Multi-Hydrogen Bonding on Quaternized-Oligourea Receptor Facilitated Its Interaction with Bacterial Cell Membranes and DNA for Broad-Spectrum Bacteria Killing
Abstract
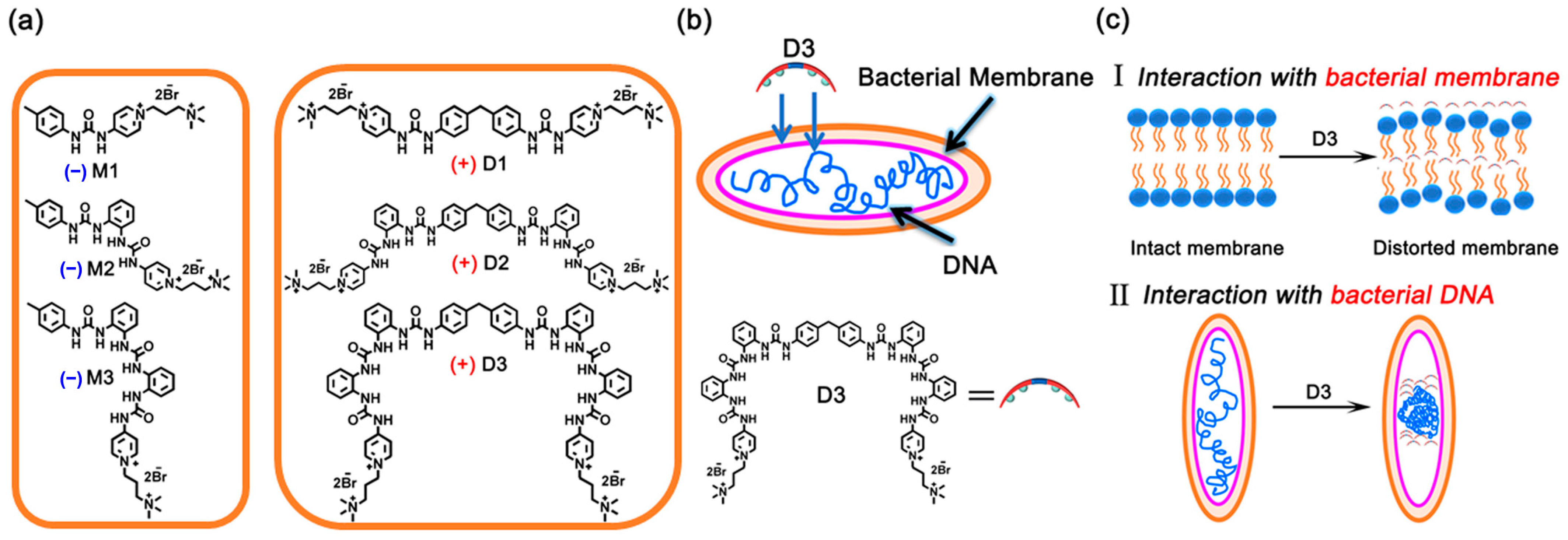
1. Introduction
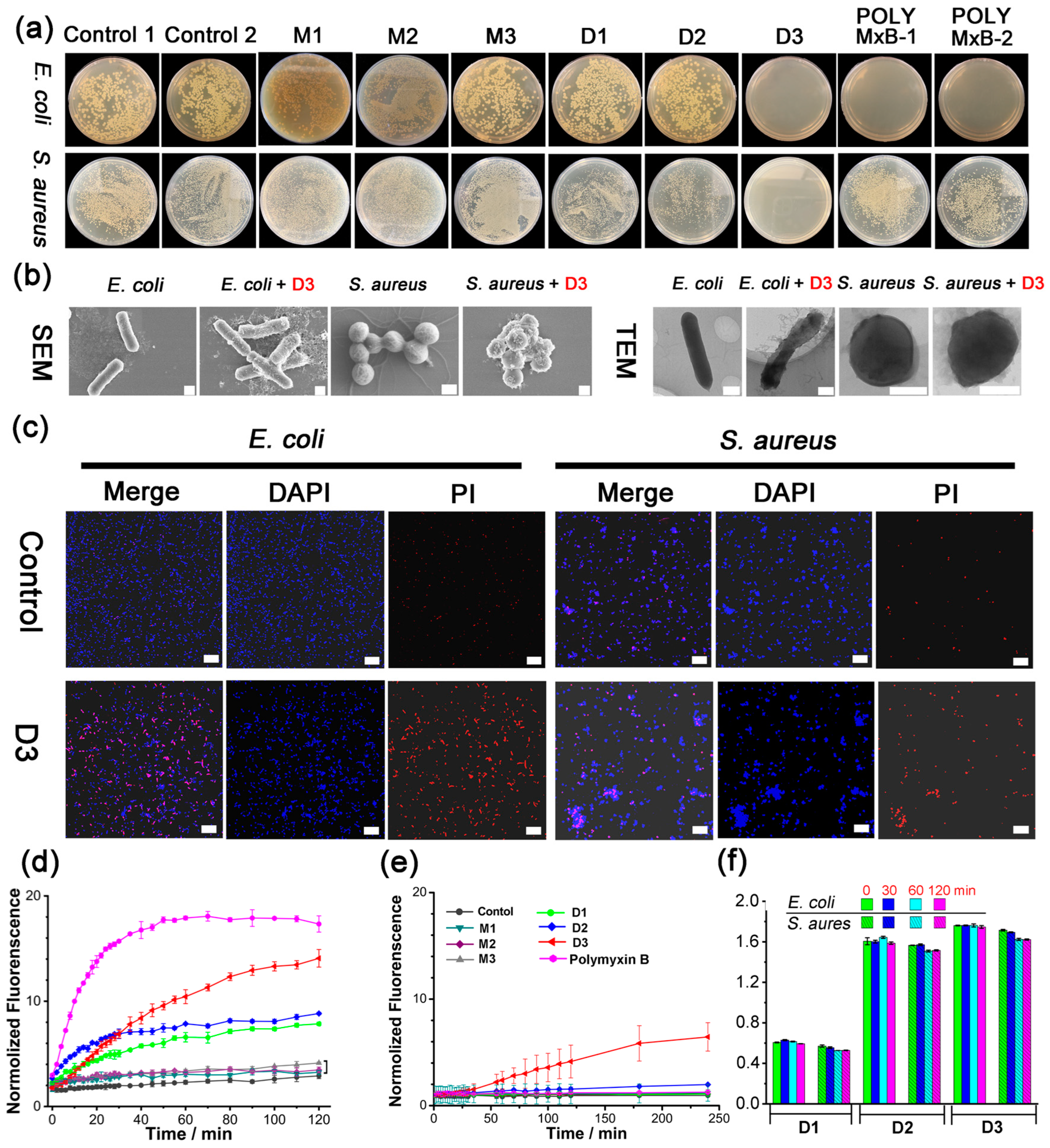
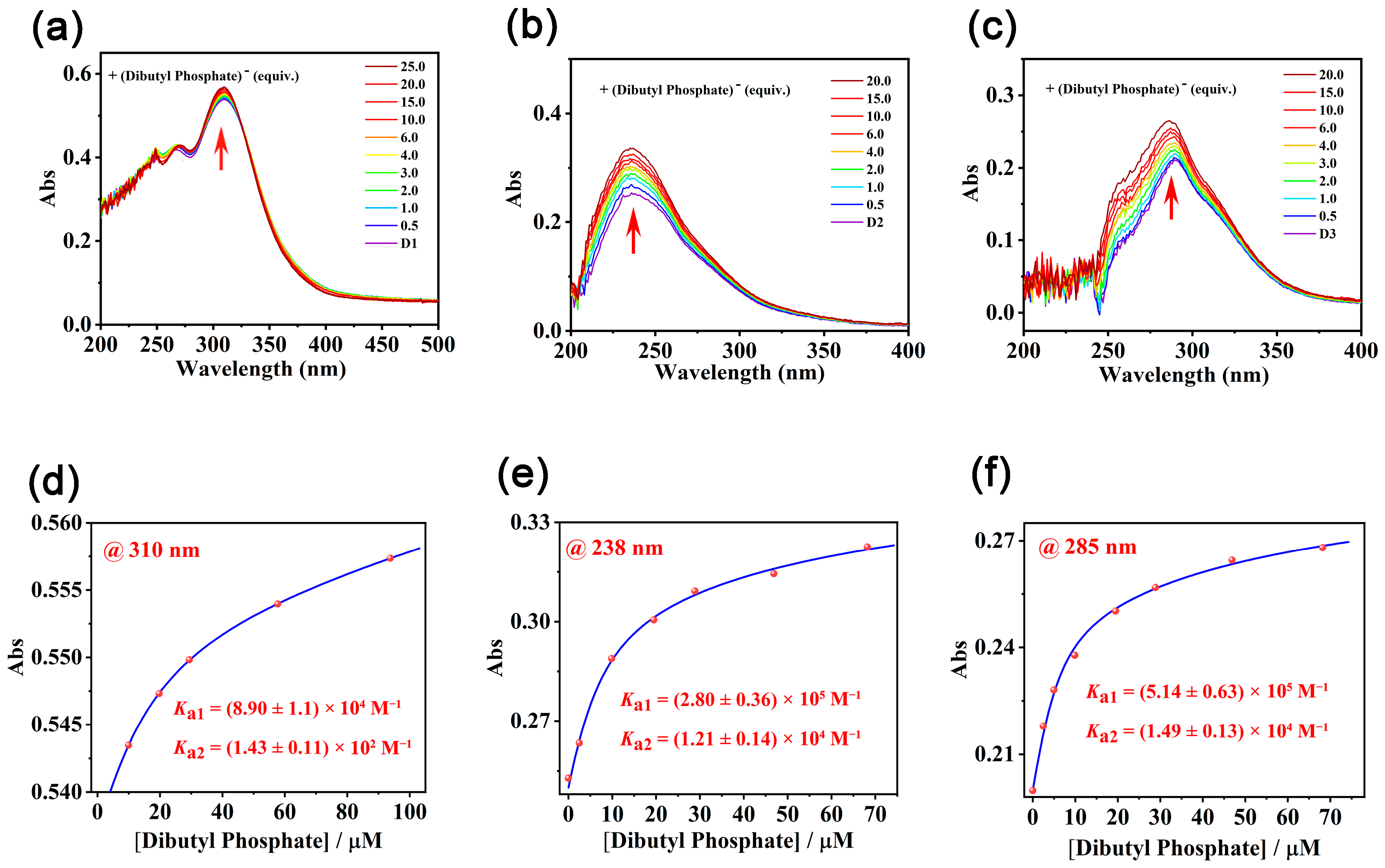
2. Results and Discussion
3. Experimental Section
3.1. Materials
3.2. Determination of MIC and MBC Values
3.3. Cell Viability Assay
3.4. DLS Study
3.5. Scanning Electron Microscopy (SEM) Study
3.6. Transmission Electron Microscopy (TEM) Study
3.7. Confocal Laser Scanning Microscopy (CLSM) Study
3.8. Propidium Iodide (PI) Uptake Assay
