Enhancement of Forskolin Production Using Aeroponic Cultivation of Coleus forskohlii and the Impact on the Plant Phytochemistry
Abstract
1. Introduction
2. Results and Discussion
2.1. Forskolin Quantification
2.2. Metabolomic Investigation
2.2.1. Comparison of the Phytochemical Composition of Root and Aerial Parts
2.2.2. Identification of Compounds Impacted by LEDs
3. Materials and Methods
3.1. Plant Cultivation
3.1.1. Aeroponic System
3.1.2. Cultivation Itinerary
3.1.3. Harvesting and Drying
3.2. Phytochemicals
3.2.1. Chemicals
3.2.2. Sample Preparation
3.2.3. HPLC-ELSD Quantification
3.2.4. UHPLC-HRMS and UHPLC-HRMS/MS Analyses
3.3. Analyses of Treatments
3.3.1. HRMS/MS Treatment
3.3.2. Statistical Treatment of HRMS Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shivaprasad, H.N.; Pandit, S.; Bhanumathy, M.; Manohar, D.; Jain, V.; Thandu, S.A.; Su, X. Ethnopharmacological and Phytomedical Knowledge of Coleus Forskohlii: An Approach towards Its Safety and Therapeutic Value. Orient. Pharm. Exp. Med. 2014, 14, 301–312. [Google Scholar] [CrossRef]
- World Flora Online: Coleus Forskohli Briq. 2024. Available online: https://www.worldfloraonline.org/taxon/wfo-0001070109 (accessed on 28 April 2024).
- Ammon, H.; Müller, A. Forskolin: From an Ayurvedic Remedy to a Modern Agent. Planta Med. 1985, 51, 473–477. [Google Scholar] [CrossRef]
- Paton, A.J.; Mwanyambo, M.; Govaerts, R.H.A.; Smitha, K.; Suddee, S.; Phillipson, P.B.; Wilson, T.C.; Forster, P.I.; Culham, A. Nomenclatural Changes in Coleus and Plectranthus (Lamiaceae): A Tale of More than Two Genera. PhytoKeys 2019, 129, 1–158. [Google Scholar] [CrossRef]
- Grayer, R.J.; Paton, A.J.; Simmonds, M.S.J.; Howes, M.J.R. Differences in Diterpenoid Diversity Reveal New Evidence for Separating the Genus: Coleus from Plectranthus. Nat. Prod. Rep. 2021, 38, 1720–1728. [Google Scholar] [CrossRef]
- Kavitha, C.; Rajamani, K.; Vadivel, E. Coleus Forskohlii: A Comprehensive Review on Morphology, Phytochemistry and Pharmacological Aspects. J. Med. Plants Res. 2010, 4, 278–285. [Google Scholar]
- Pateraki, I.; Andersen-Ranberg, J.; Hamberger, B.; Heskes, A.M.; Martens, H.J.; Zerbe, P.; Bach, S.S.; Møller, B.L.; Bohlmann, J.; Hamberger, B. Manoyl Oxide (13R), the Biosynthetic Precursor of Forskolin, Is Synthesized in Specialized Root Cork Cells in Coleus forskohlii. Plant Physiol. 2014, 164, 1222–1236. [Google Scholar] [CrossRef] [PubMed]
