Bio-Pathological Functions of Posttranslational Modifications of Histological Biomarkers in Breast Cancer
Abstract
1. Introduction
2. PTMs of Transcription Factors in Breast Cancer
2.1. ERα
2.2. PR
2.3. AR
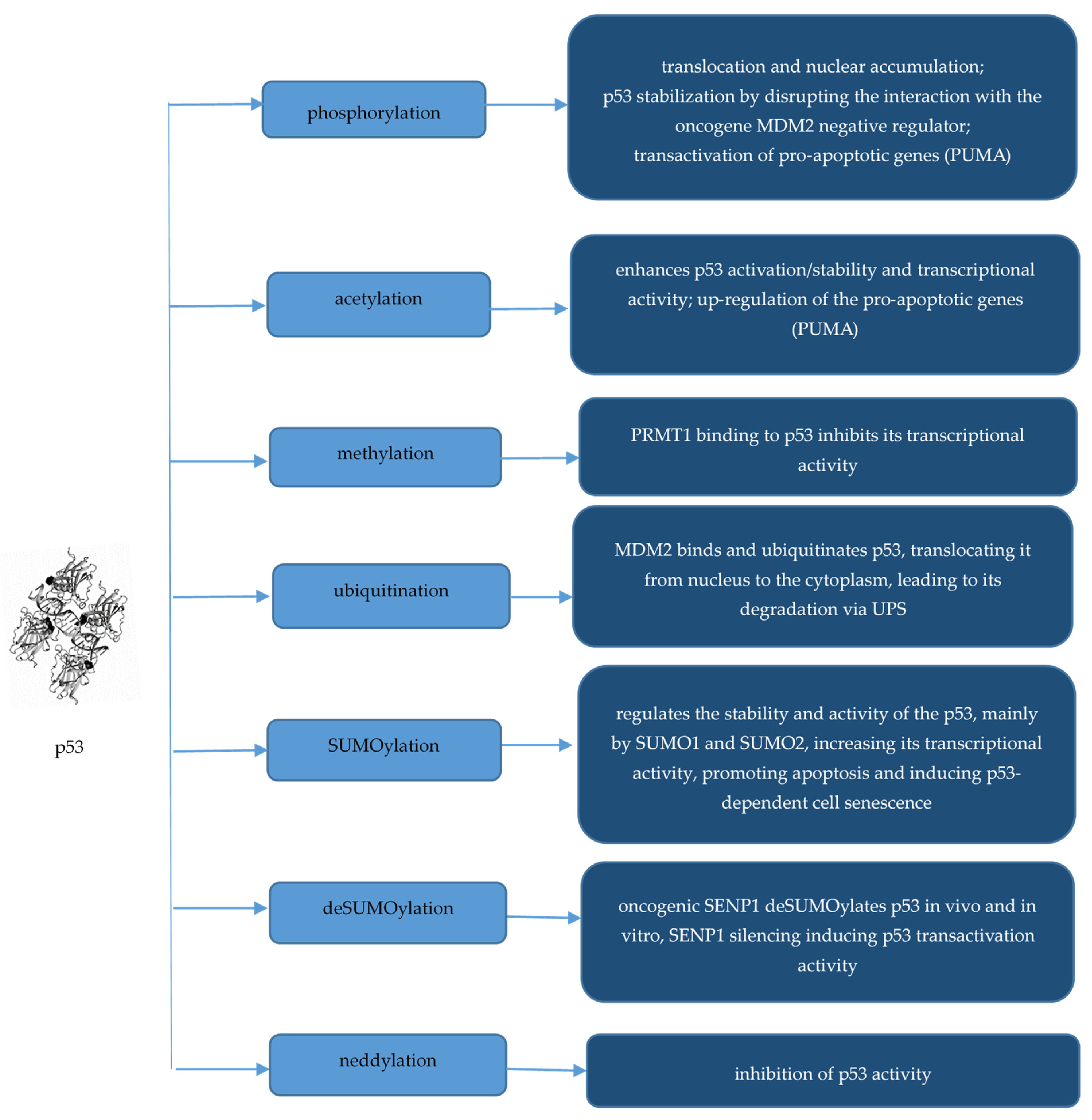
2.4. p53
2.5. Transcription Factors Involved in Epithelial–Mesenchymal Transition
3. PTMs of Proliferation Marker Ki-67
4. PTMs of Plasma Membrane Proteins
4.1. HER2
4.2. Cell Adhesion Proteins
4.3. Other Proteins
5. Role of Histone Modifications in BC
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Puntmann, V.O. How-to guide on biomarkers: Biomarker definitions, validation and applications with examples from cardiovascular disease. Postgrad. Med. J. 2009, 85, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Strimbu, K.; Tavel, J. What are Biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef]
- Afzal, S.; Hassan, M.; Ullah, S.; Abbas, H.; Tawakkal, F.; Khan, M.A. Breast Cancer; Discovery of Novel Diagnostic Biomarkers, Drug Resistance, and Therapeutic Implications. Front Mol. Biosci. 2022, 9, 783450. [Google Scholar] [CrossRef]
- Lopez-Gonzalez, L.; Sanchez Cendra, A.; Sanchez Cendra, C.; Roberts Cervantes, E.D.; Espinosa, J.C.; Pekarek, T.; Fraile-Martinez, O.; García-Montero, C.; Rodriguez-Slocker, A.M.; Jiménez-Álvarez, L.; et al. Exploring Biomarkers in Breast Cancer: Hallmarks of Diagnosis, Treatment, and Follow-Up in Clinical Practice. Medicina 2024, 60, 168. [Google Scholar] [CrossRef] [PubMed]
- Veyssière, H.; Bidet, Y.; Penault-Llorca, F.; Radosevic-Robin, N.; Durando, X. Circulating proteins as predictive and prognostic biomarkers in breast cancer. Clin. Proteom. 2022, 19, 25. [Google Scholar] [CrossRef]
- Galati, F.; Rizzo, V.; Trimboli, R.M.; Kripa, E.; Maroncelli, R.; Pediconi, F. MRI as a biomarker for breast cancer diagnosis and prognosis. BJR Open 2022, 4, 20220002. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Yuan, K.; Chen, L. Molecular biomarkers, network biomarkers, and dynamic network biomarkers for diagnosis and prediction of rare diseases. Fundam. Res. 2022, 2, 894–902. [Google Scholar] [CrossRef]
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- Nimse, S.B.; Sonawane, M.D.; Song, K.-S.; Kim, T. Biomarker detection technologies and future directions. Analyst 2016, 141, 740–755. [Google Scholar] [CrossRef]
- Vihinen, M. Functional effects of protein variants. Biochimie 2021, 180, 104–120. [Google Scholar] [CrossRef]
- Kuo, H.-T.; Hsu, H.-T.; Chang, C.-C.; Jiang, M.-C.; Yeh, C.-M.; Shen, K.-H.; Hsu, P.-C.; Tai, C.-J. High nuclear phosphorylated extracellular signal-regulated kinase expression associated with poor differentiation, larger tumor size, and an advanced stage of breast cancer. Pol. J. Pathol. 2013, 64, 163–169. [Google Scholar] [CrossRef]
- Isono, E.; Li, J.; Pulido, P.; Siao, W.; Spoel, S.H.; Wang, Z.; Zhuang, X.; Trujillo, M. Protein degrons and degradation: Exploring substrate recognition and pathway selection in plants. Plant Cell 2024, koae141. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Liu, J.; Inuzuka, H.; Wei, W. Targeted protein posttranslational modifications by chemically induced proximity for cancer therapy. J. Biol. Chem. 2023, 299, 104572. [Google Scholar] [CrossRef]
- Trivedi, R.; Nagarajaram, H. Intrinsically Disordered Proteins: An Overview. Int. J. Mol. Sci. 2022, 23, 14050. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Wang, Y.; Yang, M.; Hong, F.; Yang, S. Protein Phosphorylation in Cancer: Role of Nitric Oxide Signaling Pathway. Biomolecules 2021, 11, 1009. [Google Scholar] [CrossRef] [PubMed]
- Perez-Oquendo, M.; Gibbons, D.L. Regulation of ZEB1 Function and Molecular Associations in Tumor Progression and Metastasis. Cancers 2022, 14, 1864. [Google Scholar] [CrossRef] [PubMed]
- Rabellino, A.; Khanna, K.K. The implication of the SUMOylation pathway in breast cancer pathogenesis and treatment. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 54–70. [Google Scholar] [CrossRef] [PubMed]
- Dutta, H.; Jain, N. Post-translational modifications and their implications in cancer. Front. Oncol. 2023, 13, 1240115. [Google Scholar] [CrossRef]
- Doyle, H.A.; Mamula, M.J. Post-translational protein modifications in antigen recognition and autoimmunity. Trends Immunol. 2001, 22, 443–449. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Guadagnin, G.; Cappello, P.; Novelli, F. Post-Translational Modifications in Tumor-Associated Antigens as a Platform for Novel Immuno-Oncology Therapies. Cancers 2023, 15, 138. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Q.; Kang, Y.; Xu, S.; Pang, D. Unconventional protein post-translational modifications: The helmsmen in breast cancer. Cell Biosci. 2022, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Hamza, A.; Boyle, E.; Donu, D.; Cen, Y. Post-Translational Modifications and Diabetes. Biomolecules 2024, 14, 310. [Google Scholar] [CrossRef]
- Hermann, J.; Schurgers, L.; Jankowski, V. Identification and characterization of post-translational modifications: Clinical implications. Mol. Asp. Med. 2022, 86, 101066. [Google Scholar] [CrossRef]
- Dunphy, K.; Dowling, P.; Bazou, D.; O’Gorman, P. Current Methods of Post-Translational Modification Analysis and Their Applications in Blood Cancers. Cancers 2021, 13, 1930. [Google Scholar] [CrossRef] [PubMed]
- Hattori, T.; Koide, S. Next-generation antibodies for post-translational modifications. Curr. Opin. Struct. Biol. 2018, 51, 141–148. [Google Scholar] [CrossRef]
- Coleman, D.; Soung, Y.; Surh, Y.-J.; Cardelli, J.; Chung, J. Curcumin Prevents Palmitoylation of Integrin β4 in Breast Cancer Cells. PLoS ONE 2015, 10, e0125399. [Google Scholar] [CrossRef]
- Bollu, L.R.; Ren, J.; Blessing, A.M.; Katreddy, R.R.; Gao, G.; Xu, L.; Wang, J.; Su, F.; Weihua, Z. Involvement of de novo synthesized palmitate and mitochondrial EGFR in EGF induced mitochondrial fusion of cancer cells. Cell Cycle 2014, 13, 2415–2430. [Google Scholar] [CrossRef][Green Version]
- Zhong, Q.; Xiao, X.; Qiu, Y.; Xu, Z.; Chen, C.; Chong, B.; Zhao, X.; Hai, S.; Li, S.; An, Z.; et al. Protein posttranslational modifications in health and diseases: Functions, regulatory mechanisms, and therapeutic implications. MedComm 2023, 4, e261. [Google Scholar] [CrossRef]
- Wang, H.; Yang, L.; Liu, M.; Luo, J. Protein post-translational modifications in the regulation of cancer hallmarks. Cancer Gene Therapy 2022, 30, 529–547. [Google Scholar] [CrossRef]
- Olzscha, H. Posttranslational modifications and proteinopathies: How guardians of the proteome are defeated. Biological Chemistry 2019, 400, 895–915. [Google Scholar] [CrossRef]
- Antonarelli, G.; Pieri, V.; Porta, F.M.; Fusco, N.; Finocchiaro, G.; Curigliano, G.; Criscitiello, C. Targeting Post-Translational Modifications to Improve Combinatorial Therapies in Breast Cancer: The Role of Fucosylation. Cells 2023, 12, 840. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, F.; Zhang, X.; Lin, H.-K.; Xu, C. Insights into the post-translational modification and its emerging role in shaping the tumor microenvironment. Signal Transduct. Target. Ther. 2021, 6, 422. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, C.-W. Epigenetic modulations in triple-negative breast cancer: Therapeutic implications for tumor microenvironment. Pharmacol. Res. 2024, 204, 107205. [Google Scholar] [CrossRef] [PubMed]
- Dees, S.; Pontiggia, L.; Jasmin, J.-F.; Mercier, I. Phosphorylated STAT3 (Tyr705) as a biomarker of response to pimozide treatment in triple-negative breast cancer. Cancer Biol. Ther. 2020, 21, 506–521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, X.; Gao, J.; Sun, Y.; Liu, T.; Yan, Q.; Yang, X. The role of epithelial cell adhesion molecule N-glycosylation on apoptosis in breast cancer cells. Tumor Biol. 2017, 39, 101042831769597. [Google Scholar] [CrossRef]
- Mnatsakanyan, R.; Shema, G.; Basik, M.; Batist, G.; Borchers, C.H.; Sickmann, A.; Zahedi, R.P. Detecting post-translational modification signatures as potential biomarkers in clinical mass spectrometry. Expert Rev. Proteom. 2018, 15, 515–535. [Google Scholar] [CrossRef]
- Heo, K.-S. Regulation of post-translational modification in breast cancer treatment. BMB Rep. 2019, 52, 113–118. [Google Scholar] [CrossRef]
- Ha, J.; Kang, E.; Seo, J.; Cho, S. Phosphorylation Dynamics of JNK Signaling: Effects of Dual-Specificity Phosphatases (DUSPs) on the JNK Pathway. Int. J. Mol. Sci. 2019, 20, 6157. [Google Scholar] [CrossRef]
