Effective Synthesis of mRNA during In Vitro Transcription with Fewer Impurities Produced
Abstract
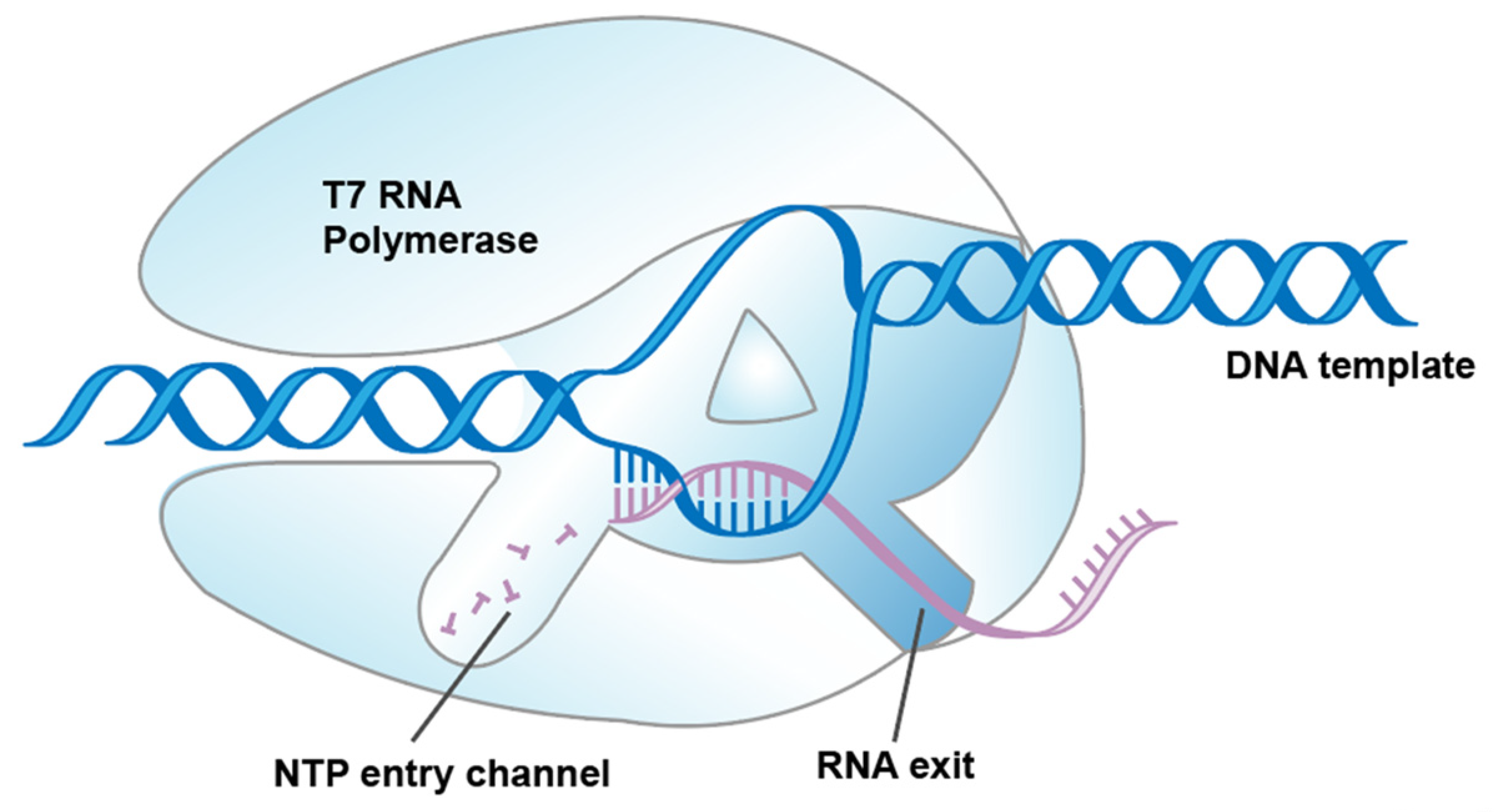
1. Introduction
2. Results and Discussion
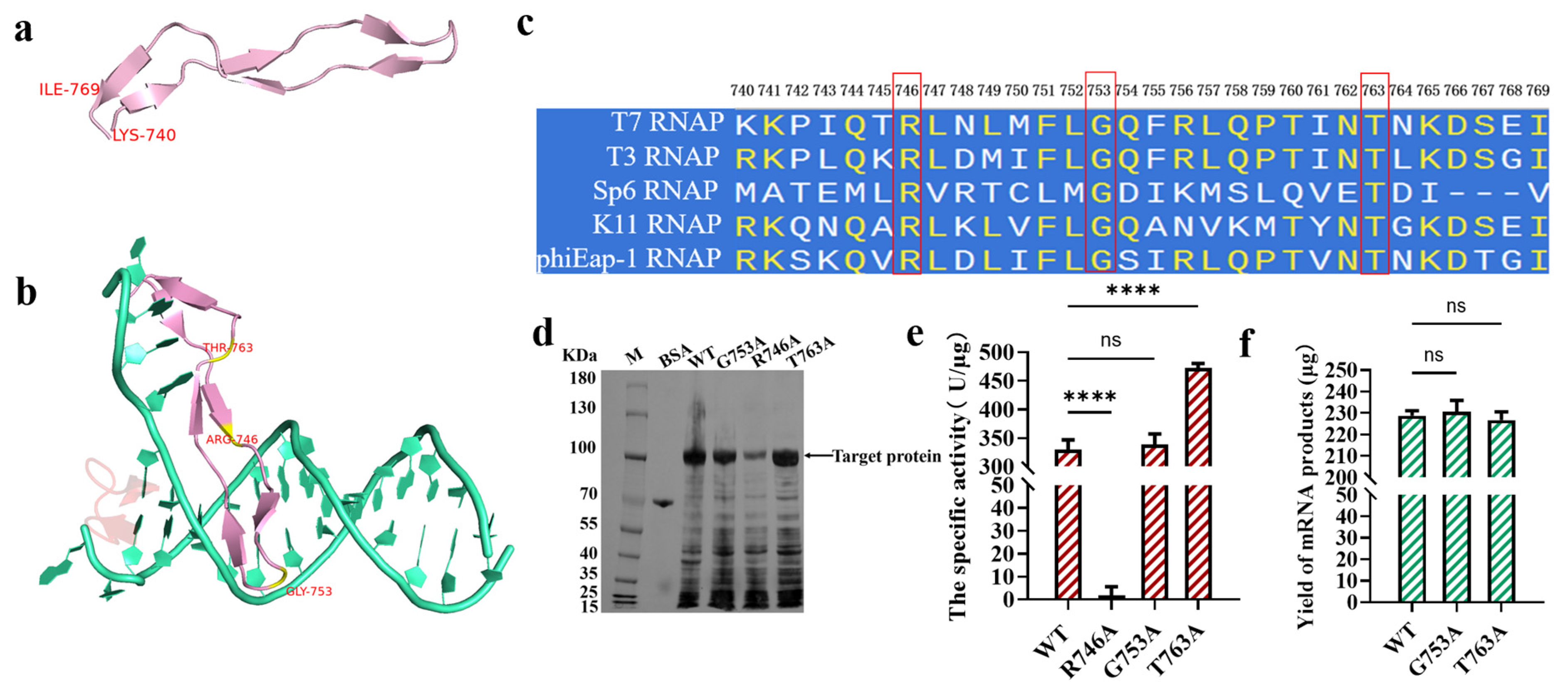
2.1. Screening Specificity Loop Substitutions via Conservative Analysis
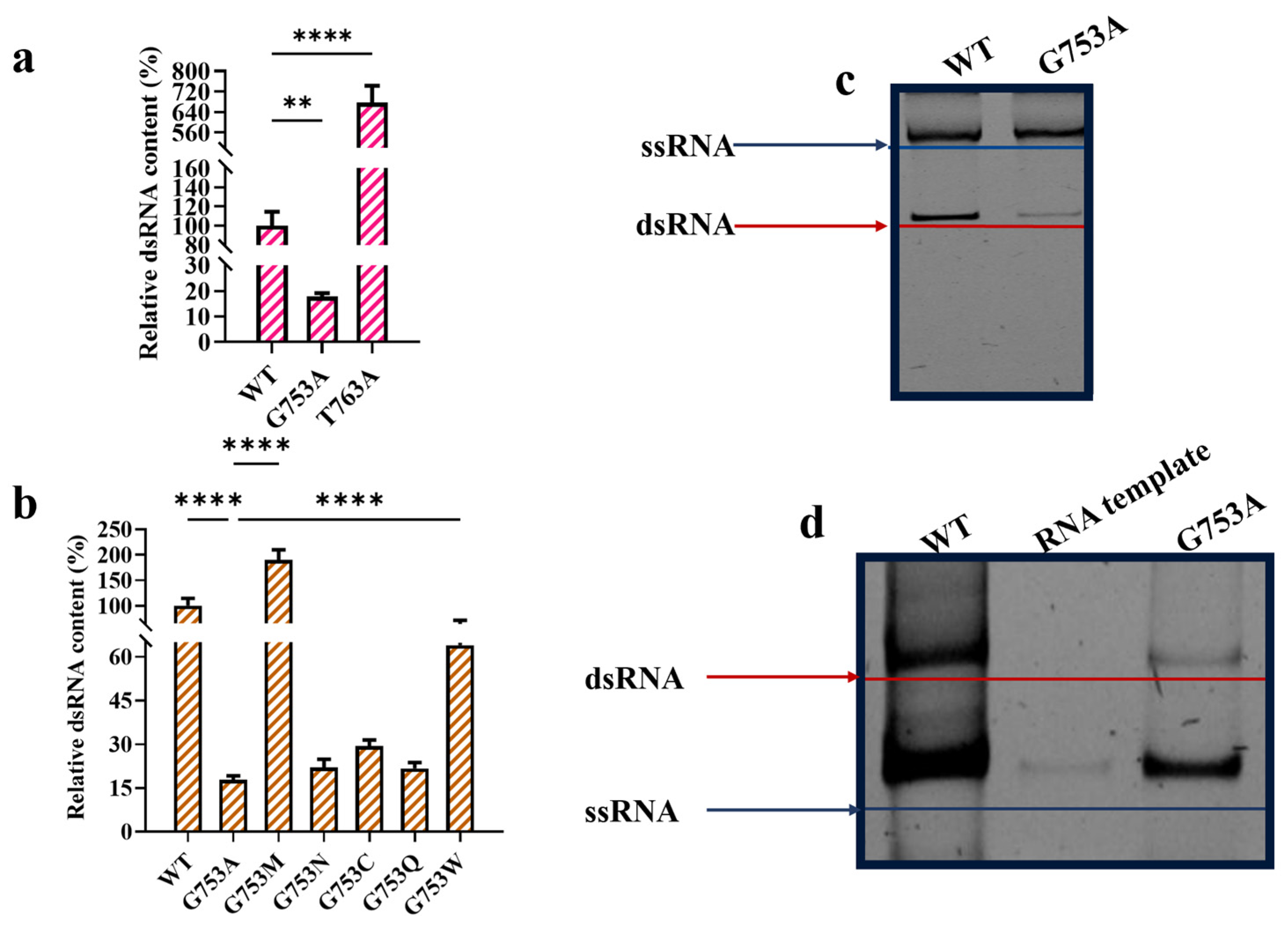
2.2. T7 RNAP Substitutions Closely Related with the Production of dsRNA Byproducts
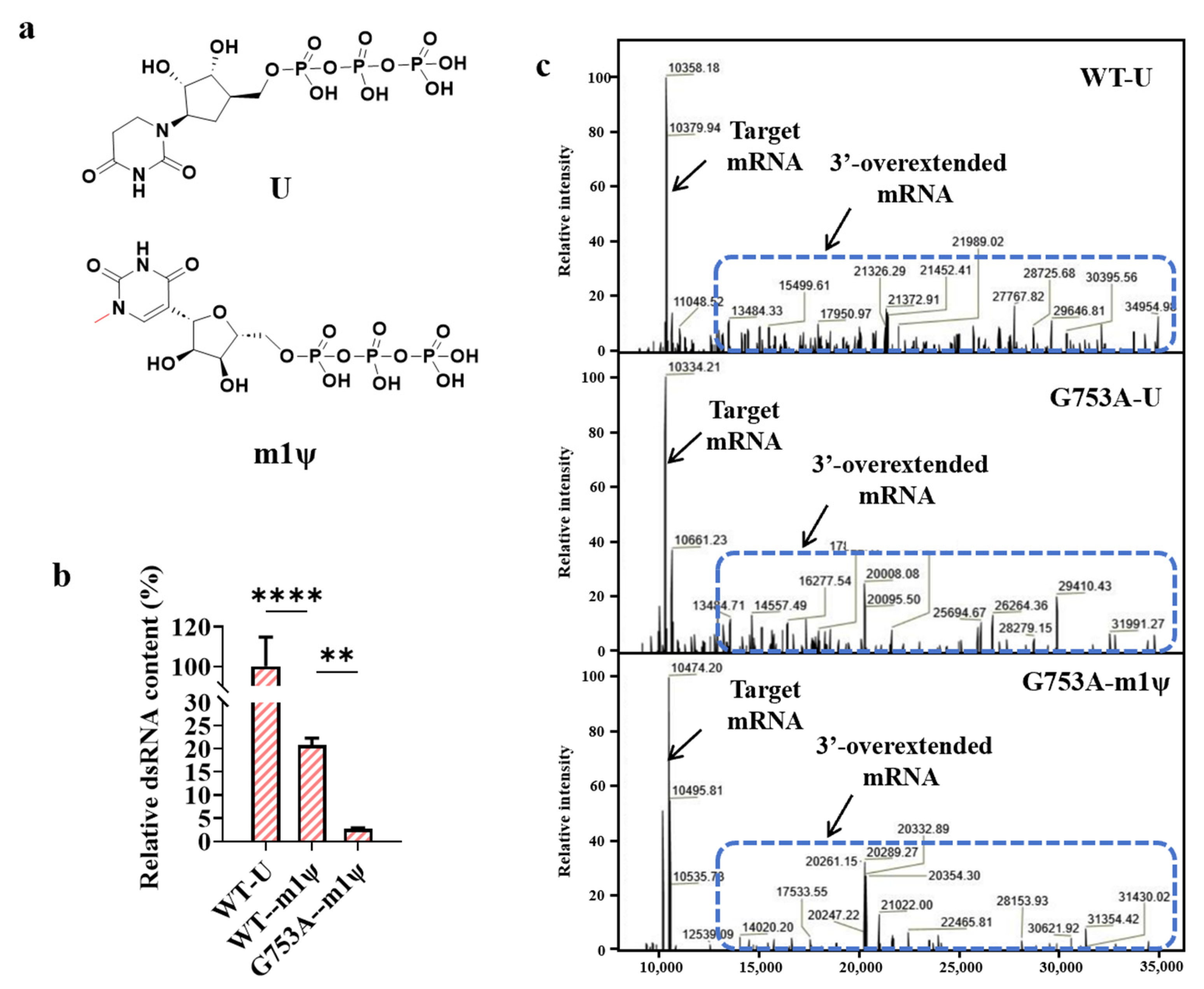
2.3. m1ψTP Further Decreases Production of dsRNA Byproducts
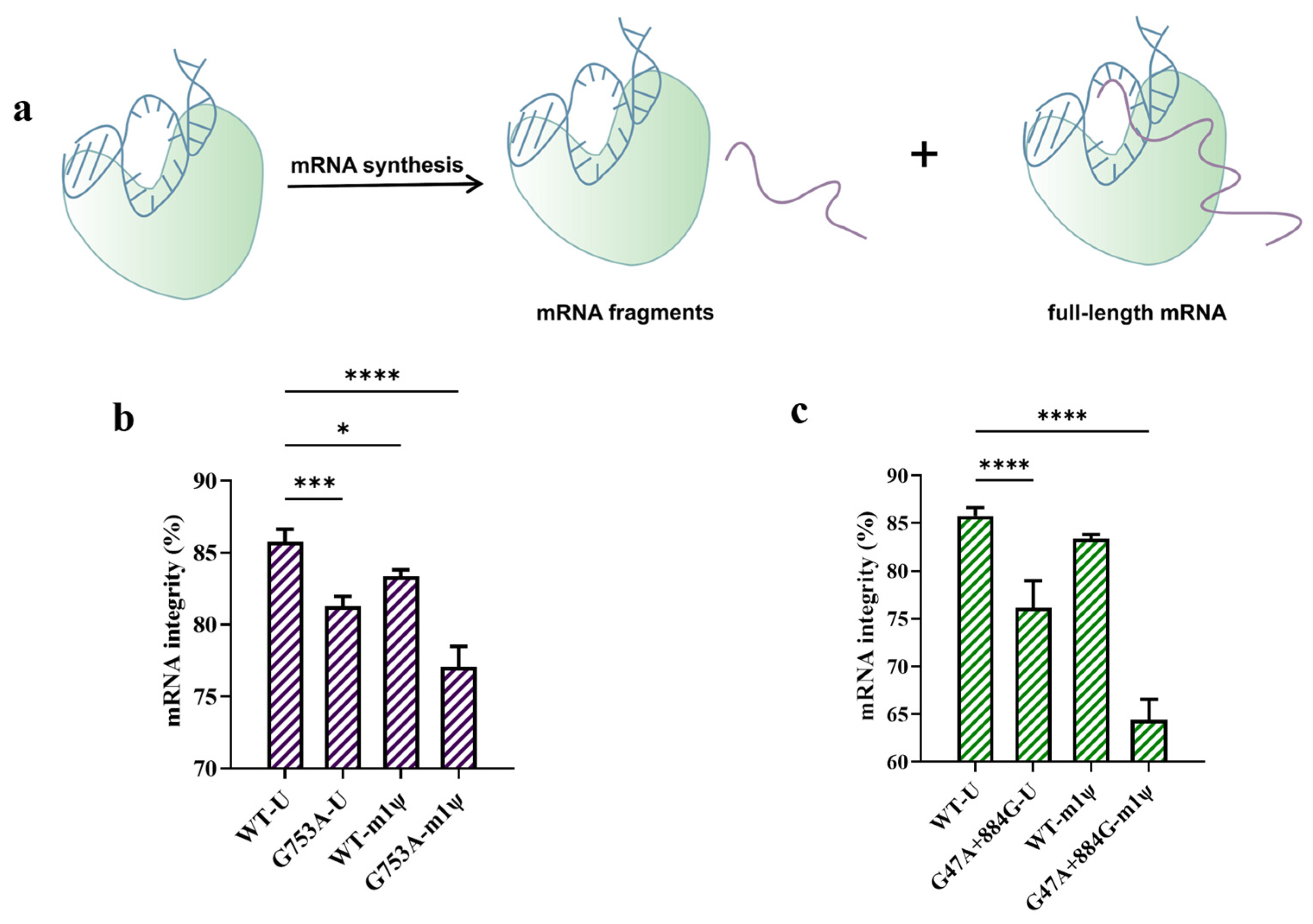
2.4. Substitution Produced More Fragmented mRNA
3. Materials and Methods
3.1. Reagents and Chemicals
3.2. Overexpression and Purification of T7 RNAP
3.3. The Measurement of Specific Activity
3.4. Measurement of dsRNA Content
3.5. An Analysis of Antisense RNA Production Transcribed Using DNA as the Template
3.6. An Analysis of Antisense RNA Production Transcribed Using RNA as the Template
3.7. 3′-Overextended mRNA Measured Using LC-MS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Park, J.W.; Lagniton, P.N.P.; Liu, Y.; Xu, R.H. mRNA vaccines for COVID-19: What, why and how. Int. J. Biol. Sci. 2021, 17, 1446–1460. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.; Liu, X.; Li, M.; Zhang, Z.; Song, L.; Zhu, B.; Wu, X.; Liu, J.; Zhao, D.; Li, Y. Advances in COVID-19 mRNA vaccine development. Signal Transduct. Target. Ther. 2022, 7, 94. [Google Scholar] [CrossRef]
- Lamb, Y.N. BNT162b2 mRNA COVID-19 Vaccine: First Approval. Drugs 2021, 81, 495–501. [Google Scholar] [CrossRef]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. mRNA vaccines for infectious diseases: Principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021, 20, 817–838. [Google Scholar] [CrossRef]
