Creation of Metal-Complex-Integrated Tensegrity Triangle DNA Crystals
Abstract
1. Introduction
2. Results and Discussion
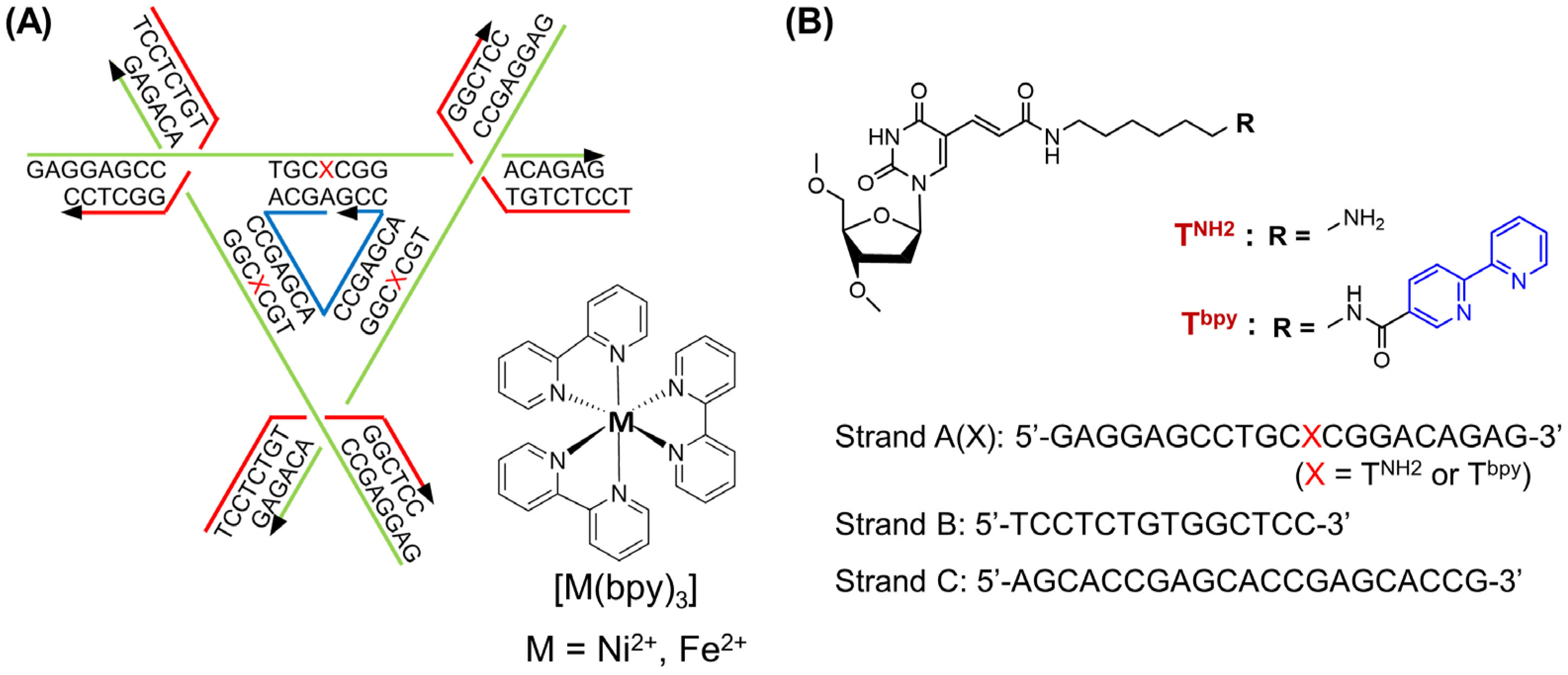
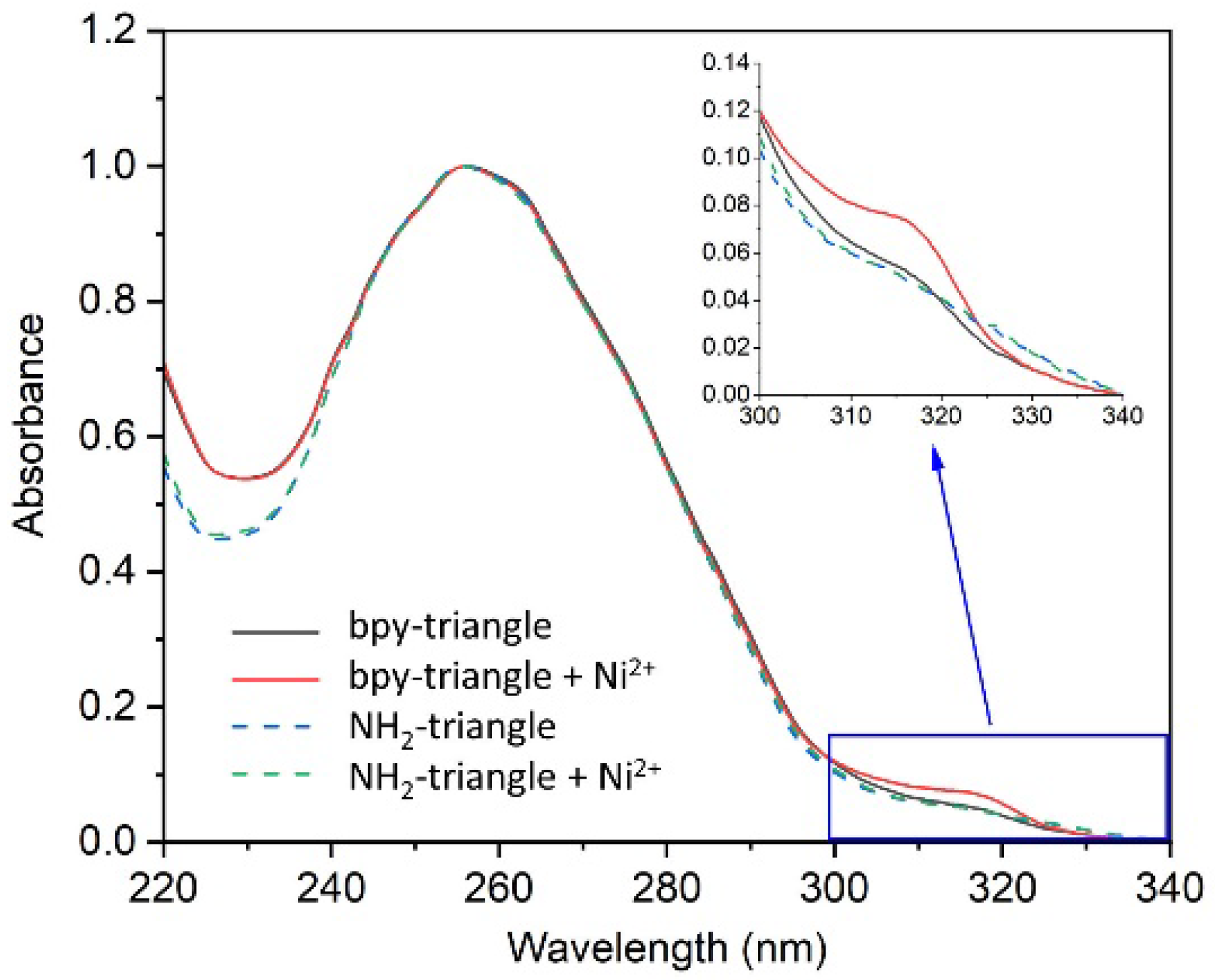
2.1. Formation of the Metal Complex in the DNA Tensegrity Triangle