- Khatun, S.; Cakılcıoglu, U.; Chatterjee, N.C. Pharmacognostic Value of Leaf Anatomy and Trichome Morphology for Identification of Forskolin in a Novel Medicinal Plant Coleus forskohlii. Biol. Divers. Conserv. 2011, 4, 165–171. [Google Scholar]
- Alasbahi, R.H.; Melzig, M.F. Plectranthus Barbatus: A Review of Phytochemistry, Ethnobotanical Uses and Pharmacology Part 2. Planta Med. 2010, 76, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Nafi’Ah, R.; Sumirtapura, Y.C.; Darijanto, S.T.; Iwo, I. Diffusion and Lipolysis Test on Forskolin Microemulsion Gel. J. Res. Pharm. 2023, 27, 1447–1450. [Google Scholar] [CrossRef]
- Ravinder, D.; Rampogu, S.; Dharmapuri, G.; Pasha, A.; Lee, K.W.; Pawar, S.C. Inhibition of DDX3 and COX-2 by Forskolin and Evaluation of Anti-Proliferative, pro-Apoptotic Effects on Cervical Cancer Cells: Molecular Modelling and in Vitro Approaches. Med. Oncol. 2022, 39, 61. [Google Scholar] [CrossRef]
- Chiadak, J.D.; Arsenijevic, T.; Verstrepen, K.; Gregoire, F.; Bolaky, N.; Delforge, V.; Flamand, V.; Perret, J.; Delporte, C. Forskolin Inhibits Lipopolysaccharide-Induced Modulation of MCP-1 and GPR120 in 3T3-L1 Adipocytes through an Inhibition of NFκB. Mediat. Inflamm. 2016, 2016, 1431789. [Google Scholar] [CrossRef] [PubMed]
- Gabetta, B.; Zini, G.; Danieli, B. Minor Diterpenoids of Coleus forskohlii. Phytochemistry 1989, 28, 859–862. [Google Scholar] [CrossRef]
- Abdel-Mogib, M.; Albar, H.; Batterjee, S. Chemistry of the Genus Plectranthus. Molecules 2002, 7, 271–301. [Google Scholar] [CrossRef]
- Schultz, C.; Bossolani, M.P.; Torres, L.M.B.; Lima-Landman, M.T.R.; Lapa, A.J.; Souccar, C. Inhibition of the Gastric H+,K+-ATPase by Plectrinone A, a Diterpenoid Isolated from Plectranthus barbatus Andrews. J. Ethnopharmacol. 2007, 111, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Falé, P.L.; Borges, C.; Madeira, P.J.A.; Ascensão, L.; Araújo, M.E.M.; Florêncio, M.H.; Serralheiro, M.L.M. Rosmarinic Acid, Scutellarein 4′-Methyl Ether 7-O-Glucuronide and (16S)-Coleon E Are the Main Compounds Responsible for the Antiacetylcholinesterase and Antioxidant Activity in Herbal Tea of Plectranthus barbatus (“falso Boldo”). Food Chem. 2009, 114, 798–805. [Google Scholar] [CrossRef]
- Pullaiah, T. Forskolin: Natural Sources, Pharmacology and Biotechnology; Springer Nature: Singapore, 2022; ISBN 978-981-19652-0-3. [Google Scholar]