- Gote, V.; Bolla, P.K.; Kommineni, N.; Butreddy, A.; Nukala, P.K.; Palakurthi, S.S.; Khan, W. A Comprehensive Review of mRNA Vaccines. Int. J. Mol. Sci. 2023, 24, 2700. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Kwon, M.; Im, S.; Lee, K.; Lee, H. mRNA vaccines: The most recent clinical applications of synthetic mRNA. Arch. Pharmacal Res. 2022, 45, 245–262. [Google Scholar] [CrossRef]
- Miao, L.; Zhang, Y.; Huang, L. mRNA vaccine for cancer immunotherapy. Mol. Cancer 2021, 20, 41. [Google Scholar] [CrossRef]
- Deng, Z.; Tian, Y.; Song, J.; An, G.; Yang, P. mRNA Vaccines: The Dawn of a New Era of Cancer Immunotherapy. Front. Immunol. 2022, 13, 887125. [Google Scholar] [CrossRef]
- Connolly, C.D.; Quinonez, S.C.; Ames, E.G. Rare disease therapeutics: The future of medical genetics in a changing landscape. Genet. Med. 2023, 25, 100339. [Google Scholar] [CrossRef]
- Borkotoky, S.; Murali, A. The highly efficient T7 RNA polymerase: A wonder macromolecule in biological realm. Int. J. Biol. Macromol. 2018, 118, 49–56. [Google Scholar] [CrossRef]
- Szabó, G.T.; Mahiny, A.J.; Vlatkovic, I. COVID-19 mRNA vaccines: Platforms and current developments. Mol. Ther. 2022, 30, 1850–1868. [Google Scholar] [CrossRef]
- Sousa, R.; Chung, Y.J.; Rose, J.P.; Wang, B.C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 Å resolution. Nature 1993, 364, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Girbig, M.; Misiaszek, A.D.; Müller, C.W. Structural insights into nuclear transcription by eukaryotic DNA-dependent RNA polymerases. Nat. Rev. Mol. Cell Biol. 2022, 23, 603–622. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I. Bacterial RNA polymerase: A promising target for the discovery of new antimicrobial agents. Curr. Opin. Investig. Drugs 2007, 8, 600–607. [Google Scholar] [PubMed]
- Temiakov, D.; Tahirov, T.H.; Anikin, M.; McAllister, W.T.; Vassylyev, D.G.; Yokoyama, S. Crystallization and preliminary crystallographic analysis of T7 RNA polymerase elongation complex. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003, 59, 185–187. [Google Scholar] [CrossRef]
- Gardner, L.P.; Mookhtiar, K.A.; Coleman, J.E. Initiation, elongation, and processivity of carboxyl-terminal mutants of T7 RNA polymerase. Biochemistry 1997, 36, 2908–2918. [Google Scholar] [CrossRef]
- Tunitskaya, V.L.; Kochetkov, S.N. Structural-functional analysis of bacteriophage T7 RNA polymerase. Biochemistry 2002, 67, 1124–1135. [Google Scholar] [CrossRef]
- Camperi, J.; Lippold, S.; Ayalew, L.; Roper, B.; Shao, S.; Freund, E.; Nissenbaum, A.; Galan, C.; Cao, Q.; Yang, F.; et al. Comprehensive Impurity Profiling of mRNA: Evaluating Current Technologies and Advanced Analytical Techniques. Anal. Chem. 2024, 96, 3886–3897. [Google Scholar] [CrossRef]
- Miller, M.; Alvizo, O.; Baskerville, S.; Chintala, A.; Chng, C.; Dassie, J.; Dorigatti, J.; Huisman, G.; Jenne, S.; Kadam, S.; et al. An Engineered T7 RNA polymerase for Efficient co-transcriptional capping with reduced dsRNA byproducts in mRNA synthesis. Faraday Discuss. 2024, 252, 431–449. [Google Scholar] [CrossRef]