- Turdo, A.; D’Accardo, C.; Glaviano, A.; Porcelli, G.; Colarossi, C.; Colarossi, L.; Mare, M.; Faldetta, N.; Modica, C.; Pistone, G.; et al. Targeting Phosphatases and Kinases: How to Checkmate Cancer. Front. Cell Dev. Biol. 2021, 9, 690306. [Google Scholar] [CrossRef]
- Xia, X.; Huang, C.; Liao, Y.; Liu, Y.; He, J.; Shao, Z.; Hu, T.; Yu, C.; Jiang, L.; Liu, J.; et al. The deubiquitinating enzyme USP15 stabilizes ERα and promotes breast cancer progression. Cell Death Dis. 2021, 12, 329. [Google Scholar] [CrossRef]
- Xia, X.; Liao, Y.; Huang, C.; Liu, Y.; He, J.; Shao, Z.; Jiang, L.; Dou, Q.P.; Liu, J.; Huang, H. Deubiquitination and stabilization of estrogen receptor α by ubiquitin-specific protease 7 promotes breast tumorigenesis. Cancer Lett. 2019, 465, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Chen, W.; Li, H.; Li, M.; Li, L. The Histone Acetylation Modifications of Breast Cancer and their Therapeutic Implications. Pathol. Oncol. Res. 2018, 24, 807–813. [Google Scholar] [CrossRef]
- Zaha, D. Significance of immunohistochemistry in breast cancer. World J. Clin. Oncol. 2014, 5, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Zhang, L.; Ruoff, R.; Ha, S.; Wang, J.; Jain, S.; Reuter, V.; Gerald, W.; Giri, D.D.; Melamed, J.; et al. Expression of androgen receptor and its phosphorylated forms in breast cancer progression. Cancer 2013, 119, 2532–2540. [Google Scholar] [CrossRef]
- Elsheikh, S.E.; Green, A.R.; Rakha, E.A.; Powe, D.G.; Ahmed, R.A.; Collins, H.M.; Soria, D.; Garibaldi, J.M.; Paish, C.E.; Ammar, A.A.; et al. Global Histone Modifications in Breast Cancer Correlate with Tumor Phenotypes, Prognostic Factors, and Patient Outcome. Cancer Res. 2009, 69, 3802–3809. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, Q.; Lin, Y.; Jiang, M.; Xiao, H.; Zhang, J.; Guo, R.; Kang, S.; Lin, Y.; Song, C. An immunohistochemical panel of three small ubiquitin-like modifier genes predicts outcomes of patients with triple-negative breast cancer. Gland Surg. 2021, 10, 1067–1084. [Google Scholar] [CrossRef] [PubMed]
- Ueki, T.; Park, J.-H.; Nishidate, T.; Kijima, K.; Hirata, K.; Nakamura, Y.; Katagiri, T. Ubiquitination and Downregulation of BRCA1 by Ubiquitin-Conjugating Enzyme E2T Overexpression in Human Breast Cancer Cells. Cancer Res. 2009, 69, 8752–8760. [Google Scholar] [CrossRef]
- Mandell, J.W. Immunohistochemical assessment of protein phosphorylation state: The dream and the reality. Histochem. Cell Biol. 2008, 130, 465–471. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, J.; Song, C.; Zhan, X. The role of protein acetylation in carcinogenesis and targeted drug discovery. Front. Endocrinol. 2022, 13, 972312. [Google Scholar] [CrossRef]
- Mohanty, S.S.; Sahoo, C.R.; Padhy, R.N. Role of hormone receptors and HER2 as prospective molecular markers for breast cancer: An update. Genes Dis. 2022, 9, 648–658. [Google Scholar] [CrossRef]
- Wong, G.L.; Manore, S.G.; Doheny, D.L.; Lo, H.-W. STAT family of transcription factors in breast cancer: Pathogenesis and therapeutic opportunities and challenges. Semin. Cancer Biol. 2022, 86, 84–106. [Google Scholar] [CrossRef]
- Gomarasca, M.; Lombardi, G.; Maroni, P. SUMOylation and NEDDylation in Primary and Metastatic Cancers to Bone. Front. Cell Dev. Biol. 2022, 10, 889002. [Google Scholar] [CrossRef]
- Rakha, E.A.; Chmielik, E.; Schmitt, F.C.; Tan, P.H.; Quinn, C.M.; Gallagy, G. Assessment of Predictive Biomarkers in Breast Cancer: Challenges and Updates. Pathobiology 2022, 89, 263–277. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, H.; Yao, J. ERα, A Key Target for Cancer Therapy: A Review. Onco. Targets Ther. 2020, 13, 2183–2191. [Google Scholar] [CrossRef] [PubMed]
- Thiebaut, C.; Konan, H.-P.; Guerquin, M.-J.; Chesnel, A.; Livera, G.; Le Romancer, M.; Dumond, H. The Role of ERα36 in Development and Tumor Malignancy. Int. J. Mol. Sci. 2020, 21, 4116. [Google Scholar] [CrossRef]
- Gong, Z.; Yang, S.; Wei, M.; Vlantis, A.C.; Chan, J.Y.K.; van Hasselt, C.A.; Li, D.; Zeng, X.; Xue, L.; Tong, M.C.F.; et al. The Isoforms of Estrogen Receptor Alpha and Beta in Thyroid Cancer. Front. Oncol. 2022, 12, 916804. [Google Scholar] [CrossRef] [PubMed]
- Isigkeit, L.; Schallmayer, E.; Busch, R.; Brunello, L.; Menge, A.; Elson, L.; Müller, S.; Knapp, S.; Stolz, A.; Marschner, J.A.; et al. Chemogenomics for NR1 nuclear hormone receptors. Nat. Commun. 2024, 15, 5201. [Google Scholar] [CrossRef] [PubMed]
- Shao, D.; Lazar, M.A. Modulating nuclear receptor function: May the phos be with you. J. Clin. Investig. 1999, 103, 1617–1618. [Google Scholar] [CrossRef] [PubMed]
- Miziak, P.; Baran, M.; Błaszczak, E.; Przybyszewska-Podstawka, A.; Kałafut, J.; Smok-Kalwat, J.; Dmoszyńska-Graniczka, M.; Kiełbus, M.; Stepulak, A. Estrogen Receptor Signaling in Breast Cancer. Cancers 2023, 15, 4689. [Google Scholar] [CrossRef]
- Li, L.; Wang, Q.; Lv, X.; Sha, L.; Qin, H.; Wang, L.; Li, L. Expression and Localization of Estrogen Receptor in Human Breast Cancer and Its Clinical Significance. Cell Biochem. Biophys. 2015, 71, 63–68. [Google Scholar] [CrossRef]
- Traboulsi, T.; El Ezzy, M.; Dumeaux, V.; Audemard, E.; Mader, S. Role of SUMOylation in differential ERα transcriptional repression by tamoxifen and fulvestrant in breast cancer cells. Oncogene 2019, 38, 1019–1037. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, N.; Yang, X.; Feng, S.; Wang, F.; Zhang, W.; He, Z. ERα promotes SUMO1 transcription by binding with the ERE and enhances SUMO1-mediated protein SUMOylation in breast cancer. Gland. Surg. 2023, 12, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Rusidzé, M.; Adlanmérini, M.; Chantalat, E.; Raymond-Letron, I.; Cayre, S.; Arnal, J.-F.; Deugnier, M.-A.; Lenfant, F. Estrogen receptor-α signaling in post-natal mammary development and breast cancers. Cell. Mol. Life Sci. 2021, 78, 5681–5705. [Google Scholar] [CrossRef]
- Gutiérrez, A.; Sambuco, L.; Álvarez, L.; Núñez, M.; Bergoc, R.; Zotta, E.; Martín, G.; Randi, A. Expression of estrogen receptor α variants and c-Fos in rat mammary gland and tumors. J. Steroid Biochem. Mol. Biol. 2020, 199, 105594. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, J.; Ying, G.; Xie, X.-Q.; Zhang, X.; Xu, W.; Zhang, X.; Song, E.; Bu, H.; Ping, Y.-F.; et al. Tamoxifen enhances stemness and promotes metastasis of ERα36+ breast cancer by upregulating ALDH1A1 in cancer cells. Cell Res. 2018, 28, 336–358. [Google Scholar] [CrossRef]
- Meitzen, J.; Luoma, J.I.; Boulware, M.I.; Hedges, V.L.; Peterson, B.M.; Tuomela, K.; Britson, K.A.; Mermelstein, P.G. Palmitoylation of Estrogen Receptors Is Essential for Neuronal Membrane Signaling. Endocrinology 2013, 154, 4293–4304. [Google Scholar] [CrossRef] [PubMed]
- Acconcia, F.; Ascenzi, P.; Bocedi, A.; Spisni, E.; Tomasi, V.; Trentalance, A.; Visca, P.; Marino, M. Palmitoylation-dependent Estrogen Receptor α Membrane Localization: Regulation by 17β-Estradiol. Mol. Biol. Cell 2005, 16, 231–237. [Google Scholar] [CrossRef]
- Garcia-Garcia, T.; Poncet, S.; Derouiche, A.; Shi, L.; Mijakovic, I.; Noirot-Gros, M.-F. Role of Protein Phosphorylation in the Regulation of Cell Cycle and DNA-Related Processes in Bacteria. Front. Microbiol. 2016, 7, 184. [Google Scholar] [CrossRef]
- Ghosh, R.; Ahmed, R.; Ahmed, H.; Chatterjee, B.P. Phosphorylated Proteins from Serum: A Promising Potential Diagnostic Biomarker of Cancer. Int. J. Mol. Sci. 2022, 23, 12359. [Google Scholar] [CrossRef]
- Hochgräfe, F.; Zhang, L.; O’Toole, S.A.; Browne, B.C.; Pinese, M.; Porta Cubas, A.; Lehrbach, G.M.; Croucher, D.R.; Rickwood, D.; Boulghourjian, A.; et al. Tyrosine Phosphorylation Profiling Reveals the Signaling Network Characteristics of Basal Breast Cancer Cells. Cancer Res. 2010, 70, 9391–9401. [Google Scholar] [CrossRef]
- Yamasaki, A.; Jin, Y.; Ohsumi, Y. Mitotic phosphorylation of the ULK complex regulates cell cycle progression. PLoS Biol. 2020, 18, e3000718. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.-L.; Chu, P.-Y.; Lai, I.R.; Wang, M.-Y.; Tseng, H.-Y.; Guan, J.-L.; Liou, J.-Y.; Shen, T.-L. An EGFR/Src-dependent β4 integrin/FAK complex contributes to malignancy of breast cancer. Sci. Rep. 2015, 5, 16408. [Google Scholar] [CrossRef]
- Ooshima, A.; Park, J.; Kim, S.-J. Phosphorylation status at Smad3 linker region modulates transforming growth factor-β-induced epithelial-mesenchymal transition and cancer progression. Cancer Sci. 2019, 110, 481–488. [Google Scholar] [CrossRef]