- Jiménez Amezcua, I.; Rivas Blas, S.; Díez Municio, M.; Soria, A.C.; Ruiz Matute, A.I.; Sanz, M.L. Development of a Multianalytical Strategy for Detection of Frauds in Coleus forskohlii Supplements. J. Chromatogr. A 2022, 1676, 4–13. [Google Scholar] [CrossRef]
- Reddy, C.S.; Praveena, C.H.; Veeresham, C. Strategies to Improve the Production of Forskolin from Hairy Root Cultures of Coleus forskohlii Briq. Int. J. Pharm. Sci. Nanotechnol. 2012, 5, 1720–1726. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.-M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and Resupply of Pharmacologically Active Plant-Derived Natural Products: A Review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef]
- Ibrahim, M.; Arafa, N.; Aly, U. Antioxidant Activity, Phenol and Flavonoid Contents of Plant and Callus Cultures of Plectranthus barbatus Andrews. Egypt. Pharm. J. 2018, 17, 32. [Google Scholar] [CrossRef]
- Erekath, S.; Seidlitz, H.; Schreiner, M.; Dreyer, C. Food for Future: Exploring Cutting-Edge Technology and Practices in Vertical Farm. Sustain. Cities Soc. 2024, 106, 105357. [Google Scholar] [CrossRef]
- Morard, P. Les Cultures Vegetales Hors Sol; Publications Agricoles: Agen, France, 1995. [Google Scholar]
- Lakhiar, I.A.; Gao, J.; Syed, T.N.; Chandio, F.A.; Buttar, N.A. Modern Plant Cultivation Technologies in Agriculture under Controlled Environment: A Review on Aeroponics. J. Plant Interact. 2018, 13, 338–352. [Google Scholar] [CrossRef]
- Misra, A.; Srivastava, S.; Srivastava, P.; Shukla, P.; Agrawal, P.K.; Rawat, A.K.S. Chemotaxonomic Variation in Forskolin Content and Its Correlation with Ecogeographical Factors in Natural Habitat of Coleus forskohlii Briq. Collected from Vidarbha (Maharashtra, India). Ind. Crops Prod. 2016, 84, 50–58. [Google Scholar] [CrossRef]
- Chaudhary, M.K.; Misra, A.; Tripathi, D.; Srivastava, P.K.; Srivastava, S. Impact of Seasonal Variation on Four Labdane-Type Diterpenoids in Coleus forskholii Briq. Nat. Prod. Res. 2024, 38, 2342–2347. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Park, J.-H.; Kim, E.-J.; Lee, J.-M.; Park, J.-W.; Kim, Y.-S.; Kim, G.-R.; Lee, J.-S.; Lee, E.-P.; You, Y.-H. White LED Lighting Increases the Root Productivity of Panax Ginseng C. A. Meyer in a Hydroponic Cultivation System of a Plant Factory. Biology 2023, 12, 1052. [Google Scholar] [CrossRef]