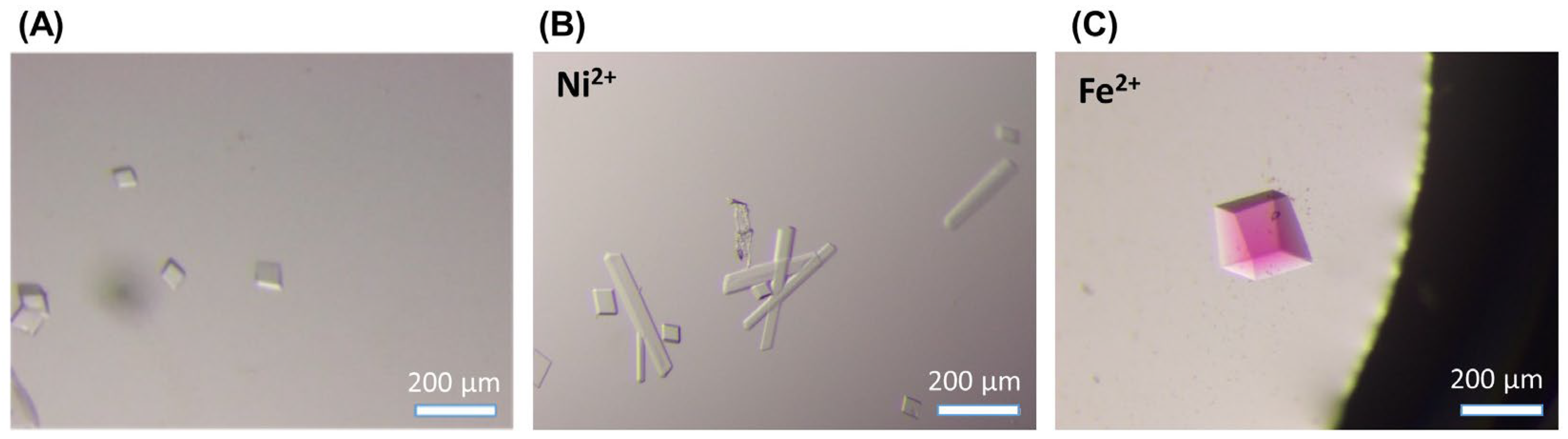
2.2. Crystallization of the Metal-Complex-Incorporated DNA Tensegrity Triangle
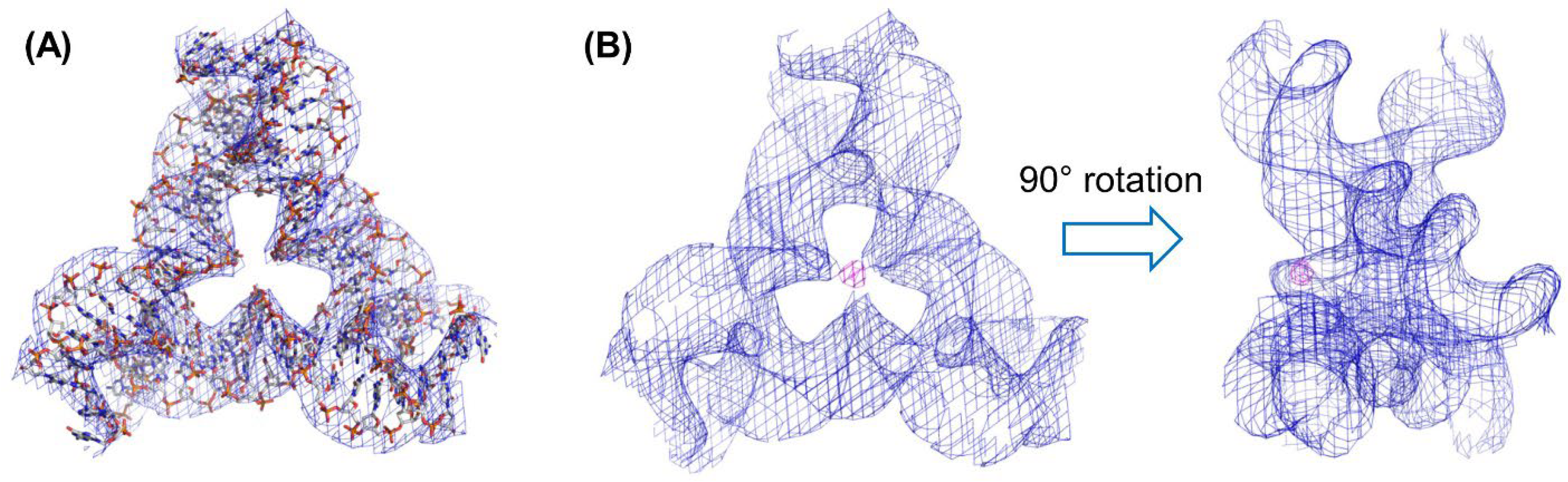
2.3. Analysis of the DNA Tensegrity Triangle Crystals with Metal Complexes
2.4. Modeling of the Metal Complex in the Tensegrity Triangle Based on the Crystal Structure
3. Materials and Methods
3.1. Material
- Strand A(X): 5′-GAGGAGCCTGCXCGGACAGAG-3′ (X = TNH2 or Tbpy);
- Strand B: 5′-TCCTCTGTGGCTCC-3′;
- Strand C: 5′-AGCACCGAGCACCGAGCACCG-3′.