- Fan, Y.; Si, W.; Ji, W.; Wang, Z.; Gao, Z.; Tian, R.; Song, W.; Zhang, H.; Niu, R.; Zhang, F. Rack1 mediates Src binding to drug transporter P-glycoprotein and modulates its activity through regulating Caveolin-1 phosphorylation in breast cancer cells. Cell Death Dis. 2019, 10, 394. [Google Scholar] [CrossRef]
- Yang, G.; Zuo, C.; Lin, Y.; Zhou, X.; Wen, P.; Zhang, C.; Xiao, H.; Jiang, M.; Fujita, M.; Gao, X.-D.; et al. Comprehensive proteome, phosphoproteome and kinome characterization of luminal A breast cancer. Front. Oncol. 2023, 13, 1127446. [Google Scholar] [CrossRef]
- Zawadzka, A.M.; Schilling, B.; Cusack, M.P.; Sahu, A.K.; Drake, P.; Fisher, S.J.; Benz, C.C.; Gibson, B.W. Phosphoprotein Secretome of Tumor Cells as a Source of Candidates for Breast Cancer Biomarkers in Plasma. Mol. Cell. Proteom. 2014, 13, 1034–1049. [Google Scholar] [CrossRef]
- Anbalagan, M.; Rowan, B.G. Estrogen receptor alpha phosphorylation and its functional impact in human breast cancer. Mol. Cell. Endocrinol. 2015, 418, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Fu, M.; Pestell, R.G. Estrogen receptor acetylation and phosphorylation in hormone responses. Breast Cancer Online 2005, 8, e46. [Google Scholar] [CrossRef][Green Version]
- Tecalco-Cruz, A.C.; Macías-Silva, M.; Ramírez-Jarquín, J.O.; Ramírez-Jarquín, U.N. Decoding the Therapeutic Implications of the ERα Stability and Subcellular Distribution in Breast Cancer. Front. Endocrinol. 2022, 13, 867448. [Google Scholar] [CrossRef] [PubMed]
- Castoria, G.; Giovannelli, P.; Lombardi, M.; De Rosa, C.; Giraldi, T.; de Falco, A.; Barone, M.V.; Abbondanza, C.; Migliaccio, A.; Auricchio, F. Tyrosine phosphorylation of estradiol receptor by Src regulates its hormone-dependent nuclear export and cell cycle progression in breast cancer cells. Oncogene 2012, 31, 4868–4877. [Google Scholar] [CrossRef] [PubMed]
- Frei, A.; MacDonald, G.; Lund, I.; Gustafsson, J.-Å.; Hynes, N.E.; Nalvarte, I. Memo interacts with c-Src to control Estrogen Receptor alpha sub-cellular localization. Oncotarget 2016, 7, 56170. [Google Scholar] [CrossRef] [PubMed]
- Kondakova, I.V.; Shashova, E.E.; Sidenko, E.A.; Astakhova, T.M.; Zakharova, L.A.; Sharova, N.P. Estrogen Receptors and Ubiquitin Proteasome System: Mutual Regulation. Biomolecules 2020, 10, 500. [Google Scholar] [CrossRef]
- Tecalco-Cruz, A.; Ramírez, J. Polyubiquitination inhibition of Estrogen receptor alpha and its implications in breast cancer. World J. Clin. Oncol. 2018, 9, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Celen, A.B.; Sahin, U. Sumoylation on its 25th anniversary: Mechanisms, pathology, and emerging concepts. FEBS J. 2020, 287, 3110–3140. [Google Scholar] [CrossRef]
- Le Romancer, M.; Treilleux, I.; Leconte, N.; Robin-Lespinasse, Y.; Sentis, S.; Bouchekioua-Bouzaghou, K.; Goddard, S.; Gobert-Gosse, S.; Corbo, L. Regulation of Estrogen Rapid Signaling through Arginine Methylation by PRMT1. Mol. Cell 2008, 31, 212–221. [Google Scholar] [CrossRef]
- Ilicic, M.; Zakar, T.; Paul, J.W. Epigenetic regulation of progesterone receptors and the onset of labour. Reprod. Fertil. Dev. 2019, 31, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Kariagina, A.; Aupperlee, M.D.; Haslam, S.Z. Progesterone Receptor Isoform Functions in Normal Breast Development and Breast Cancer. Crit. Rev.™ Eukaryot. Gene Expr. 2008, 18, 11–33. [Google Scholar] [CrossRef]
- Li, Z.; Wei, H.; Li, S.; Wu, P.; Mao, X. The Role of Progesterone Receptors in Breast Cancer. Drug Des. Dev. Ther. 2022, 16, 305–314. [Google Scholar] [CrossRef]
- Chung, H.H.; Sze, S.K.; Tay, A.S.L.; Lin, V.C.L. Acetylation at Lysine 183 of Progesterone Receptor by p300 Accelerates DNA Binding Kinetics and Transactivation of Direct Target Genes. J. Biol. Chem. 2014, 289, 2180–2194. [Google Scholar] [CrossRef]
- Bleach, R.; McIlroy, M. The Divergent Function of Androgen Receptor in Breast Cancer; Analysis of Steroid Mediators and Tumor Intracrinology. Front. Endocrinol. 2018, 9. [Google Scholar] [CrossRef]
- Wen, S.; Niu, Y.; Huang, H. Posttranslational regulation of androgen dependent and independent androgen receptor activities in prostate cancer. Asian J. Urol. 2020, 7, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Roseweir, A.K.; McCall, P.; Scott, A.; Liew, B.; Lim, Z.; Mallon, E.A.; Edwards, J. Phosphorylation of androgen receptors at serine 515 is a potential prognostic marker for triple negative breast cancer. Oncotarget 2017, 8, 37172. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, S.; Wu, L.; Lai, C.; Yang, G. Hydrogen Sulfide Represses Androgen Receptor Transactivation by Targeting at the Second Zinc Finger Module. J. Biol. Chem. 2014, 289, 20824–20835. [Google Scholar] [CrossRef] [PubMed]
- Khattak, S.; Rauf, M.A.; Khan, N.H.; Zhang, Q.-Q.; Chen, H.-J.; Muhammad, P.; Ansari, M.A.; Alomary, M.N.; Jahangir, M.; Zhang, C.-Y.; et al. Hydrogen Sulfide Biology and Its Role in Cancer. Molecules 2022, 27, 3389. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, F.; Gao, G.; Gao, X.; Wu, B.; Zheng, C.; Wang, P.; Li, Z.; Hua, H.; Li, D. Hydrogen sulfide and its donors: Novel antitumor and antimetastatic therapies for triple-negative breast cancer. Redox Biol. 2020, 34, 101564. [Google Scholar] [CrossRef]
- Shackelford, R.E.; Mohammad, I.Z.; Meram, A.T.; Kim, D.; Alotaibi, F.; Patel, S.; Ghali, G.E.; Kevil, C.G. Molecular Functions of Hydrogen Sulfide in Cancer. Pathophysiology 2021, 28, 437–456. [Google Scholar] [CrossRef]
- Cui, X.; Liu, R.; Duan, L.; Zhang, Q.; Cao, D.; Zhang, A. Exogenous hydrogen sulfide (H2S) exerts therapeutic potential in triple-negative breast cancer by affecting cell cycle and DNA replication pathway. Biomed. Pharmacother. 2023, 161, 114488. [Google Scholar] [CrossRef]
- Stoltzfus, A.T.; Ballot, J.G.; Vignane, T.; Li, H.; Worth, M.M.; Muller, L.; Siegler, M.A.; Kane, M.A.; Filipovic, M.R.; Goldberg, D.P.; et al. Chemoselective Proteomics, Zinc Fingers, and a Zinc(II) Model for H2S Mediated Persulfidation. Angew. Chem. Int. Ed. 2024, 63, e202401003. [Google Scholar] [CrossRef]
- He, B.; Zhang, Z.; Huang, Z.; Duan, X.; Wang, Y.; Cao, J.; Li, L.; He, K.; Nice, E.C.; He, W.; et al. Protein persulfidation: Rewiring the hydrogen sulfide signaling in cell stress response. Biochem. Pharmacol. 2023, 209, 115444. [Google Scholar] [CrossRef]
- Vignane, T.; Filipovic, M.R. Emerging Chemical Biology of Protein Persulfidation. Antioxid. Redox Signal. 2023, 39, 19–39. [Google Scholar] [CrossRef]
- Erdélyi, K.; Ditrói, T.; Johansson, H.J.; Czikora, Á.; Balog, N.; Silwal-Pandit, L.; Ida, T.; Olasz, J.; Hajdú, D.; Mátrai, Z.; et al. Reprogrammed transsulfuration promotes basal-like breast tumor progression via realigning cellular cysteine persulfidation. Proc. Natl. Acad. Sci. USA 2021, 118, e2100050118. [Google Scholar] [CrossRef]
- Gao, W.; Liu, Y.-F.; Zhang, Y.-X.; Wang, Y.; Jin, Y.-Q.; Yuan, H.; Liang, X.-Y.; Ji, X.-Y.; Jiang, Q.-Y.; Wu, D.-D. The potential role of hydrogen sulfide in cancer cell apoptosis. Cell Death Discov. 2024, 10, 114. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Endoh, H.; Masuhiro, Y.; Kitamoto, T.; Uchiyama, S.; Sasaki, H.; Masushige, S.; Gotoh, Y.; Nishida, E.; Kawashima, H.; et al. Activation of the Estrogen Receptor Through Phosphorylation by Mitogen-Activated Protein Kinase. Science 1995, 270, 1491–1494. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, Y.; Setlem, R.; Murakami, S.; Kraus, W. SUN-028 Role of Estrogen Receptor Alpha (ERa) Acetylation in Estrogen-Dependent Gene Regulation in Breast Cancers. J. Endocr. Soc. 2019, 3, SUN-028. [Google Scholar] [CrossRef]
- Kim, M.Y.; Woo, E.M.; Chong, Y.T.E.; Homenko, D.R.; Kraus, W.L. Acetylation of Estrogen Receptor α by p300 at Lysines 266 and 268 Enhances the Deoxyribonucleic Acid Binding and Transactivation Activities of the Receptor. Mol. Endocrinol. 2006, 20, 1479–1493. [Google Scholar] [CrossRef]
- Malbeteau, L.; Poulard, C.; Languilaire, C.; Mikaelian, I.; Flamant, F.; Le Romancer, M.; Corbo, L. PRMT1 Is Critical for the Transcriptional Activity and the Stability of the Progesterone Receptor. iScience 2020, 23, 101236. [Google Scholar] [CrossRef]