- Harde, S.M.; Singhal, R.S. Extraction of Forskolin from Coleus forskohlii Roots Using Three Phase Partitioning. Sep. Purif. Technol. 2012, 96, 20–25. [Google Scholar] [CrossRef]
- Lunz, K.; Stappen, I. Back to the Roots-an Overview of the Chemical Composition and Bioactivity of Selected Root-Essential Oils. Molecules 2021, 26, 3155. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, R.; Tyagi, B.; Ahmed, B.; Husain, A. Variation in Forskolin Content in the Roots of Coleus forskohlii. Planta Med. 1988, 54, 471–472. [Google Scholar] [CrossRef]
- Kim, S.P.; Moon, E.; Nam, S.H.; Friedman, M. Composition of Herba Pogostemonis Water Extract and Protection of Infected Mice against Salmonella Typhimurium-Induced Liver Damage and Mortality by Stimulation of Innate Immune Cells. J. Agric. Food Chem. 2012, 60, 12122–12130. [Google Scholar] [CrossRef]
- Uchida, M.; Miyase, T.; Yoshizaki, F.; Bieri, J.H.; Rüedi, P.; Eugster, C.H. 14-Hydroxytaxodion Als Hauptditerpen in Plectranthus grandidentatus GÜRKE; Isolierung von Sieben Neuen Dimeren Diterpenen Aus P. grandidentatus, P. myrianthus BRIQ. Und Coleus carnosus HASSK.: Strukturen Der Grandidone A, 7-Epi-A, B, 7-Epi-B, C, D Und 7-Epi-D. Helv. Chim. Acta 1981, 64, 2227–2250. [Google Scholar] [CrossRef]
- Ahmed, B.; Vishwakarma, R.A. Coleoside, a Monoterpene Glycoside from Coleus forskohlii. Phytochemistry 1988, 27, 3309–3310. [Google Scholar] [CrossRef]
- Al Musayeib, N.M.; Amina, M.; Al-Hamoud, G.A.; Mohamed, G.A.; Ibrahim, S.R.M.; Shabana, S. Plectrabarbene, a New Abietane Diterpene from Plectranthus barbatus Aerial Parts. Molecules 2020, 25, 2365. [Google Scholar] [CrossRef]
- Kelecom, A. Isolation, Structure Determination, and Absolute Configuration of Barbatusol, a New Bioactive Diterpene with a Rearranged Abietane Skeleton from the Labiate Coleus barbatus. Tetrahedron 1983, 39, 3603–3608. [Google Scholar] [CrossRef]
- Painuly, P.; Tandon, J.S. Triterpenes and Flavones from Coleus spicatus. J. Nat. Prod. 1983, 46, 285. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Yu, H.-M.; Shie, J.-J.; Cheng, T.-J.R.; Wu, C.-Y.; Fang, J.-M.; Wong, C.-H. Chemical Constituents of Plectranthus amboinicus and the Synthetic Analogs Possessing Anti-Inflammatory Activity. Bioorg. Med. Chem. 2014, 22, 1766–1772. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M. Cytochrome P450-Dependent Hydroxylation in the Biosynthesis of Rosmarinic Acid in Coleus. Int. J. Plant Biochem. Mol. Biol. 1997, 45, 1165–1172. [Google Scholar] [CrossRef]
