Antibacterial Activity and Multi-Targeted Mechanism of Action of Suberanilic Acid Isolated from Pestalotiopsis trachycarpicola DCL44: An Endophytic Fungi from Ageratina adenophora
Abstract
1. Introduction
2. Results
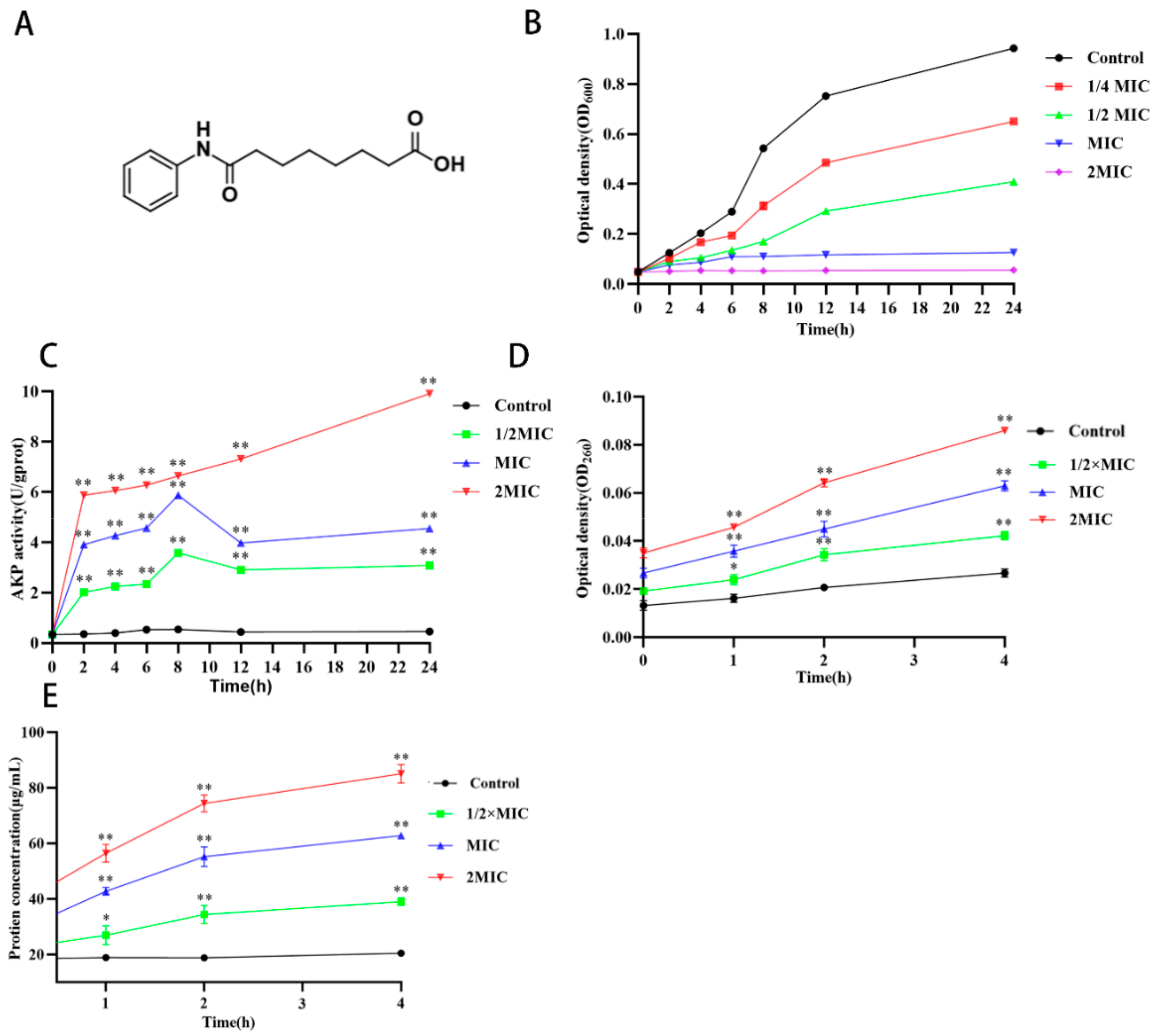
2.1. Antimicrobial Effect of Suberanilic Acid
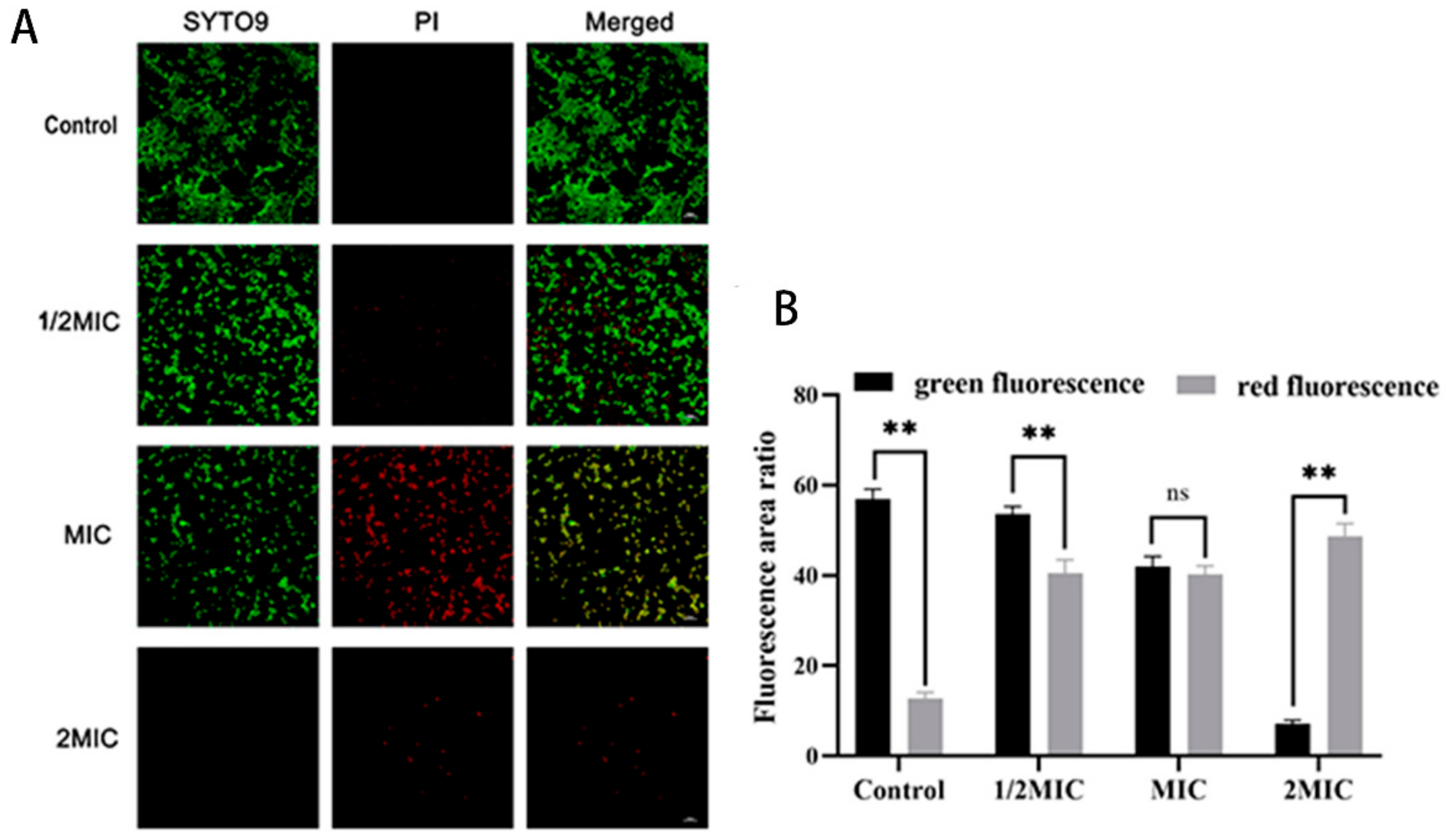
2.2. Disruption of MRSA Cell Wall Membranes by Suberanilic Acid
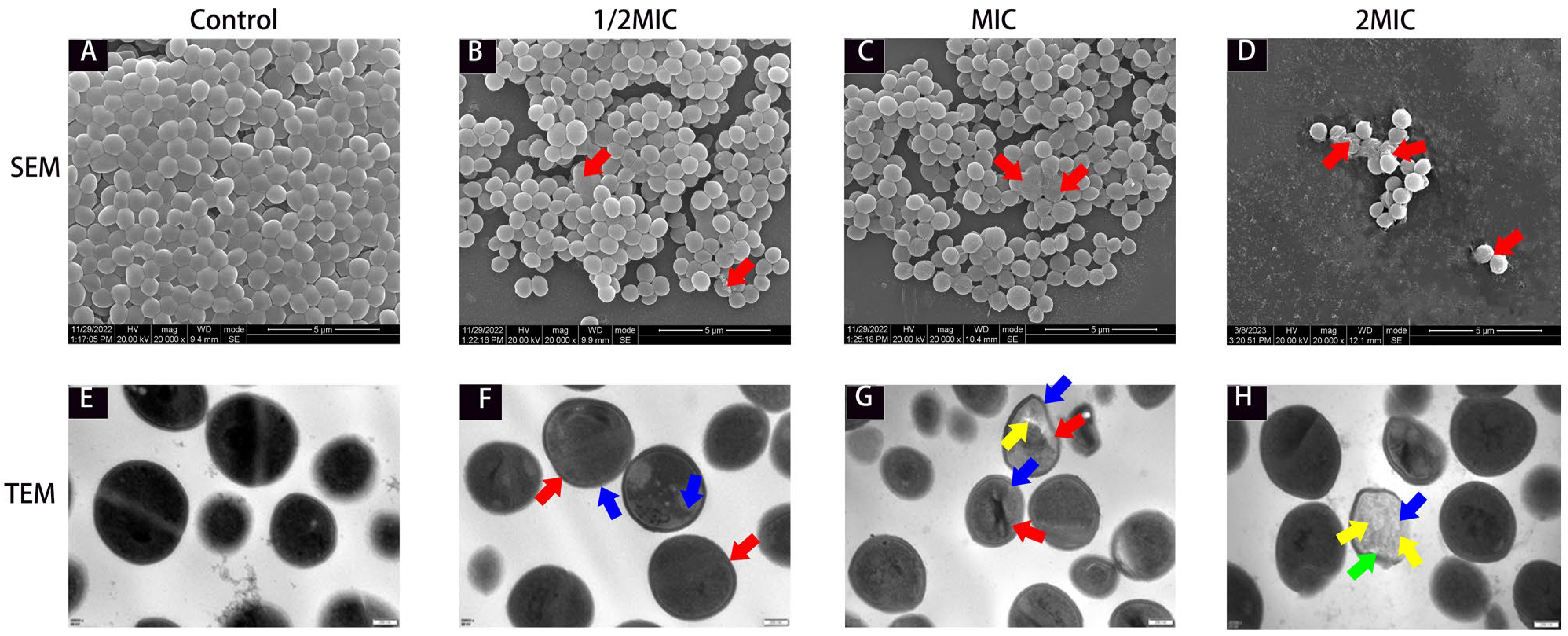
2.3. Ultrastructural Changes in MRSA in the Presence of Suberanilic Acid
2.4. TMT Proteomic Analysis of MRSA Action by Suberanilic Acid
2.4.1. Functional Classification of Differentially Expressed Proteins
2.4.2. Kyoto Encyclopedia of Genes and Genomes Analysis of Differentially Expressed Proteins
2.5. PRM Analysis
3. Discussion
4. Material and Methods
4.1. Compounds and Bacterial Cultures
4.2. Antimicrobial Activity
4.2.1. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
4.2.2. Effect of Suberanilic Acid on Growth Curves
4.2.3. Fractional Inhibition Concentration Index of Antibiotics and Suberanilic Acid
4.3. Effect of Suberanilic Acid on MRSA
4.3.1. Detection of Alkaline Phosphatase Leakage
4.3.2. Nucleic Acid and Protein Leakage Detection
4.3.3. Effect of Suberanilic Acid on MRSA Viability
4.3.4. SEM and TEM Observations
4.4. TMT Quantitative Proteomics Analysis of Differentially Expressed Proteins
4.5. PRM Analysis of Key Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Titouche, Y.; Akkou, M.; Houali, K.; Frédéric, A.; Hennekinne, J.A. Role of milk and milk products in the spread of methicillin-resistant Staphylococcus aureus in the dairy production chain. J. Food Sci. 2022, 87, 3699–3723. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, A.C.; Rodrigues, M.X.; Cirone Silva, N.C. Methicillin-resistant Staphylococcus aureus in food and the prevalence in Brazil: A review. Braz. J. Microbiol. 2020, 51, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Rembischevski, P.; Caldas, E.D. Risk perception related to food. Food Sci. Technol. 2020, 40, 779–785. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Wei, X.; Hu, Y.; Sun, C.; Wu, S. Characterization of a novel antimicrobial peptide bacipeptin against foodborne pathogens. J. Agric. Food Chem. 2017, 72, 5283–5292. [Google Scholar] [CrossRef]
