C); 138.85 (C); 149.24 (C); 168.19 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −3.13; −9.78; −12.37. ESI-HRMS (m/z) average mass for C13H19B10N4S: 373.2283 [M + H]+. Found: 373.2272 [M + H]+.
3.1.11. 2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)-4-(3,4,5-Trimethoxyphenyl)Thiazole (4i)
Yield: 0.14 g, 92%, (dichloromethane/methanol (95:5), Rf = 0.63); mp 192–194 °C. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.64–2.70 (bs, 10H, 10B-H); 3.65 (s, 3H, CH3); 3.79 (s, 6H, 2CH3); 5.42 (bs, 1H, CH-carborane); 7.10 (s, 2H, 2CH); 7.37 (s, 1H, CH); 7.39 (s, 1H, CH); 12.53 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 56.35 (2C); 60.55 (C); 61.61 (C); 73.24 (C); 103.41 (2C); 104.89 (C); 130.37 (C); 132.75 (C); 137.77 (C); 150.55 (C); 153.49 (2C); 167.62 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −2.59; −4.14; −9.22; −11.14; −12.52. ESI-HRMS (m/z) average mass for C15H26B10N3O3S: 438.2652 [M + H]+. Found: 438.2652 [M + H]+.
3.1.12. 4-(2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)Thiazol-4-yl)Benzene-1-Sulfonyl Fluoride (4j)
Yield: 0.13 g, 93%, (dichloromethane/methanol (95:5), Rf = 0.68); mp 216–218 °C. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.28–3.15 (bs, 10H, 10B-H); 5.43 (bs, 1H, CH-carborane); 7.43 (s, 1H, CH); 7.84 (s, 1H, CH); 8.11–8.21 (m, 4H, 4CH); 12.60 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 61.61 (C); 73.02 (C); 110.94 (C); 127.31 (2C); 129.58 (2C); 133.40 (C); 141.88 (C); 148.58 (C); 168.39 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −3.05; −9.87; −12.38. ESI-HRMS (m/z) average mass for C12H19B10FN3O2S2: 430.1857 [M + H]+. Found: 430.1852 [M + H]+.
3.1.13. 4-(Adamant-1-yl)-2-(2-(1,2-Dicarba-closo-Dodecaboranylmethylene)Hydrazinyl)Thiazole (4k)
Yield: 0.09 g, 60%, (dichloromethane/methanol (95:5), Rf = 0.84); mp > 260 °C. 1H NMR (DMSO-d6, 400 MHz); δ (ppm); 1.84–2.42 (bs, 10H, 10B-H); 1.63–1.71 (m, 6H, 3CH2); 1.79–1.81 (m, 6H, 3CH2); 1.96–1.99 (m, 3H, 3CH); 5.35 (bs, 1H, CH-carborane); 6.34 (s, 1H, CH); 7.30 (s, 1H, CH); 12.31 (bs, 1H, NH). 13C NMR (DMSO-d6, 100 MHz), δ (ppm): 28.29 (3C); 36.28 (3C); 36.70 (3C); 41.59 (C); 61.51 (C); 73.26 (C); 101.28 (C); 133.50 (C); 160.19 (C); 168.31 (C). 11B NMR (DMSO-d6, 224 MHz); δ (ppm); −2.58; −4.09; −9.23; −11.18; −12.49. ESI-HRMS (m/z) average mass for C16H30B10N3S: 406.3117 [M + H]+. Found: 406.3113 [M + H]+.
3.2. Biological Activity
3.2.1. Antiproliferative Activity
All cell lines were obtained from ATCC, and all chemicals were purchased from Sigma-Aldrich, St. Louis, MO, USA. Briefly, cells were cultivated in a controlled atmosphere using DMEM medium with fetal bovine serum (FBS) at a final concentration of 10% (A-172), an L15 medium with 15% FBS, cultivated without CO2 (MDA-MB-231), F12K with a heparin final concentration of 0.1 mg/mL, endothelial cell growth supplement with a final concentration of 0.03 mg/mL (HUVEC), and RPMI with 5% FBS. All media were supplemented with antibiotic and antimycotic solutions, according to the manufacturers’ instructions. MTT cytotoxicity assessments were performed as described previously [40,41]. Briefly, cell lines A172, B16F10, MDA-MB-231, and HUVEC were seeded 24 h prior to the addition of the tested compounds. The cells counted using an automated cell counter (Countess® II FL, Invitrogen by Thermo Fisher Scientific Inc., Waltham, MA, USA) were plated in 96-well plates (Nunc Edge 2F, Thermo Fisher Scientific Inc.) at a density of 0.1 × 104 per well. After an additional 72 h, cells underwent an MTT cytotoxicity assay (see below). The IC50 values were calculated using ED50plus v 1.0 freeware software (Instituto Nacional de Enfermedades Respiratorias, Mexico City, Mexico). Each cell line was tested in three independent replicates.