- Niu, Z.; Li, X.; Feng, S.; Huang, Q.; Zhuang, T.; Yan, C.; Qian, H.; Ding, Y.; Zhu, J.; Xu, W. The deubiquitinating enzyme USP1 modulates ERα and modulates breast cancer progression. J. Cancer 2020, 11, 6992–7000. [Google Scholar] [CrossRef]
- Su, Y.; Zeng, K.; Liu, S.; Wu, Y.; Wang, C.; Wang, S.; Lin, L.; Zou, R.; Sun, G.; Luan, R.; et al. Ubiquitin-specific peptidase 14 maintains estrogen receptor α; stability via its deubiquitination activity in endometrial cancer. J. Biol. Chem. 2023, 299, 102734. [Google Scholar] [CrossRef]
- Xue, Q.; Tang, D.; Chen, X.; Liu, J. Targeting deubiquitinases for cancer therapy. Clin. Transl. Discov. 2023, 3, e242. [Google Scholar] [CrossRef]
- Sentis, S.; Le Romancer, M.; Bianchin, C.; Rostan, M.-C.; Corbo, L. Sumoylation of the Estrogen Receptor α Hinge Region Regulates Its Transcriptional Activity. Mol. Endocrinol. 2005, 19, 2671–2684. [Google Scholar] [CrossRef]
- Hilmi, K.; Hussein, N.; Mendoza-Sanchez, R.; El-Ezzy, M.; Ismail, H.; Durette, C.; Bail, M.; Rozendaal, M.J.; Bouvier, M.; Thibault, P.; et al. Role of SUMOylation in Full Antiestrogenicity. Mol. Cell. Biol. 2012, 32, 3823–3837. [Google Scholar] [CrossRef]
- Faivre, E.J.; Daniel, A.R.; Hillard, C.J.; Lange, C.A. Progesterone Receptor Rapid Signaling Mediates Serine 345 Phosphorylation and Tethering to Specificity Protein 1 Transcription Factors. Mol. Endocrinol. 2008, 22, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hafiz, H.A.; Dudevoir, M.L.; Perez, D.; Abdel-Hafiz, M.; Horwitz, K.B. SUMOylation Regulates Transcription by the Progesterone Receptor A Isoform in a Target Gene Selective Manner. Diseases 2018, 6, 5. [Google Scholar] [CrossRef]
- Marquez-Lago, T.T.; Steinberg, S. Stochastic model of ERK-mediated progesterone receptor translocation, clustering and transcriptional activity. Sci. Rep. 2022, 12, 11791. [Google Scholar] [CrossRef]
- Qiu, M.; Olsen, A.; Faivre, E.; Horwitz, K.B.; Lange, C.A. Mitogen-Activated Protein Kinase Regulates Nuclear Association of Human Progesterone Receptors. Mol. Endocrinol. 2003, 17, 628–642. [Google Scholar] [CrossRef] [PubMed]
- Hagan, C.R.; Daniel, A.R.; Dressing, G.E.; Lange, C.A. Role of phosphorylation in progesterone receptor signaling and specificity. Mol. Cell. Endocrinol. 2012, 357, 43–49. [Google Scholar] [CrossRef]
- Grimm, S.L.; Ward, R.D.; Obr, A.E.; Franco, H.L.; Fernandez-Valdivia, R.; Kim, J.-S.; Roberts, J.M.; Jeong, J.-W.; DeMayo, F.J.; Lydon, J.P.; et al. A Role for Site-Specific Phosphorylation of Mouse Progesterone Receptor at Serine 191 in vivo. Mol. Endocrinol. 2014, 28, 2025–2037. [Google Scholar] [CrossRef]
- Font-Mateu, J.; Sanllehí, P.; Sot, J.; Abad, B.; Mateos, N.; Torreno-Pina, J.A.; Ferrari, R.; Wright, R.H.G.; Garcia-Parajo, M.F.; Joglar, J.; et al. A progesterone derivative linked to a stable phospholipid activates breast cancer cell response without leaving the cell membrane. Cell. Mol. Life Sci. 2024, 81, 98. [Google Scholar] [CrossRef]
- Jaiswal, B.; Agarwal, A.; Gupta, A. Lysine Acetyltransferases and Their Role in AR Signaling and Prostate Cancer. Front. Endocrinol. 2022, 13, 886594. [Google Scholar] [CrossRef]
- Wen, Y.; Hu, M.; Makino, K.; Spohn, B.; Bartholomeusz, G.; Yan, D.; Hung, M. HER-2/neu promotes androgen-independent survival and growth of prostate cancer cells through the Akt pathway. Cancer Res. 2001, 60, 6841–6845. [Google Scholar]
- Lin, H.-K.; Yeh, S.; Kang, H.-Y.; Chang, C. Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc. Natl. Acad. Sci. USA 2001, 98, 7200–7205. [Google Scholar] [CrossRef] [PubMed]
- Gioeli, D.; Ficarro, S.B.; Kwiek, J.J.; Aaronson, D.; Hancock, M.; Catling, A.D.; White, F.M.; Christian, R.E.; Settlage, R.E.; Shabanowitz, J.; et al. Androgen Receptor Phosphorylation: REGULATION AND IDENTIFICATION OF THE PHOSPHORYLATION SITES. J. Biol. Chem. 2002, 277, 29304–29314. [Google Scholar] [CrossRef] [PubMed]
- Marvalim, C.; Datta, A.; Lee, S.C. Role of p53 in breast cancer progression: An insight into p53 targeted therapy. Theranostics 2023, 13, 1421–1442. [Google Scholar] [CrossRef]
- Nagasaka, M.; Miyajima, C.; Aoki, H.; Aoyama, M.; Morishita, D.; Inoue, Y.; Hayashi, H. Insights into Regulators of p53 Acetylation. Cells 2022, 11, 3825. [Google Scholar] [CrossRef]
- Dai, C.; Gu, W. p53 post-translational modification: Deregulated in tumorigenesis. Trends Mol. Med. 2010, 16, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Abuetabh, Y.; Wu, H.H.; Chai, C.; Al Yousef, H.; Persad, S.; Sergi, C.M.; Leng, R. DNA damage response revisited: The p53 family and its regulators provide endless cancer therapy opportunities. Exp. Mol. Med. 2022, 54, 1658–1669. [Google Scholar] [CrossRef] [PubMed]
- Meek, D.W.; Anderson, C.W. Posttranslational Modification of p53: Cooperative Integrators of Function. Cold Spring Harb. Perspect. Biol. 2009, 1, a000950. [Google Scholar] [CrossRef]
- Ray, D.; Murphy, K.R.; Gal, S. The DNA binding and accumulation of p53 from breast cancer cell lines and the link with serine 15 phosphorylation. Cancer Biol. Ther. 2012, 13, 848–857. [Google Scholar] [CrossRef]
- Appella, E.; Anderson, C.W. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 2001, 268, 2764–2772. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, Y.; Luo, Q.; Li, L.; Jia, L. Neddylation: A novel modulator of the tumor microenvironment. Mol. Cancer 2019, 18, 77. [Google Scholar] [CrossRef]
- Park, J.B.; Seo, J.; Park, J.-W.; Chun, Y.-S. Neddylation blockade induces HIF-1α driven cancer cell migration via upregulation of ZEB1. Sci. Rep. 2020, 10, 18210. [Google Scholar] [CrossRef] [PubMed]
- Abida, W.M.; Nikolaev, A.; Zhao, W.; Zhang, W.; Gu, W. FBXO11 Promotes the Neddylation of p53 and Inhibits Its Transcriptional Activity. J. Biol. Chem. 2007, 282, 1797–1804. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, K.M.; Chen, Y.; Chen, Y.; Liu, A.T.; Sun, X.-X.; Dai, M.-S. The SUMO-specific protease SENP1 deSUMOylates p53 and regulates its activity. J. Cell. Biochem. 2021, 122, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-M.; Tang, Q.; Hu, X.; Zhao, J.-J.; Zhang, Y.; Ying, G.-G.; Zhang, F. Arginine Methyltransferase PRMT1 Regulates p53 Activity in Breast Cancer. Life 2021, 11, 789. [Google Scholar] [CrossRef]
- Mitsis, T.; Efthimiadou, A.; Bacopoulou, F.; Vlachakis, D.; Chrousos, G.P.; Eliopoulos, E. Transcription factors and evolution: An integral part of gene expression (Review). World Acad. Sci. J. 2020, 2, 3–8. [Google Scholar] [CrossRef]
- Kim, H.K.; Jeong, M.G.; Hwang, E.S. Post-Translational Modifications in Transcription Factors that Determine T Helper Cell Differentiation. Mol. Cells 2021, 44, 318–327. [Google Scholar] [CrossRef]
- Zhang, P.; Wei, Y.; Wang, L.; Debeb, B.G.; Yuan, Y.; Zhang, J.; Yuan, J.; Wang, M.; Chen, D.; Sun, Y.; et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat. Cell Biol. 2014, 16, 864–875. [Google Scholar] [CrossRef]
- Xu, R.; Won, J.-Y.; Kim, C.-H.; Kim, D.-E.; Yim, H. Roles of the Phosphorylation of Transcriptional Factors in Epithelial-Mesenchymal Transition. J. Oncol. 2019, 2019, 5810465. [Google Scholar] [CrossRef]
- Zhao, Z.; Rahman, M.A.; Chen, Z.G.; Shin, D.M. Multiple biological functions of Twist1 in various cancers. Oncotarget 2017, 8, 20380. [Google Scholar] [CrossRef]
- Hong, J.; Zhou, J.; Fu, J.; He, T.; Qin, J.; Wang, L.; Liao, L.; Xu, J. Phosphorylation of Serine 68 of Twist1 by MAPKs Stabilizes Twist1 Protein and Promotes Breast Cancer Cell Invasiveness. Cancer Res. 2011, 71, 3980–3990. [Google Scholar] [CrossRef]
- Li, C.-W.; Xia, W.; Lim, S.-O.; Hsu, J.L.; Huo, L.; Wu, Y.; Li, L.-Y.; Lai, C.-C.; Chang, S.-S.; Hsu, Y.-H.; et al. AKT1 Inhibits Epithelial-to-Mesenchymal Transition in Breast Cancer through Phosphorylation-Dependent Twist1 Degradation. Cancer Res. 2016, 76, 1451–1462. [Google Scholar] [CrossRef] [PubMed]
- Llorens, M.C.; Lorenzatti, G.; Cavallo, N.L.; Vaglienti, M.V.; Perrone, A.P.; Carenbauer, A.L.; Darling, D.S.; Cabanillas, A.M. Phosphorylation Regulates Functions of ZEB1 Transcription Factor. J. Cell. Physiol. 2016, 231, 2205–2217. [Google Scholar] [CrossRef]
- Smith, B.; Burton, L.; Henderson, V.; Randle, D.; Morton, D.; Smith, B.; Taliaferro-Smith, L.; Nagappan, P.; Yates, C.; Zayzafoon, M.; et al. Snail Promotes Epithelial Mesenchymal Transition in Breast Cancer Cells in Part via Activation of Nuclear ERK2. PLoS ONE 2014, 9, e104987. [Google Scholar] [CrossRef]