3.2. Synthesis, Purification, and the Identification of Strand A(Tbpy)
3.3. Assembly of DNA Triangle and Measurement of UV Spectra
3.4. Crystallization of DNA via Sitting Drop Vapor Diffusion Method
3.5. Data Collection and Crystal Structure Analysis
3.6. In Silico Molecular Modeling of Fe–Bipyridine Complex
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seeman, N.C. Nanomaterials based on DNA. Annu. Rev. Biochem. 2010, 79, 65–87. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA origami: Scaffolds for creating higher order structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Gu, H.; Li, Q.; Fan, C. Concept and development of framework nucleic acids. J. Am. Chem. Soc. 2018, 140, 17808–17819. [Google Scholar] [CrossRef] [PubMed]
- Kuan, S.L.; Bergamini, F.R.G.; Weil, T. Functional protein nanostructures: A chemical toolbox. Chem. Soc. Rev. 2018, 47, 9069–9105. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Assefi, M.; Ataeinaeini, M.; Nazari, A.; Gholipour, A.; Vertiz-Osores, J.J.; Calla-Vásquez, K.M.; Al-Naqeeb, B.Z.T.; Jassim, K.H.; Kalajahi, H.G.; Yasamineh, S.; et al. A state-of-the-art review on solid lipid nanoparticles as a nanovaccines delivery system. J. Drug. Deliv. Sci. Technol. 2023, 86, 104623. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef]
- Zhang, F.; Nangreave, J.; Liu, Y.; Yan, H. Structural DNA nanotechnology: State of the art and future perspective. J. Am. Chem. Soc. 2014, 136, 11198–11211. [Google Scholar] [CrossRef]
- Endo, M.; Sugiyama, H. Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy. Acc. Chem. Res. 2014, 47, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Krissanaprasit, A.; Key, C.M.; Pontula, S.; Labean, T.H. Self-assembling nucleic acid nanostructures functionalized with aptamers. Chem. Rev. 2021, 121, 13797–13868. [Google Scholar] [CrossRef]
- Rossi-Gendron, C.; El Fakih, F.; Bourdon, L.; Nakazawa, K.; Finkel, J.; Triomphe, N.; Chocron, L.; Endo, M.; Sugiyama, H.; Bellot, G.; et al. Isothermal self-assembly of multicomponent and evolutive DNA nanostructures. Nat. Nanotechnol. 2023, 18, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Jerson, G.; Han, J.Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef]
- Wang, Q.; Astruc, D. State of the art and prospects in metal–organic framework (MOF)-based and MOF-derived nanocatalysis. Chem. Rev. 2020, 120, 1438–1511. [Google Scholar] [CrossRef]
- Liu, D.; Wang, M.; Deng, Z.; Walulu, R.; Mao, C. Tensegrity: Construction of rigid DNA triangles with flexible four-arm DNA junctions. J. Am. Chem. Soc. 2004, 126, 2324–2325. [Google Scholar] [CrossRef]
- Zheng, J.; Birktoft, J.J.; Chen, Y.; Wang, T.; Sha, R.; Constantinou, P.E.; Ginell, S.L.; Mao, C.; Seeman, N.C. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 2009, 461, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Vecchioni, S.; Ohayon, Y.P.; Sha, R.; Woloszyn, K.; Yang, B.; Mao, C.; Seeman, N.C. 3D hexagonal arrangement of DNA tensegrity triangles. ACS Nano 2021, 15, 16788–16793. [Google Scholar] [CrossRef]
- Lu, B.; Woloszyn, K.; Ohayon, Y.P.; Yang, B.; Zhang, C.; Mao, C.; Seeman, N.C.; Vecchioni, S.; Sha, R. Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles. Angew. Chem. Int. Ed. 2023, 62, e202213451. [Google Scholar] [CrossRef]