Twenty microliters of an MTT solution (MTT: 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide, stock solution: 5 mg/mL) was added to each cell line for 2 h (A172, B16F10, MDA-MB-231, and HUVEC). After the incubation time was complete, the medium was removed and crystals of formazan were solubilized in pure isopropanol. The absorbance readout was conducted immediately using a plate reader, Multiskan Spectrum, at 570 nm (Thermo Fisher Scientific Inc., Waltham, MA, USA). The background optical density was measured in the wells filled with the culture medium, without any cells.
3.2.2. Mushroom Tyrosinase Inhibition Assay
The mushroom tyrosinase (Sigma-Aldrich) inhibition was performed following previously reported methods [43]. All the assays were carried out with solutions containing phosphate buffer (50 mM, pH 6.8), L-DOPA (0.17 mM), EDTA (0.022 mM), tyrosinase (50–100 units) and varying concentrations of compounds and were performed in triplicate at room temperature. The inhibitor solutions were prepared in DMSO with an initial concentration of 1 mM. Different aliquots were added to the solution containing the buffer, L-DOPA, and EDTA, with the enzyme being added last. The formation of dopachromone was determined by monitoring the absorbance at 475 nm with a T60U spectrophotometer (PG Instruments) equipped with quartz cells of 1 cm path length. Ascorbic acid was used as a reference inhibitor with an initial concentration of 1 mM. The IC50 values were calculated from the equation generated via exponential fit of the experimental data. The effectiveness of inhibition was expressed for the investigated compounds as the percentage of concentration necessary to achieve 50% inhibition (IC50), calculated using Equation (1):
where B0 = absorbance of L-DOPA + tyrosinase at t = 0 min, B30 = absorbance of L-DOPA + tyrosinase at t = 30 min, A0 = absorbance of L-DOPA + tyrosinase + inhibitor at t = 0 min, and A30 = absorbance of L-DOPA + tyrosinase + inhibitor at t = 30 min.
% of Inhibition = {[(B30 − B0) − (A30 − A0)]/(B30 − B0)} × 100,
3.2.3. Kinetic Analysis of the Inhibition of Tyrosinase
A series of experiments were performed to determine the inhibition kinetics of compounds by following the already reported method [44]. The inhibitor concentrations for compounds were 50 and 100 µM. The substrate L-DOPA concentration was between 100 and 250 µM in all kinetic studies. Maximal initial velocity was determined from the initial linear portion of absorbance up to ten minutes after the addition of the enzyme. The inhibition type of the enzyme, Michaelis constant (KM), and maximal velocity (Vmax) were determined by Lineweaver–Burk plots of the inverse of velocities (1/V) versus the inverse of substrate concentration 1/[L-DOPA] mM−1.
3.2.4. Reagents and Solvents
18-beta-Glycyrrhetinic acid was obtained from Acros Organic; phosphate buffer powder, cortisone, NADPH tetrasodium salt, and NAD Cofactor were obtained from Sigma-Aldrich; dimethylsulfoxide was obtained from POCh Poland; and carbenoxolone (sodium salt) was obtained from Cayman Chemical Company, Ann Arbor, Michigan, USA. Pooled human liver microsomes, mixed gender, 1 mL, 20 mg/mL, Lot No. 1410013, were obtained from XenoTech. Cortisol Elisa Ref DKO001, Lot No. 5148A was obtained from DiaMetra. The Enzyme-Linked Immunosorbent Assay (ELISA) Kit for 11-Beta-Hydroxysteroid Dehydrogenase Type 1, Lot No. L160706125, was obtained from Cloud-Clone Corp. PBS, Lot No. H161008, was obtained from Pan Biotech. Human kidney microsomes, mixed gender, 0.5 mL, 10 mg/mL, Lot No. 1710160, were obtained from XenoTech. The Enzyme-Linked Immunosorbent Assay (ELISA) Kit for 11-Beta-Hydroxysteroid Dehydrogenase Type 2, Lot No. L191113457, was obtained from Cloud-Clone Corp.