- Zhou, B.P.; Deng, J.; Xia, W.; Xu, J.; Li, Y.M.; Gunduz, M.; Hung, M.-C. Dual regulation of Snail by GSK-3β-mediated phosphorylation in control of epithelial–mesenchymal transition. Nat. Cell Biol. 2004, 6, 931–940. [Google Scholar] [CrossRef]
- Zheng, H.; Shen, M.; Zha, Y.-L.; Li, W.; Wei, Y.; Blanco, M.A.; Ren, G.; Zhou, T.; Storz, P.; Wang, H.-Y.; et al. PKD1 Phosphorylation-Dependent Degradation of SNAIL by SCF-FBXO11 Regulates Epithelial-Mesenchymal Transition and Metastasis. Cancer Cell 2014, 26, 358–373. [Google Scholar] [CrossRef] [PubMed]
- Yook, J.I.; Li, X.-Y.; Ota, I.; Fearon, E.R.; Weiss, S.J. Wnt-dependent Regulation of the E-cadherin Repressor Snail. J. Biol. Chem. 2005, 280, 11740–11748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Corsa, C.; Ponik, S.; Prior, J.; Piwnica-Worms, D.; Eliceiri, K.; Keely, P.; Longmore, G. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat. Cell Biol. 2013, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Rodriguez-Aznar, E.; Yabuta, N.; Owen, R.J.; Mingot, J.M.; Nojima, H.; Nieto, M.A.; Longmore, G.D. Lats2 kinase potentiates Snail1 activity by promoting nuclear retention upon phosphorylation. EMBO J. 2012, 31, 29–43. [Google Scholar] [CrossRef]
- Ma, J.-H.; Qin, L.; Li, X. Role of STAT3 signaling pathway in breast cancer. Cell Commun. Signal. 2020, 18, 33. [Google Scholar] [CrossRef]
- Rodríguez-Enríquez, S.; Marín-Hernández, Á.; Gallardo-Pérez, J.C.; Pacheco-Velázquez, S.C.; Belmont-Díaz, J.A.; Robledo-Cadena, D.X.; Vargas-Navarro, J.L.; Corona de la Peña, N.A.; Saavedra, E.; Moreno-Sánchez, R. Transcriptional Regulation of Energy Metabolism in Cancer Cells. Cells 2019, 8, 1225. [Google Scholar] [CrossRef]
- Proietti, C.; Salatino, M.; Rosemblit, C.; Carnevale, R.; Pecci, A.; Kornblihtt, A.R.; Molinolo, A.A.; Frahm, I.; Charreau, E.H.; Schillaci, R.; et al. Progestins Induce Transcriptional Activation of Signal Transducer and Activator of Transcription 3 (Stat3) via a Jak- and Src-Dependent Mechanism in Breast Cancer Cells. Mol. Cell. Biol. 2005, 25, 4826–4840. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jin, J.; Yang, S.; Xu, W.; Meng, X.; Deng, H.; Zhan, J.; Gao, S.; Zhang, H. GATA3 acetylation at K119 by CBP inhibits cell migration and invasion in lung adenocarcinoma. Biochem. Biophys. Res. Commun. 2018, 497, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Liu, P.; Lau, A.W.; Liu, Y.; Inuzuka, H. Acetylation-dependent regulation of essential iPS-inducing factors: A regulatory crossroad for pluripotency and tumorigenesis. Cancer Med. 2014, 3, 1211–1224. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Liu, X.; Ren, Y.; Li, J.; Li, P.; Jiao, Q.; Meng, P.; Wang, F.; Wang, Y.; Wang, Y.-s.; et al. Loss of SIRT4 promotes the self-renewal of Breast Cancer Stem Cells. Theranostics 2020, 10, 9458–9476. [Google Scholar] [CrossRef]
- Baulida, J.; Díaz, V.M.; García de Herreros, A. Snail1: A Transcriptional Factor Controlled at Multiple Levels. J. Clin. Med. 2019, 8, 757. [Google Scholar] [CrossRef]
- Chen, L.; Pan, X.-W.; Huang, H.; Gao, Y.; Yang, Q.-W.; Wang, L.-H.; Cui, X.-G.; Xu, D.-F. Epithelial-mesenchymal transition induced by GRO-α-CXCR2 promotes bladder cancer recurrence after intravesical chemotherapy. Oncotarget 2017, 8, 45274. [Google Scholar] [CrossRef]
- MacPherson, M.R.; Molina, P.; Souchelnytskyi, S.; Wernstedt, C.; Martin-Pérez, J.; Portillo, F.; Cano, A. Phosphorylation of Serine 11 and Serine 92 as New Positive Regulators of Human Snail1 Function: Potential Involvement of Casein Kinase-2 and the cAMP-activated Kinase Protein Kinase, A. Mol. Biol. Cell 2010, 21, 244–253. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, H.S.; Kim, N.H.; Ji, S.; Cha, S.Y.; Kang, J.G.; Ota, I.; Shimada, K.; Konishi, N.; Nam, H.W.; et al. Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. EMBO J. 2010, 29, 3787–3796. [Google Scholar] [CrossRef]
- Park, J.-J.; Park, M.-H.; Oh, E.H.; Soung, N.-K.; Lee, S.J.; Jung, J.-K.; Lee, O.-J.; Yun, S.J.; Kim, W.-J.; Shin, E.-Y.; et al. The p21-activated kinase 4-Slug transcription factor axis promotes epithelial−mesenchymal transition and worsens prognosis in prostate cancer. Oncogene 2018, 37, 5147–5159. [Google Scholar] [CrossRef]
- Izzo, F.; Mercogliano, F.; Venturutti, L.; Tkach, M.; Inurrigarro, G.; Schillaci, R.; Cerchietti, L.; Elizalde, P.V.; Proietti, C.J. Progesterone receptor activation downregulates GATA3 by transcriptional repression and increased protein turnover promoting breast tumor growth. Breast Cancer Res. 2014, 16, 491. [Google Scholar] [CrossRef]
- Hosokawa, H.; Kato, M.; Tohyama, H.; Tamaki, Y.; Endo, Y.; Kimura, M.Y.; Tumes, D.J.; Motohashi, S.; Matsumoto, M.; Nakayama, K.I.; et al. Methylation of Gata3 Protein at Arg-261 Regulates Transactivation of the Il5 Gene in T Helper 2 Cells. J. Biol. Chem. 2015, 290, 13095–13103. [Google Scholar] [CrossRef]
- Yamazaki, H.; Takagi, M.; Kosako, H.; Hirano, T.; Yoshimura, S.H. Cell cycle-specific phase separation regulated by protein charge blockiness. Nat. Cell Biol. 2022, 24, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Remnant, L.; Kochanova, N.Y.; Reid, C.; Cisneros-Soberanis, F.; Earnshaw, W.C. The intrinsically disorderly story of Ki-67. Open Biol. 2021, 11, 210120. [Google Scholar] [CrossRef] [PubMed]
- Soliman, N.A.; Yussif, S.M. Ki-67 as a prognostic marker according to breast cancer molecular subtype. Cancer Biol. Med. 2016, 13, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Endl, E.; Gerdes, J. Posttranslational modifications of the KI-67 protein coincide with two major checkpoints during mitosis. J. Cell. Physiol. 2000, 182, 371–380. [Google Scholar] [CrossRef]
- Mendonça, J.B.; Fernandes, P.V.; Fernandes, D.C.; Rodrigues, F.R.; Waghabi, M.C.; Tilli, T.M. Unlocking Overexpressed Membrane Proteins to Guide Breast Cancer Precision Medicine. Cancers 2024, 16, 1402. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, Y.; Moresco, J.; Tu, P.; Yates, J.; Nardulli, A. Plasma Membrane Proteomics of Human Breast Cancer Cell Lines Identifies Potential Targets for Breast Cancer Diagnosis and Treatment. PLoS ONE 2014, 9, e102341. [Google Scholar] [CrossRef]
- Burguin, A.; Furrer, D.; Ouellette, G.; Jacob, S.; Diorio, C.; Durocher, F. Trastuzumab effects depend on HER2 phosphorylation in HER2-negative breast cancer cell lines. PLoS ONE 2020, 15, e0234991. [Google Scholar] [CrossRef]
- Bradley, R.; Braybrooke, J.; Gray, R.; Hills, R.; Liu, Z.; Peto, R.; Davies, L.; Dodwell, D.; McGale, P.; Pan, H.; et al. Trastuzumab for early-stage, HER2-positive breast cancer: A meta-analysis of 13,864 women in seven randomised trials. Lancet Oncol. 2021, 22, 1139–1150. [Google Scholar] [CrossRef]
- Xia, X.; Hu, T.; He, X.; Liu, Y.; Yu, C.; Kong, W.; Liao, Y.; Tang, D.; Liu, J.; Huang, H. Neddylation of HER2 Inhibits its Protein Degradation and promotes Breast Cancer Progression. Int. J. Biol. Sci. 2023, 19, 377–392. [Google Scholar] [CrossRef]
- Ko, P.-J.; Dixon, S.J. Protein palmitoylation and cancer. EMBO Rep. 2018, 19, e46666. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xiao, M.; Mo, Y.; Wang, H.; Han, Y.; Zhao, X.; Yang, X.; Liu, Z.; Xu, B. Emerging roles of protein palmitoylation and its modifying enzymes in cancer cell signal transduction and cancer therapy. Int. J. Biol. Sci. 2022, 18, 3447–3457. [Google Scholar] [CrossRef]
- Chen, B.; Zheng, B.; DeRan, M.; Jarugumilli, G.K.; Fu, J.; Brooks, Y.S.; Wu, X. ZDHHC7-mediated S-palmitoylation of Scribble regulates cell polarity. Nat. Chem. Biol. 2016, 12, 686–693. [Google Scholar] [CrossRef]
- Li, M.; Zhang, L.; Chen, C.-W. Diverse Roles of Protein Palmitoylation in Cancer Progression, Immunity, Stemness, and Beyond. Cells 2023, 12, 2209. [Google Scholar] [CrossRef]
- Zhou, B.; Hao, Q.; Liang, Y.; Kong, E. Protein palmitoylation in cancer: Molecular functions and therapeutic potential. Mol. Oncol. 2023, 17, 3–26. [Google Scholar] [CrossRef]
- Masuda, H.; Zhang, D.; Bartholomeusz, C.; Doihara, H.; Hortobagyi, G.N.; Ueno, N.T. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res. Treat. 2012, 136, 331–345. [Google Scholar] [CrossRef]
- Song, H.; Huang, L.; Zhang, M.; Wang, X.; Song, S.; Yang, L. Transphosphorylation of EGFR at Y845 plays an important role in its autophosphorylation and kinase activity. Oncol. Rep. 2014, 31, 2393–2398. [Google Scholar] [CrossRef]