- Wen, J.; Okyere, S.K.; Wang, S.; Wang, J.; Xie, L.; Ran, Y.; Hu, Y. Endophytic fungi: An effective alternative source of plant-derived bioactive compounds for pharmacological studies. J. Fungi 2022, 8, 205. [Google Scholar] [CrossRef]
- Atiphasaworn, P.; Monggoot, S.; Gentekaki, E.; Brooks, S.; Pripdeevech, P. Antibacterial and antioxidant constituents of extracts of endophytic fungi isolated from Ocimum basilicum var. thyrsiflora Leaves. Curr. Microbiol. 2017, 74, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Stowell, J.C.; Huot, R.I.; Voast, L.V. The synthesis of N-Hydroxy-N’-phenyloctanediamide and its inhibitory effect on proliferation of AXC rat prostate cancer cells. J. Med. Chem. 1995, 38, 1411–1413. [Google Scholar] [CrossRef]
- Rocha, B.R.; Martins, G.M.; Cruz, B.C.; Lilenbaum, W. Determination of minimum inhibitory and minimum bactericidal concentrations of brazilian strains of Leptospira spp. for streptomycin sulfate. Epidemiol. Infect. 2020, 148, e235. [Google Scholar] [CrossRef]
- Katsipis, G.; Tsalouxidou, V.; Halevas, E.; Geromichalou, E.; Geromichalos, G.; Pantazaki, A.A. In vitro and in silico evaluation of the inhibitory effect of a curcumin-based oxovanadium (iv) complex on alkaline phosphatase activity and bacterial biofilm formation. Appl. Microbiol. Biotechnol. 2020, 105, 147–168. [Google Scholar] [CrossRef]
- Kusari, S.; Pandey, S.P.; Spiteller, M. Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites. Phytochemistry 2013, 91, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, S.K.; Dufossé, L.; Chhipa, H.; Saxena, S.; Mahajan, G.B.; Gupta, M.K. Fungal endophytes: A potential source of antibacterial compounds. J. Fungi 2022, 8, 164. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, B.; Yang, J.; Ma, X.; Deng, S.; Huang, Y.; Wen, Y.; Yuan, J.; Yang, X. Essential oil derived from Eupatorium adenophorum Spreng. mediates anticancer effect by inhibiting STAT3 and AKT activation to induce apoptosis in hepatocellular carcinoma. Front. Pharmacol. 2018, 9, 483. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Xie, L.; Okyere, S.K.; Wen, J.; Ran, Y.; Nong, X.; Hu, Y. Antibacterial activity of two metabolites isolated from endophytic bacteria Bacillus velezensis Ea73 in Ageratina adenophora. Front. Microbiol. 2022, 13, 860009. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Hu, L.; Shi, Z.; Sun, W.; Yue, D.; Wang, Y.; Ma, X.; Ren, Z.; Zuo, Z.; Peng, G.; et al. Two metabolites isolated from endophytic fungus Coniochaeta sp. F-8 in Ageratina adenophora exhibit antioxidative activity and cytotoxicity. Nat. Prod. Res. 2019, 35, 2840–2848. [Google Scholar] [CrossRef]
- Okyere, S.; Xie, L.; Wen, J.; Hu, Y. Bacillus toyonensis SAU-19 ameliorates hepatic insulin resistance in high-fat diet/streptozocin-induced diabetic mice. Nutrients 2021, 13, 4512. [Google Scholar] [CrossRef]
- Wen, J.; Okyere, S.K.; Wang, J.; Huang, R.; Wang, Y.; Liu, L.; Nong, X.; Hu, Y. Endophytic fungi isolated from Ageratina adenophora exhibits potential antimicrobial activity against Multidrug-resistant Staphylococcus aureus. Plants 2023, 12, 65. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, T.; Ming, Q.; Wu, L.; Rahman, K.; Qin, L. Alkaloids produced by endophytic fungi: A review. Nat. Prod. Commun. 2012, 7, 963–968. [Google Scholar] [CrossRef]
- Liu, F.; Wang, F.T.; Du, L.H.; Zhao, T.; Doyle, M.; Wang, D.Y.; Zhang, X.X.; Sun, Z.L.; Xu, W.M. Antibacterial and antibiofilm activity of phenyllactic acid against Enterobacter cloacae. Food Control. 2018, 84, 442–448. [Google Scholar] [CrossRef]
- Deng, H.T.; Kong, Y.W.; Zhu, J.Y.; Jiao, X.Y.; Tong, Y.Q.; Wan, M.; Zhao, Y.; Lin, S.; Ma, Y.; Meng, X. Proteomic analyses revealed the antibacterial mechanism of Aronia melanocarpa isolated anthocyanins against Escherichia coli O157: H7. Curr. Res. Food Sci. 2022, 5, 1559–1569. [Google Scholar] [CrossRef]