- Mu, X.; Hur, S. Immunogenicity of In Vitro-Transcribed RNA. Acc. Chem. Res. 2021, 54, 4012–4023. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Baiersdörfer, M.; Boros, G.; Muramatsu, H.; Mahiny, A.; Vlatkovic, I.; Sahin, U.; Karikó, K. A facile method for the removal of dsRNA contaminant from in vitro-transcribed mRNA. Mol. Ther. Nucleic Acids 2019, 15, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Muramatsu, H.; Ludwig, J.; Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011, 39, e142. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A. From rejection to the Nobel Prize: Karikó and Weissman’s pioneering work on mRNA vaccines, and the need for diversity and inclusion in translational immunology. Front. Immunol. 2023, 14, 1306025. [Google Scholar] [CrossRef] [PubMed]
- Yadav, T.; Kumar, S.; Mishra, G.; Saxena, S.K. Tracking the COVID-19 vaccines: The global landscape. Hum. Vaccines Immunother. 2023, 19, 2191577. [Google Scholar] [CrossRef]
- Xia, X. Detailed dissection and critical evaluation of the Pfizer/BioNTech and Moderna mRNA vaccines. Vaccines 2021, 9, 734. [Google Scholar] [CrossRef]
- Corbett, K.S.; Flynn, B.; Foulds, K.E.; Francica, J.R.; Boyoglu-Barnum, S.; Werner, A.P.; Flach, B.; O’Connell, S.; Bock, K.W.; Minai, M.; et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020, 383, 1544–1555. [Google Scholar] [CrossRef]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Anderson, B.R.; Muramatsu, H.; Nallagatla, S.R.; Bevilacqua, P.C.; Sansing, L.H.; Weissman, D.; Karikó, K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010, 38, 5884–5892. [Google Scholar] [CrossRef]