3.2.5. Inhibition of 11β-HSD1 Assays
The reduction reaction of cortisone to cortisol was carried out on 96-well microtiter plates in the presence of the enzyme 11β-HSD1 in a total volume of 100 µL. To this end, 20 μL of a cortisone/NADPH mixture with a final concentration of 200 nM/2 mM, 10 µL of a microsome (1.13 µg/mL 11β-HSD1) solution in PBS (final quantity 2.5 µg), 60 µL of phosphate buffer (pH 7.4), and 10 µL of the appropriate inhibitor solution (solvent DMSO/water 1/99, final concentration 10 µM) were placed in each well. The microplates were incubated on an orbital shaker for 150 min at 37 °C. The reaction was stopped via the addition of 10 µL of a solution containing 100 µM 18β-glycyrrhetinic acid in PBS. The quantity of cortisol formed from cortisone was determined using a cortisol ELISA kit, DiaMetra.
3.2.6. Inhibition of 11β-HSD2 Assays
The oxidation reaction of cortisol to cortisone was carried out on 96-well microtiter plates in the presence of the enzyme 11β-HSD2 in a total volume of 100 µL. To this end, 60 µL of phosphate buffer (pH 7.4), 10 µL of a microsomes(0.12 µg/mL 11β-HSD1) solution in PBS (final quantity 2.5 µg), 20 µL of substrate mixture cortisol/NAD (final concentration 200 nM/2 mM), and 10 µL of the appropriate inhibitor solution (solvent DMSO/water 1/99, final concentration 10 µM) were placed in each well. The microplates were incubated on an orbital shaker for 150 min at 37 °C. The reaction was stopped by the addition of 10 µL solution containing 100 µM carbenoxolone in PBS. The quantity of unreacted cortisol was determined using a cortisol ELISA kit DiaMetra.
3.3. Molecular Docking Study
The ligand molecules were drawn by using the Avogadro 1.1.1 software [45] and initially optimized within the UFF force field [46] (5000 steps, steepest descent algorithm). Further geometrical optimization was carried out at the level of the ab initio calculations at the DFT/B3LYP/6-31G(d,p) level of theory [47,48,49] by using the Gaussian09 package [50]. Flexible, optimized ligands molecules were docked into the binding pocket of (1) the tyrosinase structures found in the 2y9x PDB entry (tyrosinase from Agaricus bisporus, X-ray resolution: 0.278 nm). All four protein structures present in this PDB record were used in the four independent docking procedures; (2) the 11β-HSD1 protein structures found in the following PDB entries: 3crz, 3qqp, 4bb5, 4c7j, and 4hfr. These five structures were used for five independent docking procedures.
The AutoDock Vina 1.1.2 software [51] was applied for docking simulations. Due to the lack of boron-related parameters in nearly all available docking programs, we applied the procedure proposed and validated in [52], relying on directly adopting the parameters corresponding to the carbon atoms, characterized by the same molecular topology as the boron atoms of interest. Additionally, in order to increase the accuracy, the atomic partial charges for the o-carborane moiety were assigned according to the Mulliken scheme [53], on the basis of the previous DFT calculations. This protocol was initially validated for the case of human dihydrofolate reductase complexed with 2,4-diamino-5-(1-o-carboranylmethyl)-6-methylpyrimidine (PDB: 2c2s). Both the default parameters (i.e., boron treated as carbon) and the parameters with boron-characteristic atomic charges derived from the DFT calculations provided a similar, satisfactory quality of docking. The graphical illustration of the validation results is given in the Supporting Information as Figure S1. In the context of other, non-carborane ligands, the validation methodology and the corresponding results are given in our previous works [39].