- Tewtrakul, S.; Miyashiro, H.; Nakamura, N.; Hattori, M.; Kawahata, T.; Otake, T.; Yoshinaga, T.; Fujiwara, T.; Supavita, T.; Yuenyongsawad, S.; et al. HIV-1 Integrase Inhibitory Substances from Coleus parvifolius. Phytother. Res. 2003, 17, 232–239. [Google Scholar] [CrossRef]
- Bantis, F.; Ouzounis, T.; Radoglou, K. Artificial LED Lighting Enhances Growth Characteristics and Total Phenolic Content of Ocimum basilicum, but Variably Affects Transplant Success. Sci. Hortic. 2016, 198, 277–283. [Google Scholar] [CrossRef]
- Teixeira, A.P.; Batista, O.; Fátima Simões, M.; Nascimento, J.; Duarte, A.; de la Torre, M.C.; Rodríguez, B. Abietane Diterpenoids from Plectranthus grandidentatus. Phytochemistry 1997, 44, 325–327. [Google Scholar] [CrossRef]
- Horvath, T.; Linden, A.; Yoshizaki, F.; Eugster, C.H.; Rüedi, P. Abietanes and a Novel 20-Norabietanoid from Plectranthus cyaneus (Lamiaceae). Helv. Chim. Acta 2004, 87, 2346–2353. [Google Scholar] [CrossRef]
- Kelecom, A. An Abietane Diterpene from the Labiate Coleus barbatus. Phytochemistry 1984, 23, 1677–1679. [Google Scholar] [CrossRef]
- Grob, K.; Rüedi, P.; Eugster, C.H. Drüsenfarbstoffe Aus Labiaten: Strukturen von 16 Diterpenen (Coleone Und Royleanone) Aus Coleus coerulescens GÜRKE. Helv. Chim. Acta 1978, 61, 871–884. [Google Scholar] [CrossRef]
- Schmid, J.M.; Rüedi, P.; Eugster, C.H. Diterpenoide Drüsenfarbstoffe Aus Labiaten: 22 Neue Coleone Und Royleanone Aus Plectranthus lanuginosus. Helv. Chim. Acta 1982, 65, 2136–2163. [Google Scholar] [CrossRef]
- Künzle, J.M.; Rüedi, P.; Eugster, C.H. Isolierung und Strukturaufklarung von 36 Diterpenoiden aus Trichomenvon Plectranthus edulis. Helv. Chim. Acta 1987, 70, 1911–1929. [Google Scholar] [CrossRef]
- Zhu, Z.-Y.; Nayeshiro, H.; Prewo, R.; Rueedi, P.; Eugster, C.H. Fredericon A, B, C, and D: Novel and Highly Functionalized Abietanoids from Leaf-Glands of Coleus fredericii G. Tayl. ChemInform 1988, 19. [Google Scholar] [CrossRef]
- Mei, S.-X.; Jiang, B.; Niu, X.-M.; Li, M.-L.; Yang, H.; Na, Z.; Lin, Z.-W.; Li, C.-M.; Sun, H.-D. Abietane Diterpenoids from Coleus xanthanthus. J. Nat. Prod. 2002, 65, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Koukoulitsa, C.; Karioti, A.; Bergonzi, M.C.; Pescitelli, G.; Di Bari, L.; Skaltsa, H. Polar Constituents from the Aerial Parts of Origanum vulgare L. ssp. Hirtum Growing Wild in Greece. J. Agric. Food Chem. 2006, 54, 5388–5392. [Google Scholar] [CrossRef]