- Weissman, D. mRNA transcript therapy. Expert Rev. Vaccines 2015, 14, 265–281. [Google Scholar] [CrossRef]
- Rohner, E.; Yang, R.; Foo, K.S.; Goedel, A.; Chien, K.R. Unlocking the promise of mRNA therapeutics. Nat. Biotechnol. 2022, 40, 1586–1600. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Klein, L.J.; Ha, S.; Rustandi, R.R. High-Resolution capillary electrophoresis separation of large RNA under non-aqueous conditions. J. Chromatogr. A 2020, 1618, 460875. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhang, X.; Zou, Y.; Li, J.; Wang, C.; He, Y.; Jin, Q.; Ye, J. Effective Synthesis of High-Integrity mRNA Using In Vitro Transcription. Molecules 2024, 29, 2461. [Google Scholar] [CrossRef] [PubMed]
- Daniel, S.; Kis, Z.; Kontoravdi, C.; Shah, N. Quality by Design for enabling RNA platform production processes. Trends Biotechnol. 2022, 40, 1213–1228. [Google Scholar] [CrossRef]
- Kis, Z. Stability Modelling of mRNA Vaccine Quality Based on Temperature Monitoring throughout the Distribution Chain. Pharmaceutics 2022, 14, 430. [Google Scholar] [CrossRef]
- Oivanen, M.; Kuusela, S.; Lönnberg, H. Kinetics and Mechanisms for the Cleavage and Isomerization of the Phosphodiester Bonds of RNA by Brønsted Acids and Bases. Chem. Rev. 1998, 98, 961–990. [Google Scholar] [CrossRef]
- Wayment-Steele, H.K.; Kim, D.S.; Choe, C.A.; Nicol, J.J.; Wellington-Oguri, R.; Watkins, A.M.; Parra Sperberg, R.A.; Huang, P.S.; Participants, E.; Das, R. Theoretical basis for stabilizing messenger RNA through secondary structure design. Nucleic Acids Res. 2021, 49, 10604–10617. [Google Scholar] [CrossRef]