- Erni, B. The bacterial phosphoenolpyruvate: Sugar phosphotransferase system (PTS): An interface between energy and signal transduction. J. Iran. Chem. Soc. 2013, 10, 593–630. [Google Scholar] [CrossRef]
- Bi, X.; Wang, Y.; Hu, X.; Liao, X. iTRAQ-based proteomic analysis of sublethally injured Escherichia coli O157:H7 cells induced by high pressure carbon dioxide. Front. Microbiol. 2017, 8, 2544. [Google Scholar] [CrossRef]
- Kang, S.; Shi, C.; Chang, J.; Kong, F.; Li, M.; Guan, B.; Zhang, Z.; Shi, X.; Zhao, H.; Peng, Y.; et al. Label free-based proteomic analysis of the food spoiler Pseudomonas fluorescens response to lactobionic acid by SWATH-MS. Food.Control. 2021, 123, 107834. [Google Scholar] [CrossRef]
- Kang, S.M.; Kong, F.H.; Liang, X.N.; Li, M.H.; Yang, N.; Cao, X.Y.; Yang, M.; Tao, D.; Yue, X.; Zheng, Y. Label-free quantitative proteomics reveals the Multitargeted antibacterial mechanisms of lactobionic acid against Methicillin resistant Staphylococcus aureus (MRSA) using SWATH-MS technology. J. Agric. Food Chem. 2019, 67, 12322–12332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wan, H.; Gao, Z.; Wei, Y.; Wang, W.; Liu, G.; Shtykova, E.; Xu, J.; Dong, Y. Insights into the catalytic mechanism of 16S rRNA methyltransferase RsmE (m3U1498) from crystal and solution structures. J. Mol. Biol. 2012, 423, 576–589. [Google Scholar] [CrossRef]
- Alex, E. Ribosome inhibitor combats bacterial drug resistance. Nat. Rev. Drug Discov. 2023, 22, 267. [Google Scholar] [CrossRef]
- Ning, Y.W.; Fu, Y.N.; Hou, L.L.; Ma, M.G.; Wang, Z.X.; Li, X.F.; Jia, Y.M. iTRAQ-based quantitative proteomic analysis of synergistic antibacterial mechanism of phenyllactic acid and lactic acid against Bacillus cereus. Food Res. Int. 2021, 139, 109562. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Gan, R.-Y.; Zhang, D.; Farha, A.K.; Habimana, O.; Mavumengwana, V.; Li, H.-B.; Wang, X.-H.; Corke, H. Large-scale screening of 239 traditional Chinese medicinal plant extracts for their antibacterial activities against Multidrug-resistant Staphylococcus aureus and cytotoxic activities. Pathogens 2020, 9, 185. [Google Scholar] [CrossRef]