- Kelecom, A.; Dos Santos, T.C.; Medeiros, W.L.B. Secoabietane Diterpenes from Coleus barbatus. Phytochemistry 1987, 26, 2337–2340. [Google Scholar] [CrossRef]
- Miyase, T.; Yoshizaki, F.; Kabengele, N.; Rüedi, P.; Eugster, C.H. Strukturen von 13 Diterpenen (Coleonen) Aus Blattdrüsen von Solenostemon sylvaticus Und Coleus garckeanus (Labiatae). Helv. Chim. Acta 1979, 62, 2374–2383. [Google Scholar] [CrossRef]
- Schmid, R.; Heuckeroth, S.; Korf, A.; Smirnov, A.; Myers, O.; Dyrlund, T.S.; Bushuiev, R.; Murray, K.J.; Hoffman, N.; Lu, M.; et al. Integrative Analysis of Multimodal Mass Spectrometry Data in MZmine 3. Nat. Biotechnol. 2023, 41, 447–449. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Duy Nguyen, D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-Based Molecular Networking in the GNPS Analysis Environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Rutz, A.; Sorokina, M.; Galgonek, J.; Mietchen, D.; Willighagen, E.; Gaudry, A.; Graham, J.G.; Stephan, R.; Page, R.; Vondrášek, J.; et al. The LOTUS Initiative for Open Knowledge Management in Natural Products Research. eLife 2022, 11, e70780. [Google Scholar] [CrossRef] [PubMed]


| Cultivation Itinerary | A (Control) | B (LEDs) | C (Biostimulant) | D (Hydric Stress) |
|---|---|---|---|---|
| Dried root biomass (g) (Relative to the control) | 296.0 - | 690.8 (+133%) | 264.0 (−11%) | 305.2 (+3%) |
| Quantity of forskolin produced (mg) (Relative to the control) | 229.9 ± 17.7 a - | 710.1 ± 21.3 b (+209%) | 229.4 ± 8.9 a (±0%) | 212.4 ± 4.9 a (−8%) |
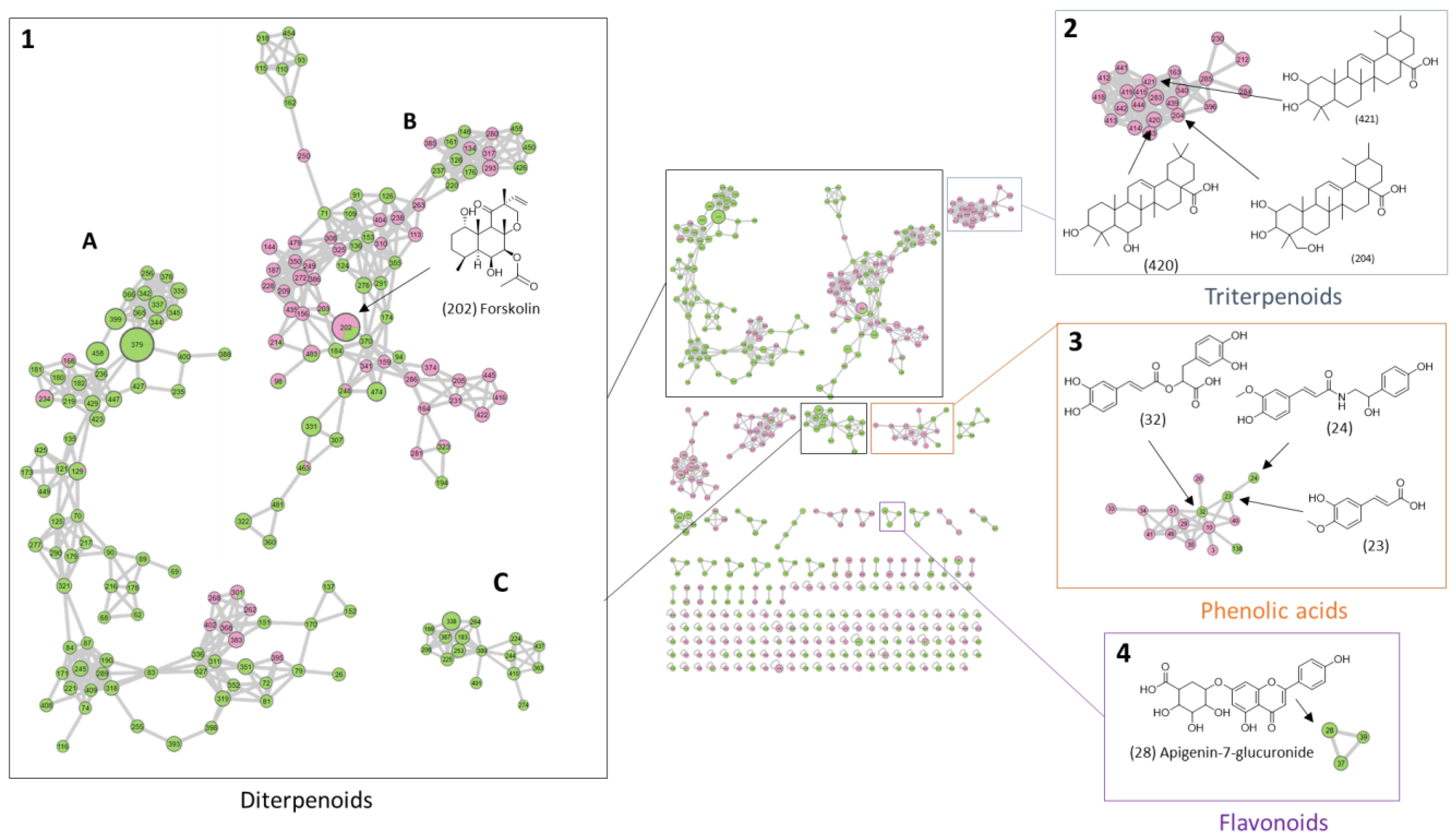
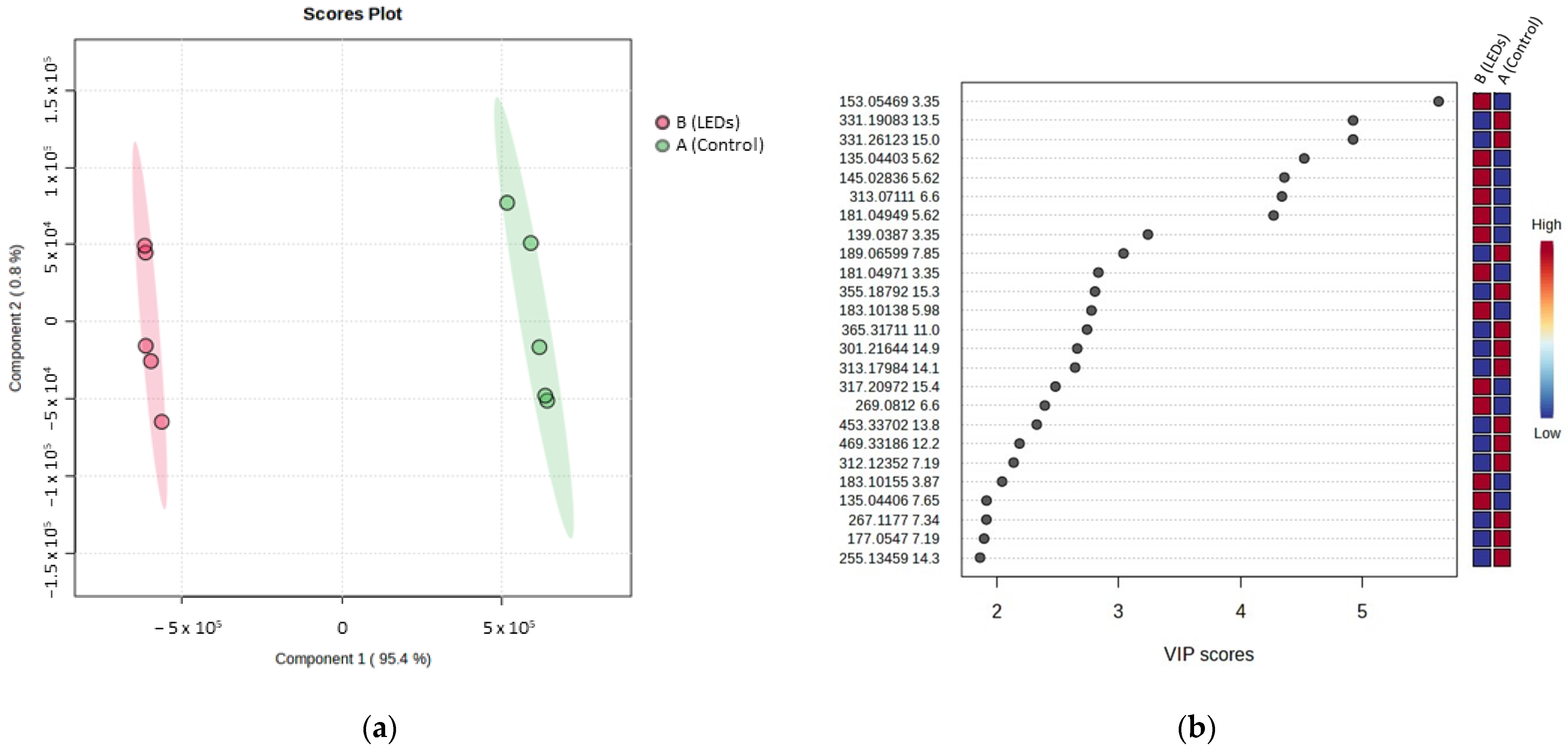
| Compound | tR | VIP Position | m/z | Adduct or Fragment | Itinerary | Calculated Molecular Formula [M] | Putative Annotation | Molecular Family |
|---|---|---|---|---|---|---|---|---|
| R1 | 3.35 | 1 | 153.0546 | Fragments | B | C9H10O5 | Vanillylmandelic acid | Phenolic acid |
| 8 | 139.0387 | |||||||
| 10 | 181.0497 | |||||||
| R2 | 3.87 | 21 | 183.1015 | [M + H]+ | B | C10H14O3 | - | - |
| R3 | 5.62 | 7 | 181.0494 | [M + H]+ | B | C9H8O4 | Caffeic acid | Phenolic acids |
| 5 | 145.0283 | Fragments | ||||||
| 4 | 135.0440 | |||||||
| R4 | 5.98 | 12 | 183.1014 | [M + H]+ | B | C10H14O3 | - | - |