- Forconi, M.; Herschlag, D. Metal Ion-Based RNA Cleavage as a Structural Probe. Methods Enzym. 2009, 468, 91–106. [Google Scholar] [CrossRef]
- Mu, X.; Greenwald, E.; Ahmad, S.; Hur, S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018, 46, 5239–5249. [Google Scholar] [CrossRef]
- Yu, B.; Chen, Y.; Yan, Y.; Lu, X.; Zhu, B. DNA-terminus-dependent transcription by T7 RNA polymerase and its C-helix mutants. Nucleic Acids Res. 2024, 52, 8443–8453. [Google Scholar] [CrossRef]
- Durniak, K.J.; Bailey, S.; Steitz, T.A. The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation. Science 2008, 322, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Sari, Y.; Sousa Rosa, S.; Jeffries, J.; Marques, M.P.C. Comprehensive evaluation of T7 promoter for enhanced yield and quality in mRNA production. Sci. Rep. 2024, 14, 9655. [Google Scholar] [CrossRef]
- Temiakov, D.; Mentesana, P.E.; Ma, K.; Mustaev, A.; Borukhov, S.; McAllister, W.T. The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc. Natl. Acad. Sci. USA 2000, 97, 14109–14114. [Google Scholar] [CrossRef] [PubMed]
- Sousa, R.; Mukherjee, S. T7 RNA polymerase. Prog. Nucleic Acid Res. Mol. Biol. 2003, 73, 1–41. [Google Scholar] [CrossRef]
- McGraw, N.J.; Bailey, J.N.; Cleaves, G.R.; Dembinski, D.R.; Gocke, C.R.; Joliffe, L.K.; MacWright, R.S.; McAllister, W.T. Sequence and analysis of the gene for bacteriophage T3 RNA polymerase. Nucleic Acids Res. 1985, 13, 6753–6766. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kotani, H.; Ishizaki, Y.; Hiraoka, N.; Obayashi, A. Nucleotide sequence and expression of the cloned gene of bacteriophage SP6 RNA polymerase. Nucleic Acids Res. 1987, 15, 2653–2664. [Google Scholar] [CrossRef]