The procedure of docking was carried out within the cuboid regions of dimensions of 18 × 18 × 18 Å3 (tyrosinase) or 22 × 22 × 22 Å3 (11β-HSD1), which cover all the originally co-crystallized ligands present in the considered PDB structures as well as the closest amino acid residues that exhibit contact with those ligands. All the default procedures and algorithms implemented in AutoDock Vina were applied during docking procedure. The predicted binding energies were averaged over all four or five considered protein structures. The calculations of the RMSD value for each pose of the docked ligand (after aligning with respect to the given structure) were carried out as well in order to ensure that the partial energies contributing the final, averaged, binding energy value corresponded to the structurally analogous orientations. Only the lowest average energy values, corresponding to the most favorable complexes, were considered during subsequent analysis.
4. Conclusions
The presented study depicted the synthesis of 11 carborane–thiazole conjugates, as well as their in-depth evaluation involving the assessment of enzymatic activity, antiproliferative activity, and molecular docking study.
The antiproliferative activity was tested for all compounds with the use of three cancer cell lines (A172, B16F10 and MDA-MB-231) and with a healthy control cell line (HUVEC). The results suggest that the newly synthesized carborane derivatives were most active against the murine melanoma cells. Moreover, in addition to the efficacy, the selectivity (affecting the safety profile) of the evaluated compounds was also the most beneficial for the murine melanoma cells.
Furthermore, compounds 4a–4g were investigated for their activity against the two isoforms of 11β-hydroxysteroid dehydrogenase, namely 11β-HSD1 and 11β-HSD2. The results show that in the same way as the reference inhibitor of carbenoxolone, the newly synthesized compounds unselectively inhibit both isoforms of the enzyme, with bias towards the 11β-HSD1 isoform. While the lowest IC50 value could be observed for the compound 4b, the greatest percentage of 11β-HSD1 inhibition could be observed for the compound 4g, which is especially advantageous.
The analysis of mushroom tyrosinase inhibition determined compounds 4h and 4f to be the two most active of the analyzed compounds. The recorded IC50 values were particularly low and surpassed the results recorded for the reference inhibitor, kojic acid, by over 6.4-fold, while they were over 34.2 times lower than the results recorded for the other reference inhibitor, ascorbic acid.
Molecular docking analysis suggested that the o-carborane group is not involved in any type of non-polar interactions when considering the interactions with tyrosinase. Instead, it maintains close contact with the hydrophilic sidechains of Glu322 and Asn81, as well as with the backbone fragment of His85. What is more, this fragment of molecule upholds a fixed position across the whole set of docked molecules, indicating the conserved character of interactions. In the case of the binding energy, the most important is the effect exerted by the nucleophilic substituent of the phenyl group such as the –CN moiety, which is present in the compound 4h. This ensures the close proximity between the molecule and copper ions present within the protein, as well as preventing the unfavorable reorientation of the phenyl group with its substituent towards the Phe264 and Val248.
The pattern of interactions obtained for 11β-HSD1 is qualitatively different and indicates that less polar contacts are preferred in the case of the o-carborane group. The divergences in binding affinities are a consequence of interactions of various chemical characters, involving Tyr177, Pro178, and, possibly, a solvent.
Considering all the results, compound 4h excels as the most promising carborane–thiazole conjugate and is set for further detailed evaluation. Moreover, it should be noted that the presented study demonstrated for the first time that carborane–thiazole derivatives are endowed with the ability to inhibit both tyrosinase and 11β-hydroxysteroid dehydrogenase type 1. This stands out as an unprecedented opportunity for simultaneous treatment of melanoma and comorbid diseases such as type II diabetes mellitus. This seems especially timely, as type II diabetes mellitus has recently been associated with more advanced forms of melanoma and an increased risk for melanoma recurrence [13,54,55]. These findings provide valuable insights for the rational design and optimization of novel tyrosinase and 11β-HSD1 inhibitors with enhanced therapeutic potential.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29194716/s1, Figure S1: The graphical illustration of the validation results.