- Suzuki, T.; Ohashi, Y. Combination effect of antibiotics against bacteria isolated from keratitis using fractional inhibitory concentration index. Cornea 2013, 32, e156–e160. [Google Scholar] [CrossRef]
- Martin, S.J.; Pendland, S.L.; Chen, C.; Schreckenberger, P.; Danziger, L.H. In vitro synergy testing of macrolide-quinolone combinations against 41 clinical isolates of Legionella. Antimicrob. Agents Chemother. 1996, 40, 1419–1421. [Google Scholar] [CrossRef]
- Wang, F.; Wei, F.; Song, C.; Jiang, B.; Tian, S.; Yi, J.; Yu, C.; Song, Z.; Sun, L.; Bao, Y.; et al. Dodartia orientalis l. essential oil exerts antibacterial activity by mechanisms of disrupting cell structure and resisting biofilm. Ind. Crops Prod. 2017, 109, 358–366. [Google Scholar] [CrossRef]
- Chen, J.; Tang, C.; Zhang, R.; Ye, S.; Zhao, Z.; Huang, Y.; Xu, X.; Lan, W.; Yang, D. Metabolomics analysis to evaluate the antibactrial activity of essential oil from the leaves of Cinnamomum camphora (Linn.) Presl. J. Ethnopharmacol. 2020, 253, 112652. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Liu, L.; Liu, Y.; Wang, X. Ferulic acid inactivates Shigella flexneri through cell membrane destructieon, biofilm retardation, and altered gene expression. J. Agric. Food Chem. 2020, 68, 7121–7131. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Wang, G.; Tam, Y.T.; Tam, C. Membrane-active epithelial keratin 6A fragments (KAMPs) are unique human Antimicrobial Peptides with a Non-αβ Structure. Front. Microbiol. 2016, 7, 1799. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gao, M.; Wang, L.; Li, S.; He, J.; Qin, A.; Ren, L.; Wang, Y.; Tang, B.Z. Aggregation-induced emission probe for study of the bactericidal mechanism of antimicrobial peptides. ACS Appl. Mater. Interfaces 2018, 10, 11436–11442. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, X.; Huang, A. Proteomic analysis and food-grade enzymes of Moringa oleifer Lam. a Lam. flower. Int. J. Biol. Macromol. 2018, 115, 883–890. [Google Scholar] [CrossRef]