- Dietz, A.; Weisser, H.J.; Kössel, H.; Hausmann, R. The gene for Klebsiella bacteriophage K11 RNA polymerase: Sequence and comparison with the homologous genes of phages T7, T3, and SP6. Mol. Gen. Genet. MGG 1990, 221, 283–286. [Google Scholar] [CrossRef]
- Li, E.; Wei, X.; Ma, Y.; Yin, Z.; Li, H.; Lin, W.; Wang, X.; Li, C.; Shen, Z.; Zhao, R.; et al. Isolation and characterization of a bacteriophage phiEap-2 infecting multidrug resistant Enterobacter aerogenes. Sci. Rep. 2016, 6, 28338. [Google Scholar] [CrossRef]
- Cheetham, G.M.T.; Jeruzalmi, D.; Steitz, T.A. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature 1999, 399, 80–83. [Google Scholar] [CrossRef]
- Brieba, L.G.; Gopal, V.; Sousa, R. Scanning mutagenesis reveals roles for helix n of the bacteriophage T7 RNA polymerase thumb subdomain in transcription complex stability, pausing, and termination. J. Biol. Chem. 2001, 276, 10306–10313. [Google Scholar] [CrossRef]
- Dousis, A.; Ravichandran, K.; Hobert, E.M.; Moore, M.J.; Rabideau, A.E. An engineered T7 RNA polymerase that produces mRNA free of immunostimulatory byproducts. Nat. Biotechnol. 2023, 41, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Blauch, L.R.; Szymanski, M.R.; Das, R.; Tang, S.K.Y.; Yin, Y.W.; Fire, A.Z. Transcription polymerase-catalyzed emergence of novel RNA replicons. Science 2020, 368, eaay0688. [Google Scholar] [CrossRef] [PubMed]