Author Contributions
Conceptualization, B.D. and K.Z.Ł.; methodology, B.D., J.C., R.S., D.K., K.J., P.S., D.R., W.P. and K.Z.Ł.; software, W.P.; validation, B.D., J.C., R.S., D.K., K.J., P.S., W.P., B.B. and K.Z.Ł.; formal analysis, B.D., J.C., R.S., D.K., K.J., P.S., W.P., A.B.-Ł., B.B. and K.Z.Ł.; investigation, B.D., J.C., R.S., D.K., K.J., P.S., D.R., W.P. and K.Z.Ł.; data curation, J.C., W.P. and K.Z.Ł.; writing—original draft preparation, B.D., J.C., D.K.-S., W.P., A.B.-Ł. and K.Z.Ł.; writing—review and editing, B.D., J.C., D.K.-S., W.P., A.B.-Ł., K.Z.Ł., B.B. and A.B.O.; visualization, B.D., J.C., W.P. and K.Z.Ł.; supervision, K.Z.Ł.; project administration, K.Z.Ł.; funding acquisition, K.Z.Ł. and R.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Nicolaus Copernicus University (project No. 540/2020).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in this article/the Supplementary Materials; further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Global Cancer Burden Growing, Amidst Mounting Need for Services. Available online: https://www.who.int/news/item/01-02-2024-global-cancer-burden-growing--amidst-mounting-need-for-services (accessed on 17 August 2024).
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer Statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49, Erratum in CA Cancer J. Clin. 2024, 74, 203. https://doi.org/10.3322/caac.21830. [Google Scholar] [CrossRef] [PubMed]
- Melanoma of the Skin—Cancer Stat Facts. Available online: https://seer.cancer.gov/statfacts/html/melan.html (accessed on 17 August 2024).
- Brożyna, A.A.; Jóźwicki, W.; Roszkowski, K.; Filipiak, J.; Slominski, A.T. Melanin content in melanoma metastases affects the outcome of radiotherapy. Oncotarget 2016, 7, 17844–17853. [Google Scholar] [CrossRef] [PubMed]
- Slominski, R.M.; Sarna, T.; Płonka, P.M.; Raman, C.; Brożyna, A.A.; Slominski, A.T. Melanoma, Melanin, and Melanogenesis: The Yin and Yang Relationship. Front. Oncol. 2022, 12, 842496. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045, Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Gautier, J.F.; Chon, S. Assessment of Insulin Secretion and Insulin Resistance in Human. Diabetes Metab. J. 2021, 45, 641–654. [Google Scholar] [CrossRef]
- Zakir, M.; Ahuja, N.; Surksha, M.A.; Sachdev, R.; Kalariya, Y.; Nasir, M.; Kashif, M.; Shahzeen, F.; Tayyab, A.; Khan, M.S.M.; et al. Cardiovascular Complications of Diabetes: From Microvascular to Macrovascular Pathways. Cureus 2023, 15, e45835. [Google Scholar] [CrossRef]
- Chapman, K.; Holmes, M.; Seckl, J. 11β-hydroxysteroid dehydrogenases: Intracellular gate-keepers of tissue glucocorticoid action. Physiol. Rev. 2013, 93, 1139–1206. [Google Scholar] [CrossRef]
- Geer, E.B.; Islam, J.; Buettner, C. Mechanisms of glucocorticoid-induced insulin resistance: Focus on adipose tissue function and lipid metabolism. Endocrin. Metab. Clin. 2014, 43, 75–102. [Google Scholar] [CrossRef]
- Tomlinson, J.W.; Sherlock, M.; Hughes, B.; Hughes, S.V.; Kilvington, F.; Bartlett, W.; Courtney, R.; Rejto, P.; Carley, W.; Stewart, P.M. Inhibition of 11beta-hydroxysteroid dehydrogenase type 1 activity in vivo limits glucocorticoid exposure to human adipose tissue and decreases lipolysis. J. Clin. Endocrinol. Metab. 2007, 92, 857–864. [Google Scholar] [CrossRef]
- Qiang, J.K.; Lipscombe, L.L.; Lega, I.C. Association between diabetes, obesity, aging, and cancer: Review of recent literature. Transl. Cancer Res. 2020, 9, 5743–5759. [Google Scholar] [CrossRef]