| Name of Antibiotic | MIC (µg/mL) | Drug Combination MIC | FICI | Result | |
|---|---|---|---|---|---|
| Antibiotics (µg/mL) | Suberanilic Acid (µg/mL) | ||||
| Oxacillin | 128 | 32 | 32 | 1.25 | I |
| Linezolid | 2 | 0.125 | 32 | 0.5625 | P |
| Vancomycin | 0.5 | 1 | 32 | 2.25 | I |
| Clindamycin | 256 | 256 | 32 | 0.75 | P |
| Sarafloxacin | 64 | 128 | 32 | 2.125 | I |
| Florfenicol | 32 | 16 | 32 | 0.75 | P |
| Ampicillin | 64 | 32 | 32 | 0.75 | P |
| Gentamycin | 64 | 32 | 32 | 1 | A |
| Doxycycline | 0.5 | 0.5 | 32 | 1.125 | I |
| Tobramycin | 256 | 16 | 32 | 0.5625 | P |
| Enrofloxacin | 0.25 | 0.125 | 32 | 1 | A |
| Amikacin | 256 | 64 | 32 | 2.25 | I |
| Protein ID | Gene | Description | Fold Change | Change | p-Value |
|---|---|---|---|---|---|
| Metabolism | |||||
| Q2FK94 | aldA | Putative aldehyde dehydrogenase AldA | 1.948 | Up | 2.01521 × 10−5 |
| Q5HGY8 | pdhD | Dihydrolipoyl dehydrogenase | 1.534 | Up | 0.000545771 |
| A0A033V2M2 | pfkA | ATP-dependent 6-phosphofructokinase | 0.828 | Down | 0.020923953 |
| P64225 | gcvT | Aminomethyltransferase | 1.857 | Up | 0.000233977 |
| Q6GG12 | icd | Isocitrate dehydrogenase (NADP) | 2.548 | Up | 2.69982 × 10−6 |
| Q6GFK5 | fumC | Fumarate hydratase class II | 1.462 | Up | 8.04802 × 10−5 |
| Q6GHI9 | sucD | Succinate–CoA ligase subunit alpha | 1.307 | Up | 0.004931368 |
| A0A8E6CNC9 | sucB | Dihydrolipoyllysine-residue succinyltransferase | 1.852 | Up | 0.000103329 |
| A0A0U1MI53 | pdhA | Pyruvate dehydrogenase E1 component subunit alpha | 1.496 | Up | 0.001443745 |
| P65884 | purA | Adenylosuccinate synthetase | 0.576 | Down | 0.000180435 |
| Q6GAW6 | argH | Argininosuccinate lyase | 0.404 | Down | 5.68 × 10−5 |
| A0A2S6D4J7 | glmS | Glutamine-fructose-6-phosphate aminotransferase | 0.799 | Down | 0.000120906 |