| R5 | 6.60 | 6 | 313.0711 | [M + H]+ | B | C17H12O6 | 4-(3,4-Dihydroxyphenyl)-6,7-dihydroxynaphthalene-2-carboxylic | Phenylpropanoids |
| 17 | 269.0812 | Fragment | ||||||
| R6 | 7.19 | 20 | 312.1235 | [M − H2O + H] | A | C18H19NO5 | N-Feruloyloctopamine | Cinnamic acid amides |
| 24 | 177.0547 | Fragment | ||||||
| R7 | 7.34 | 23 | 267.1177 | - | A | - | - | - |
| R8 | 7.65 | 22 | 135.0440 | Fragment | B | C18H16O8 | Rosmarinic acid | Phenolic acids |
| R9 | 7.85 | 9 | 189.0659 | - | A | - | - | - |
| R10 | 11.00 | 13 | 365.3171 | [M + H]+ | A | C22H40N2O2 | - | - |
| R11 | 12.20 | 19 | 469.3319 | [M + H]+ | A | - | - | - |
| R12 | 13.50 | 2 | 331.1908 | [M + H]+ | A | C20H26O4 | 14-Hydroxytaxodione (abietane diterpene) | Diterpenes |
| R13 | 13.80 | 18 | 453.3370 | [M − 2H2O + H]+ | A | C30H48O5 | Tormentic acid | Triterpenes |
| R14 | 14.10 | 15 | 313.1798 | [M + H]+ | A | C20H24O3 | Abietane diterpene derivative | Diterpenes |
| R15 | 14.30 | 25 | 255.1353 | [M + Na]+ | A | C15H20O2 | - | - |
| R16 | 14.90 | 14 | 301.2164 | [M + H]+ | A | C20H28O2 | Sugiol / barbatusol | Diterpenes |
| R17 | 15.00 | 3 | 331.2612 | [M + Na]+ | A | C20H36O2 | - | - |
| R18 | 15.30 | 11 | 355.1879 | - | A | - | - | Fatty acid- |
| R19 | 15.40 | 16 | 317.2097 | [M + Na]+ | B | C18H30O3 | Fatty acid derivative | Fatty acids |
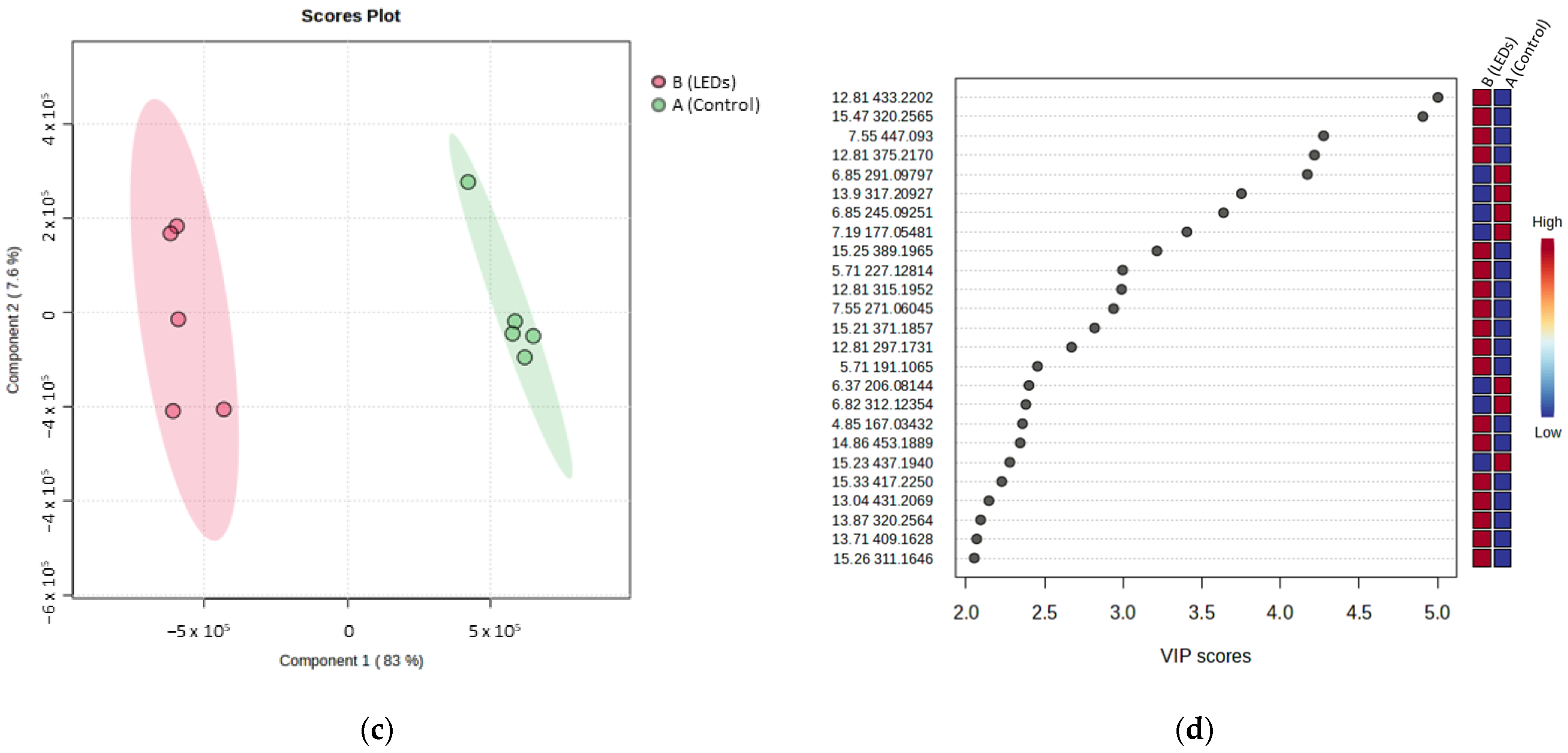
| Compound | tR | VIP Position | m/z | Adduct or Fragment | Itinerary | Calculated Molecular Formula [M] | Putative Annotation | Molecular Family |
|---|---|---|---|---|---|---|---|---|
| AP1 | 4.85 | 18 | 167.0343 | Fragment | B | C10H10O6 | 4,6-Dimethoxyisophtalic acid | Phenolic acid |
| AP2 | 5.71 | 10 | 227.1281 | Fragments | B | C18H28O9 | Hydroxyjasmonic acid hexose | Fatty acyl glycoside |
| 15 | 191.1065 | |||||||
| AP3 | 6.37 | 16 | 206.0814 | Fragment | A | C12H13NO5 | 2-[(2-Carboxyacetyl)amino]-3-phenylpropanoic | Dipeptides |
| AP4 | 6.82 | 17 | 312.1235 | [M + H]+ | A | C18H17NO4 | Hydroxycinnamic acid amide derivative | Cinnamic acid amides |
| AP5 | 6.85 | 5 | 291.0979 | [M + H]+ | A | C14H13N2O4 | N-Malonyl-D-tryptophan | Dipeptides |
| 7 | 245.0925 | Fragment | ||||||
| AP6 | 7.19 | 8 | 177.0548 | Fragment | A | C18H19NO5 | N-Feruloyloctopamine | Cinnamic acid amides |
| AP7 | 7.55 | 3 | 447.093 | [M + H]+ | B | C21H18O11 | Apigenin-glucuronide | Flavonoids |
| 12 | 271.0604 | Fragment | ||||||
| AP10 | 12.81 | 1 | 433.2202 | [M + Na]+ | B | C22H34O7 | Forskolin | Diterpenes |