- Nagore, E.; Martinez-Garcia, M.A.; Gomez-Olivas, J.D.; Manrique-Silva, E.; Martorell, A.; Bañuls, J.; Carrera, C.; Ortiz, P.; Gardeazabal, J.; Boada, A.; et al. Relationship between type 2 diabetes mellitus and markers of cutaneous melanoma aggressiveness: An observational multicentric study in 443 patients with melanoma. Br. J. Dermatol. 2021, 185, 756–763. [Google Scholar] [CrossRef]
- Silva, L.R.; Nunes, J.A.; Zhan, P.; Łączkowski, K.Z.; Cardoso, S.H.; da Silva-Júnior, E.F. Natural coumarin derivatives targeting melanoma. Curr. Med. Chem. 2024, 31, 871–886. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.A.; Araújo, R.S.A.; Silva, F.N.d.; Cytarska, J.; Łączkowski, K.Z.; Cardoso, S.H.; Mendonça-Júnior, F.J.B.; Silva-Júnior, E.F.d. Coumarin-based compounds as inhibitors of tyrosinase/tyrosine hydroxylase: Synthesis, kinetic studies, and in silico approaches. Int. J. Mol. Sci. 2023, 24, 5216. [Google Scholar] [CrossRef]
- Cytarska, J.; Szulc, J.; Kołodziej-Sobczak, D.A.; Nunes, J.A.; da Silva-Junior, E.F.; Łączkowski, K. Cyrene™ as a tyrosinase inhibitor and anti-browni ng agent. Food Chem. 2024, 442, 138430. [Google Scholar] [CrossRef]
- Baumgart, S.; Kupczyk, D.; Archała, A.; Koszła, O.; Sołek, P.; Płaziński, W.; Płazińska, A.; Studzińska, R. Synthesis of novel 2-(cyclopentylamino)thiazol-4(5H)-one derivatives with potential anticancer, antioxidant, and 11β-HSD inhibitory activities. Int. J. Mol. Sci. 2023, 24, 7252. [Google Scholar] [CrossRef] [PubMed]
- Studzińska, R.; Kołodziejska, R.; Płaziński, W.; Kupczyk, D.; Kosmalski, T.; Jasieniecka, K.; Modzelewska-Banachiewicz, B. Synthesis of the N-methyl derivatives of 2-aminothiazol-4(5H)-one and their interactions with 11βHSD1: Molecular modeling and in vitro studies. Chem. Biodivers. 2019, 16, e1900065. [Google Scholar] [CrossRef] [PubMed]
- Studzińska, R.; Kupczyk, D.; Płazińska, A.; Kołodziejska, R.; Kosmalski, T.; Modzelewska-Banachiewicz, B. Thiazolo [3,2-α]pyrimidin-5-one derivatives as a novel class of 11β-hydroxysteroid dehydrogenase inhibitors. Bioorg Chem. 2018, 81, 21–26. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, G.; He, Y.; Jing Wang, J.J.; Zhao, Y. Tyrosinase inhibitory mechanism and anti-browning properties of novel kojic acid derivatives bearing aromatic aldehyde moiety. Curr. Res. Food Sci. 2023, 6, 100421. [Google Scholar] [CrossRef]
- Grimes, R.N. Carboranes, 3rd ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Marfavi, A.; Kavianpour, P.; Rendina, L.M. Carboranes in drug discovery, chemical biology and molecular imaging. Nat. Rev. Chem. 2022, 6, 486–504. [Google Scholar] [CrossRef]
- Stockmann, P.; Gozzi, M.; Kuhnert, R.; Sárosi, M.B.; Hey-Hawkins, E. New keys for old locks: Carborane-containing drugs as platforms for mechanism-based therapies. Chem. Soc. Rev. 2019, 48, 3497–3512. [Google Scholar] [CrossRef]
- Barth, R.F.; Yang, W.; Al-Madhoun, A.S.; Johnsamuel, J.; Byun, Y.; Chandra, S.; Smith, D.R.; Tjarks, W.; Eriksson, S. Boron-containing nucleosides as potential delivery agents for neutron capture therapy of brain tumors. Cancer Res. 2004, 64, 6287–6295. [Google Scholar] [CrossRef] [PubMed]
- Kabalka, G.W.; Wu, Z.Z.; Yao, M.L.; Natarajan, N. The syntheses and in vivo biodistribution of novel boronated unnatural amino acids. Appl. Radiat. Isot. 2004, 61, 1111–1115. [Google Scholar] [CrossRef]