- Ohayon, Y.P.; Hernandez, C.; Chandrasekaran, A.R.; Wang, X.; Abdallah, H.O.; Jong, M.A.; Mohsen, M.G.; Sha, R.; Birktoft, J.J.; Lukeman, P.S.; et al. Designing higher resolution self-assembled 3D DNA crystals via strand terminus modifications. ACS Nano 2019, 13, 7957–7965. [Google Scholar] [CrossRef]
- Li, Z.; Liu, L.; Zheng, M.; Zhao, J.; Seeman, N.C.; Mao, C. Making engineered 3D DNA crystals robust. J. Am. Chem. Soc. 2019, 141, 15850–15855. [Google Scholar] [CrossRef]
- Eki, H.; Abe, K.; Sugiyama, H.; Endo, M. Nanoscopic observation of a DNA crystal surface and its dynamic formation and degradation using atomic force microscopy. Chem. Commun. 2021, 57, 1651–1654. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Kristiansen, M.; Sha, R.; Birktoft, J.J.; Hernandez, C.; Mao, C.; Seeman, N.C. A device that operates within a self-assembled 3D DNA crystal. Nat. Chem. 2017, 9, 824–827. [Google Scholar] [CrossRef]
- Stahl, E.; Praetorius, F.; de Oliveira Mann, C.C.; Hopfner, K.P.; Dietz, H. Impact of heterogeneity and lattice bond strength on DNA triangle crystal growth. ACS Nano 2016, 10, 9156–9164. [Google Scholar] [CrossRef]
- Wang, X.; Sha, R.; Kristiansen, M.; Hernandez, C.; Hao, Y.; Mao, C.; Canary, J.W.; Seeman, N.C. An Organic Semiconductor Organized into 3D DNA Arrays by “Bottom-up” Rational Design. Angew. Chem. Int. Ed. 2017, 56, 6445–6448. [Google Scholar] [CrossRef] [PubMed]
- Stulz, E. Nanoarchitectonics with porphyrin functionalized DNA. Acc. Chem. Res. 2017, 50, 823–831. [Google Scholar] [CrossRef]
- Vecchioni, S.; Lu, B.; Livernois, W.; Ohayon, Y.P.; Yoder, J.B.; Yang, C.; Woloszyn, K.; Bernfeld, W.; Anantram, M.P.; Canary, J.W.; et al. Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction. Adv. Mater. 2023, 35, 2210938. [Google Scholar] [CrossRef] [PubMed]
- Duprey, J.-L.H.A.; Takezawa, Y.; Shionoya, M. Metal-locked DNA three-way junction. Angew. Chem. Int. Ed. 2013, 52, 1212–1216. [Google Scholar] [CrossRef]
- Lin, J.; Qin, B.; Fang, Z. Nickel Bipyridine (Ni(bpy)3Cl2) Complex Used as Molecular Catalyst for Photocatalytic CO2 Reduction. Catal. Lett. 2019, 149, 25–33. [Google Scholar] [CrossRef]
- Kabsch, W. xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix. Refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abe, K.; Eki, H.; Hirose, Y.; Park, S.; Chinnathambi, S.; Namasivayam, G.P.; Takeda, K.; Sugiyama, H.; Endo, M. Creation of Metal-Complex-Integrated Tensegrity Triangle DNA Crystals. Molecules 2024, 29, 4674. https://doi.org/10.3390/molecules29194674
Abe K, Eki H, Hirose Y, Park S, Chinnathambi S, Namasivayam GP, Takeda K, Sugiyama H, Endo M. Creation of Metal-Complex-Integrated Tensegrity Triangle DNA Crystals. Molecules. 2024; 29(19):4674. https://doi.org/10.3390/molecules29194674
Chicago/Turabian StyleAbe, Katsuhiko, Haruhiko Eki, Yuki Hirose, Soyoung Park, Shanmugavel Chinnathambi, Ganesh Pandian Namasivayam, Kazuki Takeda, Hiroshi Sugiyama, and Masayuki Endo. 2024. "Creation of Metal-Complex-Integrated Tensegrity Triangle DNA Crystals" Molecules 29, no. 19: 4674. https://doi.org/10.3390/molecules29194674
APA StyleAbe, K., Eki, H., Hirose, Y., Park, S., Chinnathambi, S., Namasivayam, G. P., Takeda, K., Sugiyama, H., & Endo, M. (2024). Creation of Metal-Complex-Integrated Tensegrity Triangle DNA Crystals. Molecules, 29(19), 4674. https://doi.org/10.3390/molecules29194674