| 4 | 375.2170 | [M − H2O + H]+ | ||||||
| 11 | 315.1952 | Fragments | ||||||
| 14 | 297.1731 | |||||||
| AP11 | 13.04 | 22 | 431.2069 | [M + H]+ | B | C24H30O7 | Cycloabetiane derivative | Diterpenes |
| AP12 | 13.71 | 24 | 409.1628 | [M + Na]+ | B | C22H26O6 | Coleon Z | Diterpenes |
| AP13 | 13.87 | 23 | 320.2564 | [M + Na]+ | B | C18H35NO2 | N-Acyl amines derivatives | Fatty amides |
| AP14 | 13.9 | 6 | 317.2092 | [M + H]+ | A | C20H28O3 | 11,20-Dihydroxysugiol/ royleanone | Diterpenes |
| AP15 | 14.86 | 19 | 453.1889 | [M + Na]+ | B | C24H30O7 | Cycloabetiane derivative | Diterpenes |
| AP16 | 15.21 | 13 | 371.1857 | [M + H]+ | B | C22H26O5 | Abetiane diterpene derivative | Diterpenes |
| AP17 | 15.23 | 20 | 437.1940 | [M + Na]+ | A | C24H30O6 | 6,7-Secoabietane diterpene derivative | Diterpenes |
| AP18 | 15.25 | 9 | 389.1965 | [M + H]+ | B | C22H28O6 | Abetiane diterpene derivative | Diterpenes |
| 15.26 | 25 | 311.1646 | Fragment | |||||
| AP19 | 15.33 | 21 | 417.2250 | [M + Na]+ | B | C22H34O6 | 9-Deoxyforskolin | Diterpenes |
| AP20 | 15.47 | 2 | 320.2565 | [M + Na]+ | B | C18H35NO2 | N-Acyl amines derivatives | Fatty amides |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Cabec, A.; Campos, P.-E.; Yzebe, O.; Pelé, R.; Colas, C.; Destandau, E. Enhancement of Forskolin Production Using Aeroponic Cultivation of Coleus forskohlii and the Impact on the Plant Phytochemistry. Molecules 2024, 29, 4215. https://doi.org/10.3390/molecules29174215
Le Cabec A, Campos P-E, Yzebe O, Pelé R, Colas C, Destandau E. Enhancement of Forskolin Production Using Aeroponic Cultivation of Coleus forskohlii and the Impact on the Plant Phytochemistry. Molecules. 2024; 29(17):4215. https://doi.org/10.3390/molecules29174215
Chicago/Turabian StyleLe Cabec, Audrey, Pierre-Eric Campos, Olivier Yzebe, Ronan Pelé, Cyril Colas, and Emilie Destandau. 2024. "Enhancement of Forskolin Production Using Aeroponic Cultivation of Coleus forskohlii and the Impact on the Plant Phytochemistry" Molecules 29, no. 17: 4215. https://doi.org/10.3390/molecules29174215
APA StyleLe Cabec, A., Campos, P.-E., Yzebe, O., Pelé, R., Colas, C., & Destandau, E. (2024). Enhancement of Forskolin Production Using Aeroponic Cultivation of Coleus forskohlii and the Impact on the Plant Phytochemistry. Molecules, 29(17), 4215. https://doi.org/10.3390/molecules29174215