- Olejniczak, A.B.; Adamska, A.M.; Paradowska, E.; Studzinska, M.; Suski, P.; Leśnikowski, Z.J. Modification of selected anti-HCMV drugs with lipophilic boron cluster modulator. Acta Pol. Pharm. 2013, 70, 489–504. [Google Scholar] [PubMed]
- Fink, K.; Uchman, M. Boron clusters compounds as new chemical leads antimicrobial therapy. Coord. Chem. Rev. 2021, 431, 213684. [Google Scholar] [CrossRef]
- Smith, N.; Quan, D.; Nagalingam, G.; Triccas, J.A.; Rendina, L.M.; Rutledge, P.J. Carborane clusters increase the potency of bis-substituted cyclam derivatives against Mycobacterium tuberculosis. RSC Med. Chem. 2022, 13, 1234–1238. [Google Scholar] [CrossRef]
- Bogucka-Kocka, A.; Kołodziej, P.; Makuch-Kocka, A.; Różycka, D.; Rykowski, S.; Nekvinda, J.; Grüner, B.; Olejniczak, A.B. Nematicidal activity of naphthalimide-boron cluster conjugates. Chem. Commun. 2022, 58, 2528–2531. [Google Scholar] [CrossRef]
- Kugler, M.; Nekvinda, J.; Holub, J.; El Anwar, S.; Das, V.; Šícha, V.; Pospíšilová, K.; Fábry, M.; Král, V.; Brynda, J.; et al. Inhibitors of CA IX Enzyme Based on Polyhedral Boron Compounds. Chembiochem 2021, 22, 2741–2761. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Yasui, Y.; Maruyama, M.; Minegishi, H.; Ban, H.S.; Sato, S. Development of hypoxia-inducible factor (HIF)-1α inhibitors: Effect of ortho-carborane substituents on HIF transcriptional activity under hypoxia. Bioorg. Med. Chem. Lett. 2013, 23, 806–810. [Google Scholar] [CrossRef]
- Li, G.; Azuma, S.; Minegishi, H.; Nakamura, H. Synthesis and biological evaluation of meta-carborane-containing phenoxyacetanilides as inhibitors of hypoxia-inducible factor (HIF)-1 transcriptional activity. J. Organomet. Chem. 2015, 798, 189–195. [Google Scholar] [CrossRef]
- Asawa, Y.; Katsuragi, K.; Sato, A.; Yoshimori, A.; Tanuma, S.I.; Nakamura, H. Structure-based drug design of novel carborane-containing nicotinamide phosphoribosyltransferase inhibitors. Bioorg. Med. Chem. 2019, 27, 2832–2844. [Google Scholar] [CrossRef]
- Neumann, W.; Xu, S.; Sárosi, M.B.; Scholz, M.S.; Crews, B.C.; Ghebreselasie, K.; Banerjee, S.; Marnett, L.J.; Hey-Hawkins, E. nido-Dicarbaborate Induces Potent and Selective Inhibition of Cyclooxygenase-2. ChemMedChem 2016, 11, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Useini, L.; Mojić, M.; Laube, M.; Lönnecke, P.; Dahme, J.; Sárosi, M.B.; Mijatović, S.; Maksimović-Ivanić, D.; Pietzsch, J.; Hey-Hawkins, E. Carboranyl Analogues of Mefenamic Acid and Their Biological Evaluation. ACS Omega 2022, 7, 24282–24291. [Google Scholar] [CrossRef] [PubMed]
- Kuhnert, R.; Sárosi, M.B.; George, S.; Lönnecke, P.; Hofmann, B.; Steinhilber, D.; Murganic, B.; Mijatovic, S.; Maksimovic-Ivanic, D.; Hey-Hawkins, E. CarbORev-5901, The First Carborane-Based Inhibitor of the 5-Lipoxygenase Pathway. ChemMedChem 2017, 12, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Kuhnert, R.; Kuhnert, L.; Sárosi, M.B.; George, S.; Draca, D.; Paskas, S.; Hofmann, B.; Steinhilber, D.; Honscha, W.; Mijatović, S.; et al. Borcalein: A Carborane-Based Analogue of Baicalein with 12-Lipoxygenase-Independent Toxicity. ChemMedChem 2022, 17, e202100588. [Google Scholar] [CrossRef]
- Austin, C.J.; Kahlert, J.; Issa, F.; Reed, J.H.; Smith, J.R.; Ioppolo, J.A.; Ong, J.A.; Jamie, J.F.; Hibbs, D.; Rendina, L.M. The first indoleamine-2,3-dioxygenase-1 (IDO1) inhibitors containing carborane. Dalton Trans. 2014, 43, 10719–10724. [Google Scholar] [CrossRef]