- Song, H.; Li, C.-W.; Labaff, A.M.; Lim, S.-O.; Li, L.-Y.; Kan, S.-F.; Chen, Y.; Zhang, K.; Lang, J.; Xie, X.; et al. Acetylation of EGF receptor contributes to tumor cell resistance to histone deacetylase inhibitors. Biochem. Biophys. Res. Commun. 2011, 404, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Oved, S.; Mosesson, Y.; Zwang, Y.; Santonico, E.; Shtiegman, K.; Marmor, M.D.; Kochupurakkal, B.S.; Katz, M.; Lavi, S.; Cesareni, G.; et al. Conjugation to Nedd8 Instigates Ubiquitylation and Down-regulation of Activated Receptor Tyrosine Kinases. J. Biol. Chem. 2006, 281, 21640–21651. [Google Scholar] [CrossRef]
- Kozyulina, P.Y.; Loskutov, Y.V.; Kozyreva, V.K.; Rajulapati, A.; Ice, R.J.; Jones, B.C.; Pugacheva, E.N. Prometastatic NEDD9 Regulates Individual Cell Migration via Caveolin-1–Dependent Trafficking of Integrins. Mol. Cancer Res. 2015, 13, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Isaji, T.; Zhang, G.; Qi, F.; Duan, C.; Fukuda, T.; Gu, J. EpCAM associates with integrin and regulates cell adhesion in cancer cells. Biochem. Biophys. Res. Commun. 2020, 522, 903–909. [Google Scholar] [CrossRef]
- Soysal, S.D.; Muenst, S.; Barbie, T.; Fleming, T.; Gao, F.; Spizzo, G.; Oertli, D.; Viehl, C.T.; Obermann, E.C.; Gillanders, W.E. EpCAM expression varies significantly and is differentially associated with prognosis in the luminal B HER2+, basal-like, and HER2 intrinsic subtypes of breast cancer. Br. J. Cancer 2013, 108, 1480–1487. [Google Scholar] [CrossRef] [PubMed]
- Gires, O.; Pan, M.; Schinke, H.; Canis, M.; Baeuerle, P.A. Expression and function of epithelial cell adhesion molecule EpCAM: Where are we after 40 years? Cancer Metastasis Rev. 2020, 39, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Qiu, S.; Hu, H.; Li, W.; Li, X. Role of Caveolae family-related proteins in the development of breast cancer. Front Mol. Biosci. 2023, 10, 1242426. [Google Scholar] [CrossRef] [PubMed]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.B.G.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Hang, Q.; Isaji, T.; Lu, J.; Fukuda, T.; Gu1, J. Importance of membrane-proximal N-glycosylation on integrin α1 in its activation and complex formation. FASEB J. 2016, 30, 4120–4131. [Google Scholar] [CrossRef]
- Liu, X.; Gao, J.; Sun, Y.; Zhang, D.; Liu, T.; Yan, Q.; Yang, X. Mutation of N-linked glycosylation in EpCAM affected cell adhesion in breast cancer cells. Biol. Chem. 2017, 398, 1119–1126. [Google Scholar] [CrossRef]
- Hugonnet, M.; Singh, P.; Haas, Q.; von Gunten, S. The Distinct Roles of Sialyltransferases in Cancer Biology and Onco-Immunology. Front. Immunol. 2021, 12, 799861. [Google Scholar] [CrossRef]
- Yeh, Y.-C.; Ling, J.-Y.; Chen, W.-C.; Lin, H.-H.; Tang, M.-J. Mechanotransduction of matrix stiffness in regulation of focal adhesion size and number: Reciprocal regulation of caveolin-1 and β1 integrin. Sci. Rep. 2017, 7, 15008. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.L.; Pan, Y.-H.; Huang, Q.-Y.; Shi, Y.-B.; Huang, Q.-Y.; Hu, Z.-Z.; Xiong, L. Caveolin-1: A multifaceted driver of breast cancer progression and its application in clinical treatment. Onco. Targets Ther. 2019, 12, 1539–1552. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Park, J.H.; Lee, H.J.; Jo, U.; Park, J.K.; Seo, J.H.; Kim, Y.H.; Kim, I.; Park, K.H. Caveolin-1 Modulates Docetaxel-Induced Cell Death in Breast Cancer Cell Subtypes through Different Mechanisms. Cancer Res. Treat. 2016, 48, 715–726. [Google Scholar] [CrossRef] [PubMed]
- El-Schich, Z.; Zhang, Y.; Göransson, T.; Dizeyi, N.; Persson, J.; Johansson, E.; Caraballo, R.; Elofsson, M.; Shinde, S.; Sellergren, B.; et al. Sialic Acid as a Biomarker Studied in Breast Cancer Cells Using Fluorescent Molecularly Imprinted Polymers; MIPs: San Jose, CA, USA, 2020. [Google Scholar]
- Zhou, X.; Chi, K.; Zhang, C.; Liu, Q.; Yang, G. Sialylation: A Cloak for Tumors to Trick the Immune System in the Microenvironment. Biology 2023, 12, 832. [Google Scholar] [CrossRef] [PubMed]
- Dobie, C.; Skropeta, D. Insights into the role of sialylation in cancer progression and metastasis. Br. J. Cancer 2021, 124, 76–90. [Google Scholar] [CrossRef]
- Yuan, Y.; Wu, L.; Shen, S.; Wu, S.; Burdick, M.M. Effect of alpha 2,6 sialylation on integrin-mediated adhesion of breast cancer cells to fibronectin and collagen IV. Life Sci. 2016, 149, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xu, Z.; Peng, Y.; Wang, J.; Xiang, Y. Integrin β4 as a Potential Diagnostic and Therapeutic Tumor Marker. Biomolecules 2021, 11, 1197. [Google Scholar] [CrossRef] [PubMed]
- Alsaleem, M.; Toss, M.S.; Joseph, C.; Aleskandarany, M.; Kurozumi, S.; Alshankyty, I.; Ogden, A.; Rida, P.C.G.; Ellis, I.O.; Aneja, R.; et al. The molecular mechanisms underlying reduced E-cadherin expression in invasive ductal carcinoma of the breast: High throughput analysis of large cohorts. Mod. Pathol. 2019, 32, 967–976. [Google Scholar] [CrossRef]
- Pinho, S.; Oliveira, P.; Cabral, J.; Carvalho, S.; Huntsman, D.; Gärtner, F.; Seruca, R.; Reis, C.; Oliveira, C. Loss and Recovery of Mgat3 and GnT-III Mediated E-cadherin N-glycosylation Is a Mechanism Involved in Epithelial-Mesenchymal-Epithelial Transitions. PLoS ONE 2012, 7, e33191. [Google Scholar] [CrossRef]
- Tellez, T.; Garcia-Aranda, M.; Redondo, M. The Role of Clusterin in Carcinogenesis and its Potential Utility as Therapeutic Target. Curr. Med. Chem. 2016, 23, 4297–4308. [Google Scholar] [CrossRef]
- Merlotti, A.; Malizia, A.L.; Michea, P.; Bonte, P.-E.; Goudot, C.; Carregal, M.S.; Nuñez, N.; Sedlik, C.; Ceballos, A.; Soumelis, V.; et al. Aberrant fucosylation enables breast cancer clusterin to interact with dendritic cell-specific ICAM-grabbing non-integrin (DC-SIGN). Oncoimmunology 2019, 8, e1629257. [Google Scholar] [CrossRef]
- Janiszewska, E.; Kmieciak, A.; Kacperczyk, M.; Witkowska, A.; Kratz, E.M. The Influence of Clusterin Glycosylation Variability on Selected Pathophysiological Processes in the Human Body. Oxidative Med. Cell. Longev. 2022, 2022, 7657876. [Google Scholar] [CrossRef]
- Ding, P.; Ma, Z.; Fan, Y.; Feng, Y.; Shao, C.; Pan, M.; Zhang, Y.; Huang, D.; Han, J.; Hu, Y.; et al. Emerging role of ubiquitination/deubiquitination modification of PD-1/PD-L1 in cancer immunotherapy. Genes Dis. 2023, 10, 848–863. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hsu, J.-M.; Sun, L.; Chan, L.-C.; Li, C.-W.; Hsu, J.L.; Wei, Y.; Xia, W.; Hou, J.; Qiu, Y.; et al. Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res. 2019, 29, 83–86. [Google Scholar] [CrossRef]
- Ma, Y.; Marinkova, R.; Nenkov, M.; Jin, L.; Huber, O.; Sonnemann, J.; Peca, N.; Gaßler, N.; Chen, Y. Tumor-Intrinsic PD-L1 Exerts an Oncogenic Function through the Activation of the Wnt/β-Catenin Pathway in Human Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2022, 23, 11031. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, R.B. The ubiquitin system: From cell signalling to disease biology and new therapeutic opportunities. Cell Death Differ. 2021, 28, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, Z.; Yang, Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer 2020, 19, 146. [Google Scholar] [CrossRef]
- Mohtar, M.A.; Syafruddin, S.E.; Nasir, S.N.; Low, T.Y. Revisiting the Roles of Pro-Metastatic EpCAM in Cancer. Biomolecules 2020, 10, 255. [Google Scholar] [CrossRef]
- Munz, M.; Fellinger, K.; Hofmann, T.; Schmitt, B.; Gires, O. Glycosylation is crucial for stability of tumour and cancer stem cell antigen EpCAM. FBL 2008, 13, 5195–5201. [Google Scholar] [CrossRef]
- Sun, L.; Guo, S.; Xie, Y.; Yao, Y. The characteristics and the multiple functions of integrin β1 in human cancers. J. Transl. Med. 2023, 21, 787. [Google Scholar] [CrossRef]
- Zhuo, Y.; Chammas, R.; Bellis, S.L. Sialylation of β1 Integrins Blocks Cell Adhesion to Galectin-3 and Protects Cells against Galectin-3-induced Apoptosis. J. Biol. Chem. 2008, 283, 22177–22185. [Google Scholar] [CrossRef]
- Díaz, M.I.; Díaz, P.; Bennett, J.C.; Urra, H.; Ortiz, R.; Orellana, P.C.; Hetz, C.; Quest, A.F.G. Caveolin-1 suppresses tumor formation through the inhibition of the unfolded protein response. Cell Death Dis. 2020, 11, 648. [Google Scholar] [CrossRef]
- Zhuang, J.; Huo, Q.; Yang, F.; Xie, N. Perspectives on the Role of Histone Modification in Breast Cancer Progression and the Advanced Technological Tools to Study Epigenetic Determinants of Metastasis. Front. Genet. 2020, 11, 603552. [Google Scholar] [CrossRef]