| Q6GJB4 | ilvE | Probable branched-chain-amino-acid aminotransferase | 0.681 | Down | 0.000162841 |
| A0A850FXK9 | cysK | Cysteine synthase | 0.707 | Down | 0.015964978 |
| Q6GDG7 | arcA | Arginine deiminase | 0.301 | Down | 0.001221858 |
| Q6GIC7 | argG | Argininosuccinate synthase | 0.147 | Down | 7.32963 × 10−6 |
| Q6GAW6 | argH | Argininosuccinate lyase | 0.404 | Down | 5.68 × 10−5 |
| Membrane transport | |||||
| Q6GH18 | pstS | Phosphate-binding protein PstS | 1.544 | Up | 0.009992197 |
| A0A660A2T8 | cycB | Maltodextrin-binding protein | 0.799 | Down | 0.001032071 |
| A0A0U1MSA4 | modA | Molybdate ABC transporter substrate-binding protein | 0.621 | Down | 0.00703426 |
| A0A2S6DD31 | hrtA | Putative hemin import ATP-binding protein HrtA | 0.723 | Down | 0.008434244 |
| Signal transduction | |||||
| Q5HEP0 | vraR | Response regulator protein VraR | 0.816 | Down | 0.017176601 |
| Translation | |||||
| A6QJ83 | rpsQ | 30S ribosomal protein S17 | 1.443 | Up | 0.023323817 |
| A8Z339 | rpmD | 50S ribosomal protein L30 | 1.511 | Up | 0.010285859 |
| Q6GJD4 | rpmG3 | 50S ribosomal protein L33 3 | 0.555 | Down | 0.01660196 |
| P66645 | rpsI | 30S ribosomal protein S9 | 1.272 | Up | 0.001495131 |
| A0A850G4M4 | rplC | 50S ribosomal protein L3 | 1.326 | Up | 6.12434 × 10−6 |
| No. | Accession | Gene | TMT Fold Change | PRM | ||
|---|---|---|---|---|---|---|
| p-Value | Fold Change | Trend | ||||
| 1 | Q2FK94 | aldA | 1.948 ↑ | 0.002506652 | 3.468 ↑ | Consistency |
| 2 | Q5HGY8 | pdhD | 1.534 ↑ | 0.024147859 | 1.965 ↑ | Consistency |
| 3 | P64225 | gcvT | 1.857 ↑ | 0.007774184 | 3.685 ↑ | Consistency |
| 4 | Q6GG12 | icd | 2.548 ↑ | 0.000980574 | 3.874 ↑ | Consistency |
| 5 | Q6GFK5 | fumC | 1.462 ↑ | 0.000550982 | 2.589 ↑ | Consistency |
| 6 | A0A0U1MI53 | pdhA | 1.496 ↑ | 0.00642461 | 2.194 ↑ | Consistency |
| 7 | P65884 | purA | 0.576 ↓ | 0.023770336 | 0.390 ↓ | Consistency |
| 8 | A0A2S6D4J7 | glmS | 0.799 ↓ | 0.932842293 | 1.017 ↓ | Consistency |
| 9 | Q6GJB4 | ilvE | 0.681 ↓ | 0.088087529 | 0.723 ↓ | Consistency |