- Rosada, B.; Bekier, A.; Cytarska, J.; Płaziński, W.; Zavyalova, O.; Sikora, A.; Dzitko, K.; Łączkowski, K.Z. Benzo[b]thiophene-thiazoles as potent anti-Toxoplasma gondii agents: Design, synthesis, tyrosinase/tyrosine hydroxylase inhibitors, molecular docking study, and antioxidant activity. Eur. J. Med. Chem. 2019, 184, 111765. [Google Scholar] [CrossRef]
- Piechowska, K.; Świtalska, M.; Cytarska, J.; Jaroch, K.; Łuczykowski, K.; Chałupka, J.; Wietrzyk, J.; Misiura, K.; Bojko, B.; Kruszewski, S.; et al. Discovery of tropinone-thiazole derivatives as potent caspase 3/7 activators, and noncompetitive tyrosinase inhibitors with high antiproliferative activity: Rational design, one-pot tricomponent synthesis, and lipophilicity determination. Eur. J. Med. Chem. 2019, 175, 162–171. [Google Scholar] [CrossRef]
- Piechowska, K.; Mizerska-Kowalska, M.; Zdzisińska, B.; Cytarska, J.; Baranowska-Łączkowska, A.; Jaroch, K.; Łuczykowski, K.; Płaziński, W.; Bojko, B.; Kruszewski, S.; et al. Tropinone-derived alkaloids as potent anticancer agents: Synthesis, tyrosinase inhibition, mechanism of action, DFT calculation, and molecular docking studies. Int. J. Mol. Sci. 2020, 21, 9050. [Google Scholar] [CrossRef]
- Dozzo, P.; Kasar, R.A.; Kahl, S.B. Simple, High-yield methods for the synthesis of aldehydes directly from o-, m-, and p-carborane and their further conversions. Inorg. Chem. 2005, 44, 8053–8057. [Google Scholar] [CrossRef]
- Saeed, A.; Mahesar, P.A.; Channar, P.A.; Abbas, Q.; Larik, F.A.; Hassan, M.; Raza, H.; Seo, S.Y. Synthesis, molecular docking studies of coumarinyl-pyrazolinyl substituted thiazoles as non-competitive inhibitors of mushroom tyrosinase. Bioorg Chem. 2017, 74, 187–196. [Google Scholar] [CrossRef]
- Shin, N.H.; Ryu, S.Y.; Choi, E.J.; Kang, S.H.; Chang, I.M.; Min, K.R.; Kim, Y. Oxyresveratrol as the potent inhibitor on dopa oxidase activity of mushroom tyrosinase. Biochem. Biophys. Res. Commun. 1998, 243, 801–803. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Rappe, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A., III; Skiff, W.M. UFF, a Full Periodic Table Force Field for Molecular Mechanics and Dynamics Simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Krishnan, R.; Binkley, J.S.; Seeger, R.; Pople, J.A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 1980, 72, 650–654. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian09 Package, version 09; Gaussian, Inc.: Wallingford, CT, USA, 2016.
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Tiwari, R.; Mahasenan, K.; Pavlovicz, R.; Li, C.; Tjarks, W. Carborane clusters in computational drug design: A comparative docking evaluation using AutoDock, FlexX, Glide, and Surflex. J. Chem. Inf. Model. 2009, 49, 1581–1589. [Google Scholar] [CrossRef]
- Mulliken, R.S. Electronic Population Analysis on LCAO–MO Molecular Wave Functions. I. J. Chem. Phys. 1955, 23, 1833–1840. [Google Scholar] [CrossRef]
- Sraker, R.J.; Tortorello, G.N.; Sharon, C.E.; Keele, L.J.; Chu, E.Y.; Miura, J.T.; Karakousis, G.C.; Ming, M.E. Association of type II diabetes mellitus with characteristics and outcomes for patients undergoing sentinel lymph node biopsy for cutaneous melanoma. J. Surg. Oncol. 2022, 126, 1263–1271. [Google Scholar] [CrossRef]
- Kaneko, A.; Kanemaru, H.; Mizuhashi, S.; Kimura, T.; Kuriyama, H.; Sawamura, S.; Kajihara, I.; Makino, K.; Miyashita, A.; Aoi, J.; et al. Relationship between Type 2 diabetes mellitus and aggressiveness of melanoma. J. Dermatol. Sci. 2022, 106, 65–67. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).