- Cheung, P.; Tanner, K.G.; Cheung, W.L.; Sassone-Corsi, P.; Denu, J.M.; Allis, C.D. Synergistic Coupling of Histone H3 Phosphorylation and Acetylation in Response to Epidermal Growth Factor Stimulation. Mol. Cell 2000, 5, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, H.; Sui, S.; Wang, Q.; Xu, S.; Pang, D. Targeting Histone Modifications in Breast Cancer: A Precise Weapon on the Way. Front. Cell Dev. Biol. 2021, 9, 736935. [Google Scholar] [CrossRef]
- Fu, X.; Tan, W.; Song, Q.; Pei, H.; Li, J. BRCA1 and Breast Cancer: Molecular Mechanisms and Therapeutic Strategies. Front. Cell Dev. Biol. 2022, 10, 813457. [Google Scholar] [CrossRef] [PubMed]
- Onyiba, C.I.; Scarlett, C.J.; Weidenhofer, J. The Mechanistic Roles of Sirtuins in Breast and Prostate Cancer. Cancers 2022, 14, 5118. [Google Scholar] [CrossRef] [PubMed]
- Geffen, Y.; Anand, S.; Akiyama, Y.; Yaron, T.M.; Song, Y.; Johnson, J.L.; Govindan, A.; Babur, Ö.; Li, Y.; Huntsman, E.; et al. Pan-cancer analysis of post-translational modifications reveals shared patterns of protein regulation. Cell 2023, 186, 3945–3967. [Google Scholar] [CrossRef]

| Hormone Receptors | Role | PTMs | Modifying Enzymes | PTM Sites | Effects of PTMs |
|---|---|---|---|---|---|
| Estrogen receptor alpha (ERα encoded by ESR1 gene) | nuclear receptor; transcription factor involved in cell proliferation, differentiation, and homeostasis; related to risk of BC, EC, and OC [77]; expressed in 70% of BC cases [41] | phosphorylation | c-Src kinase and ERK1/2 signaling pathways [77] | Ser118/167 [77]; Tyr537 [80,81] | alteration of ERα function (chromatin interaction, gene expression, BC growth, morphology, migration, invasion, and response to endocrine therapy) [77]; ERα phosphorylation by Src regulates its hormone-dependent nuclear export and cell cycle progression in BC cells [80] |
| RAS-RAF-MAPK cascade [103] | Ser118 [103] | ER activation [103] | |||
| acetylation | lysine acetyltransferases p300/CBP [104] | Lys266/268 | enhances E2-responsive genes transcription and signaling in BC [104,105] | ||
| methylation | PRMT1 | Arg260 [85] | regulates ERα/PR cross-talk and activates a signaling complex in aggressive human BC [106] | ||
| ubiquitination | ubiquitin ligases (CHIP, E3 ubiquitin-protein ligase MDM2, and E6AP) | K/Lys48 | degradation of ERα in UPS, changes in ERα function [82] | ||
| deubiquitintion | deubiquitinases USP7 [41], USP1 [107], USP15 [40], USP14 [108], MINDY1 [109] | K/Lys48-linked ubiquitination | stabilizes ERα/prevents ERα degradation in BC and promotes BC progression in ERα+ BC [40,41] and EC [108], inhibits ERα K48-linked poly-ubiquitination [107] | ||
| SUMOylation | SUMO1 [110] | SUMOylated lysines are located in the hinge region of ERα [110] | regulates ERα-dependent transcription in MCF7 BC cell line [110]; full anti-estrogenicity in the absence of accelerated receptor turnover [111] | ||
| S-palmitoylation | palmitoylacyltransferases ZDHHC-7 ZHHHC-21 for sex steroid hormones | Cys447 | induces ERα localization to the plasma membrane and ERα-CAV1 interaction, enabling rapid E2 signaling for cell proliferation by ERK/MAPK, PI3K/AKT activation [67] | ||
| Progesterone receptor (PR) | nuclear receptor; transcription factor; modulates ERα action in BC; activates cytosolic signaling pathways; upregulated target gene of ER; prognostic biomarker in BC, especially in HR+ BC [88,112]; PR-A is involved in cell adhesion, morphology, invasiveness and metastasis, resistance to apoptosis and tamoxifen; PR-B is required for mammary gland development and expansion [113] | phosphorylation | MAPK CDK2 PKA CK2 EGFR-induced phosphorylation/ERK-mediated, c-Src [112,114] | Ser294/345 [112,115]; Ser190/294/554/676 [116]; Ser191A [117] | regulates PR cell cycle and growth-promoting target genes [112]; activates PR hormone-dependent transcription [116]; PR phosphorylation targets it for ubiquitination and proteasomal destruction in cytosol [114] |
| acetylation | p300 acetyltransferase | Lys183 | enhances PR-dependent initiation and reinitiation process via accelerated binding of its direct target genes; enhances Ser294 phosphorylation levels and PR activity, suppressing SUMOylation of PR [88] | ||
| methylation | PRMT1 | Arg637 | stabilizes PR and accelerates its recycling and transcriptional activity; methylate ERαArg260, involved in ERα/PR cross talk [106] | ||
| ubiquitination | PR phosphorylation via EGFR pathway leads to PR ubiquitination [114] | EC cells undergo loss of PR and do not respond to progestin therapy [114] | |||
| SUMOylation/ deSUMOylation | SENP1 deSUMOylates PRA | SUMOylation attenuates PR activity on all target genes [113] | |||
| palmitoylation | palmitoylacyltransferases ZDHHC-7 ZHHHC-21 for sex steroid hormones | a small fraction of the PR is palmitoylated and anchored to the cell membrane (mbPR), into a complex with ERα [118] | mbPR, upon progestin exposure, activates the Src/RAS/ERK kinase pathway, leading to phosphorylation of nPR by the ERK signaling pathway, leading to intracellular PR phosphorylation, gene regulation, and entry into cell cycle in the absence of detectable intracellular progestin [118] | ||
| Androgen receptor (AR) | ligand-dependent/ligand-independent nuclear transcription factor, detected in 90% of primary BC, and 75% of metastasis [90]; prognostic and predictive biomarker in BC [44], involved in transcription, cellular proliferation, apoptosis in prostate cells [119] | phosphorylation | ERK1/2 and CDK1 [90] | increased on Ser213/650 in primary ER-negative BC, ductal carcinomas and metastases, compared to benign tissue [44]; Ser515 [92] | role in signal transduction; predictive value in BC prognosis [44] |
| PI3K/AKT signaling [120] | Ser 213/791 | AKT is an activator of AR required for HER2 signaling to androgen-independent survival and growth of PC cells [120] | |||
| Ser210/790 | AKT suppresses androgen-induced apoptosis by phosphorylating and inhibiting AR [121] | ||||
| Ser16/81/256/308/424/650/94 in COS1 fibroblast-like cells and LNCaP cells [122] | analysis of cross-talk between growth factor signaling and androgen in prostate development, physiology, and cancer [122] | ||||
| acetylation | KATs (CBP, P300, PCAF, TIP60, MOF, HBO1 ARD1) [119] | plays an important role in directing its cellular activities, modulating its stability, nuclear localization, and transcriptional activity [119] | |||
| Transcription Factors | Role | PTMs | Modifying Enzymes | PTM Sites | Effects of PTMs |
|---|---|---|---|---|---|
| TWIST1 | regulator in embryonic development; implicated in tumor initiation, stemness, angiogenesis, spreading, and chemoresistance; promotes EMT in BC [139] | phosphorylation | MAPKs, JNK, ERK1/2, p38 | Ser68 | induces TWIST stabilization, EMT, invasiveness, metastasis in BC [140] |
| AKT1 | Ser42/123, Thr121 | sustains β-TRCP-mediated TWIST1 ubiquitination and degradation; suppresses EMT; AKT1 inhibition stabilizes TWIST1 and enhances EMT in BC cells [141] | |||
| SMAD3 | phosphorylation | MAPKs, CDKs, GSK3β | enhances or suppresses TGF-β signaling involved in apoptosis, growth arrest, differentiation, and EMT [73] | ||
| Zinc finger E-box binding homeobox 1/2 (ZEB1/2) | frequently expressed in carcinomas, being involved in normal development and tumor metastasis [142] | phosphorylation | IGF1/MEK/ERK pathways; PKC | Thr867 hr851, Ser852, Ser853 | ZEB downregulation/transcriptional repression of ZEB1 [16] |
| autophosphorylation | ATM hyperactivation | Ser585 | ZEB1 upregulation in radioresistant BC cells | ||
| SNAIL/SNAI1 transcriptional repressor 1 | plays a role in the control of EMT and fibroblast activation [155] | phosphorylation | PAK1 | Ser246 | induces SNAIL activation and accumulation in the nucleus; promotes EMT [143] |
| PI3K | Ser246 | increased SNAIL activation [156] | |||
| PKD1 | Ser11 | nuclear export (destabilization) of SNAIL; reduces its transcriptional repressive activity, favors its ubiquitination and degradation and suppresses EMT [145] | |||
| PKA, CK2 | Ser11/92 | SNAIL stability [157] | |||
| GSK3β | Ser92/96/100/104 | SNAIL localization in cytoplasm; β-TRCP-mediated ubiquitination and proteasomal degradation [144,146] | |||
| LATS2 | Thr203 | increases SNAIL nucleolar retention and activation [148] | |||
| ERK2 | Ser82/104 | increases SNAIL nuclear accumulation and activation; suppresses E-cadherin; induces EMT [147] | |||