- Nance, K.D.; Meier, J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Z.; Asahara, H.; Tzertzinis, G.; Roy, B. Synthesis of low immunogenicity RNA with high-temperature in vitro transcription. RNA 2020, 26, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Piao, X.; Yadav, V.; Wang, E.; Chang, W.; Tau, L.; Lindenmuth, B.E.; Wang, S.X. Double-stranded RNA reduction by chaotropic agents during in vitro transcription of messenger RNA. Mol. Ther. Nucleic Acids 2022, 29, 618–624. [Google Scholar] [CrossRef]
- Chen, T.H.; Potapov, V.; Dai, N.; Ong, J.L.; Roy, B. N1-methyl-pseudouridine is incorporated with higher fidelity than pseudouridine in synthetic RNAs. Sci. Rep. 2022, 12, 13017. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, W.; Geng, Q.; Ji, G.; Li, J.; Wang, D.; He, Y.; Jin, Q.; Ye, J. Effective Synthesis of mRNA during In Vitro Transcription with Fewer Impurities Produced. Molecules 2024, 29, 4713. https://doi.org/10.3390/molecules29194713
He W, Geng Q, Ji G, Li J, Wang D, He Y, Jin Q, Ye J. Effective Synthesis of mRNA during In Vitro Transcription with Fewer Impurities Produced. Molecules. 2024; 29(19):4713. https://doi.org/10.3390/molecules29194713
Chicago/Turabian StyleHe, Wei, Qi Geng, Guiying Ji, Ji Li, Dan Wang, Yucai He, Qiuheng Jin, and Jianren Ye. 2024. "Effective Synthesis of mRNA during In Vitro Transcription with Fewer Impurities Produced" Molecules 29, no. 19: 4713. https://doi.org/10.3390/molecules29194713
APA StyleHe, W., Geng, Q., Ji, G., Li, J., Wang, D., He, Y., Jin, Q., & Ye, J. (2024). Effective Synthesis of mRNA during In Vitro Transcription with Fewer Impurities Produced. Molecules, 29(19), 4713. https://doi.org/10.3390/molecules29194713