| 10 | Q6GDG7 | arcA | 0.301 ↓ | 0.001021429 | 0.154 ↓ | Consistency |
| 11 | Q6GIC7 | argG | 0.147 ↓ | 0.001274761 | 0.012 ↓ | Consistency |
| 12 | Q6GH18 | pstS | 1.544 ↑ | 0.093222288 | 1.783 ↑ | Consistency |
| 13 | A0660A2T8 | cycB | 0.799 ↓ | 0.199271957 | 0.797 ↓ | Consistency |
| 14 | Q5HEP0 | vraR | 0.816 ↓ | 0.965351503 | 1.006 ↓ | Consistency |
| 15 | A6QJ83 | rpsQ | 1.443 ↑ | 0.008784577 | 2.012 ↑ | Consistency |
| 16 | A8Z339 | rpmD | 1.511 ↑ | 0.004055361 | 1.613 ↑ | Consistency |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, J.; Okyere, S.K.; Wang, S.; Wang, J.; Huang, R.; Tang, Z.; Wang, X.; Shao, C.; Hu, Y. Antibacterial Activity and Multi-Targeted Mechanism of Action of Suberanilic Acid Isolated from Pestalotiopsis trachycarpicola DCL44: An Endophytic Fungi from Ageratina adenophora. Molecules 2024, 29, 4205. https://doi.org/10.3390/molecules29174205
Wen J, Okyere SK, Wang S, Wang J, Huang R, Tang Z, Wang X, Shao C, Hu Y. Antibacterial Activity and Multi-Targeted Mechanism of Action of Suberanilic Acid Isolated from Pestalotiopsis trachycarpicola DCL44: An Endophytic Fungi from Ageratina adenophora. Molecules. 2024; 29(17):4205. https://doi.org/10.3390/molecules29174205
Chicago/Turabian StyleWen, Juan, Samuel Kumi Okyere, Shu Wang, Jianchen Wang, Ruya Huang, Ziyao Tang, Xiaoxuan Wang, Chenyang Shao, and Yanchun Hu. 2024. "Antibacterial Activity and Multi-Targeted Mechanism of Action of Suberanilic Acid Isolated from Pestalotiopsis trachycarpicola DCL44: An Endophytic Fungi from Ageratina adenophora" Molecules 29, no. 17: 4205. https://doi.org/10.3390/molecules29174205
APA StyleWen, J., Okyere, S. K., Wang, S., Wang, J., Huang, R., Tang, Z., Wang, X., Shao, C., & Hu, Y. (2024). Antibacterial Activity and Multi-Targeted Mechanism of Action of Suberanilic Acid Isolated from Pestalotiopsis trachycarpicola DCL44: An Endophytic Fungi from Ageratina adenophora. Molecules, 29(17), 4205. https://doi.org/10.3390/molecules29174205