| CK1 | Ser104/107 | mediates subsequent phosphorylation by GSK3β of S100/96/92, followed by TRCP-mediated ubiquitination and proteasomal degradation | |||
| monoubiquitination | ubiquitin-editing enzyme A2 | K296, K234, K235 | monoubiquitinated SNAIL has a reduced affinity for GSK3β enzyme; stabilizes in nucleus and escape from degradation by GSK3β-mediated degradation [138] | ||
| O-GlcNAc glycation | Ser112 | increases SNAIL stability by suppressing its degradation; disrupts CK1-mediated phosphorylation at S104/107, inhibiting GSK3β-mediated degradation [158] | |||
| SLUG/SNAIL2 (encoded by SNAI2 gene) | has a zinc finger domain with transcriptional repressor activity [138] | phosphorylation | PAK4 | Ser158/254 | increases transcriptional activity and stabilization as a repressor of the CDH1 promoter in PC and accelerates cancer progression [159] |
| GATA3 binding protein 3 (GATA3) | overexpressed in BC; involved in mammary gland development; biomarker for diagnosing BC; tumor suppressor in BC; regulates BC progression depending on cellular context [160] | function modulated by PTMs: acetylation | acetylated by acetyltransferase CBP at K119; deacetylated by HDAC1, HDAC2, HDAC3 | inhibits cell migration and invasion in lung adenocarcinoma, downregulating EMT-related transcription factors (SLUG, ZEB1, and ZEB2) [152] | |
| methylation | Arg261 | regulates transactivation of the Il5 gene in T helper 2 (Th2) cells [161] | |||
| progestin-induces GATA3 phosphorylation | Ser308 | induces its proteasome-mediated degradation, decreasing GATA3 levels in BC cells upon PR activation for progestin stimulation of in vitro cell proliferation and in vivo tumor growth [160] | |||
| SRY-box transcription factor 2 (SOX2), octamer-binding transcription factor 4 (OCT4), Krűppel-like factor 4 (KLF4), c-MYC | involved in the generation of induced pluripotent stem cells (iPSCs) cells by reprogramming adult cells [153] | acetylation | PKB/AKT, p300 | stabilizes KLF4 by inactivating GSK3 [153] | |
| Classical Biomarkers | Role | PTMs | Modifying Enzymes | PTMs Sites | Effects of PTMs |
|---|---|---|---|---|---|
| Human epidermal growth factor receptor 2 (HER2/ErbB2 encoded by ERBB2 gene) | transmembrane glycoprotein with tyrosine kinase activity, overexpressed in 20–25% of BC [168] | phosphorylation | Y877 | in HER2-negative BC cell lines, makes HER2 recognizable by trastuzumab that decreases cell proliferation in HER2-/pHER2+ tumors as in HER2-positive BC cell lines [168,169] | |
| neddylation | NEDD8 and NAE1 | neddylation inhibits HER2 degradation and promotes BC progression; inhibition of neddylation promotes HER2 degradation by improving its ubiquitination [170] | |||
| Epidermal growth factor receptor (EGFR/ErbB1/ HER1) | mbEGFR glycoprotein can be internalized in the nucleus of the cell, acting in gene transcription, increasing proliferation, and favoring resistance to chemotherapy [171]; also identified as mtEGFR [27]; half of cases of TNBC and IBC overexpress EGFR that regulates EMT, migration, and invasion [176] | palmitoylation of mtEGFR [27] | promotes mitochondrial fusion and cell survival [27] | ||
| phosphorylation | MEK1/2, ERK1/2 | Tyr1068 | |||
| transphosphorylation | Src | Y845 other: Y992/1045/1068/1173 | leads to autophosphorylation and kinase activity for Ras/MAPK, PI3/AKT, and JAK-STAT3/5 [177] | ||
| Y1045 | regulates EGFR ubiquitination and degradation [177] | ||||
| acetylation | CBP acetyltransferase | affects its Tyr phosphorylation, inducing cancer cell resistance to HDAC inhibitors [178] | |||
| ubiquitination and neddylation | c-Cbl ubiquitin ligase, ubiquitin-like molecule NEDD8 | induces lysosomal degradation; desensitizes growth-factor-activated receptor tyrosine kinase [179] | |||
| Other Biomarkers | Role | PTMs | Modifying Enzymes | PTMs Sites | Effects of PTMs |
|---|---|---|---|---|---|
| Epithelial cell adhesion molecule (EpCAM) | surface protein; biomarker for epithelial tumors and CTCs; upregulated in solid epithelial cancers and stem cells; plays a role in cell adhesion, migration, proliferation, and differentiation [207] | deglycosylation | promotes apoptosis, downregulating Bcl-2 and upregulating Bax and caspase-3 through the ERK1/2 and c-Jun N-terminal kinase mitogen-activated protein kinase signaling pathways [35] | ||
| O-linked glycosylation | Thr171, Thr172 | ||||
| N-linked glycosylation | Asn74, Asn111, Asn198 | Asn198 is essential for stability maintenance of EpCAM; affects expression level and the molecule lifespan in the plasma membrane [208] | |||
| Integrin β1 (ITGβ1) | cell adhesion glycoproteins that connect ECM to the cytoskeleton, involved in cancer progression, cell motility, adhesion, migration, proliferation, differentiation, and chemotherapy resistance [209] | α2,6 sialylation | β-galactosamide α-2,6-sialyltranferase I (ST6Gal-I) | plays critical roles in integrin activation [186]; regulator of cell adhesion and contributes to invasion of BC cells to ECM [195]; integrins are highly α2,6 sialylated on the MDA-MB-231 BC cell line but not on MCF7 BC cells [195]; in human colon tumors, sialylation of ITGβ1 blocks cell adhesion to GAL3 and protects cells against GAL3-induced apoptosis [210] | |
| N-glycosylation | 12 potential N-glycosylation sites, including 7–8, 9–12 | ITGβ1 activation positively regulates cell migration, critical for PPIs with other cell membrane proteins (syndecan 4, EGFR) [186] | |||
| Integrin β4 (ITGβ4) | transmembrane adhesion molecule; plays a role in invasion, proliferation, EMT, and angiogenesis; putative tumor marker [196] | phosphorylation by EGFR/Src-signaling | enhances malignant potential of BC [72] | ||
| palmitoylation | catalyzed by ZDHHC3 | promotes invasive BC cell migration [26] | |||
| Scribble (SCRIB) | cell junction and tumor-suppressor protein with a key role in maintaining polarity of epithelial cells; overexpressed and mislocated in human cancers, being involved in the regulation of Hippo pathway in BC cells [173] | palmitoylation | important for SCRIB membrane trafficking, cell polarity, and tumor suppression, emphasizing the importance of palmitoylation for cell polarity and tumorigenesis [173]. | ||
| Caveolin 1 (CAV1) | membrane-associated scaffolding protein that acts as a tumor suppressor in early stages of cancer and promoter of metastasis; downregulated in human tumors, cancer cell lines and oncogene-transformed cells [211] | phosphorylation by Src tyrosine kinase mediated by RACK1 | RACK1 and Src silencing enhances drug sensitivity in MDR cells [74] | ||
| Clusterin (CLU1)/apolipoprotein J (APOJ) | glycoprotein/chaperone involved in apoptosis, cell cycle regulation and DNA repair [199]; activity depends on its degree of glycosylation [201] | fucosylation | plays a role in BC progression via proangiogenic cytokines and TNF-α production by intratumoral macrophages [200] | ||
| Programmed cell death ligand 1 (PD-L1) | transmembrane protein overexpressed in many cancers, known for its oncogenic function exerted by activation of Wnt/β-catenin pathway [203,204] | palmitoylation | affects its stability; targeting PD-L1 palmitoylation sensitized tumor cells to T-cell killing and inhibited tumor growth [203]. | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neagu, A.-N.; Josan, C.-L.; Jayaweera, T.M.; Morrissiey, H.; Johnson, K.R.; Darie, C.C. Bio-Pathological Functions of Posttranslational Modifications of Histological Biomarkers in Breast Cancer. Molecules 2024, 29, 4156. https://doi.org/10.3390/molecules29174156
Neagu A-N, Josan C-L, Jayaweera TM, Morrissiey H, Johnson KR, Darie CC. Bio-Pathological Functions of Posttranslational Modifications of Histological Biomarkers in Breast Cancer. Molecules. 2024; 29(17):4156. https://doi.org/10.3390/molecules29174156
Chicago/Turabian StyleNeagu, Anca-Narcisa, Claudiu-Laurentiu Josan, Taniya M. Jayaweera, Hailey Morrissiey, Kaya R. Johnson, and Costel C. Darie. 2024. "Bio-Pathological Functions of Posttranslational Modifications of Histological Biomarkers in Breast Cancer" Molecules 29, no. 17: 4156. https://doi.org/10.3390/molecules29174156
APA StyleNeagu, A.-N., Josan, C.-L., Jayaweera, T. M., Morrissiey, H., Johnson, K. R., & Darie, C. C. (2024). Bio-Pathological Functions of Posttranslational Modifications of Histological Biomarkers in Breast Cancer. Molecules, 29(17), 4156. https://doi.org/10.3390/molecules29174156