3.9. DNA Release Study
3.10. Determination of the Binding Constants by UV-Vis Titrations
3.11. DNA Binding Study
3.12. CD Experiments
3.13. Bacterial DNA Extraction Process
3.14. Agarose Gel Electrophoresis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaufmann, S.H.E.; Dorhoi, A.; Hotchkiss, R.S.; Bartenschlager, R. Host-Directed Therapies for Bacterial and Viral Infections. Nat. Rev. Drug Discov. 2018, 17, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Udaondo, Z.; Matilla, M.A. Mining for Novel Antibiotics in the Age of Antimicrobial Resistance. Microb. Biotechnol. 2020, 13, 1702–1704. [Google Scholar] [CrossRef]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Shatalin, K.; Nuthanakanti, A.; Kaushik, A.; Shishov, D.; Peselis, A.; Shamovsky, I.; Pani, B.; Lechpammer, M.; Vasilyev, N.; Shatalina, E.; et al. Inhibitors of Bacterial H2S Biogenesis Targeting Antibiotic Resistance and Tolerance. Science 2021, 372, 1169–1175. [Google Scholar] [CrossRef]
- Randall, J.R.; Davies, B.W. Mining for Novel Antibiotics. Curr. Opin. Microbiol. 2021, 63, 66–69. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial Peptides: Application Informed by Evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Fox, J.L. Antimicrobial Peptides Stage a Comeback. Nat. Biotechnol. 2013, 31, 379–382. [Google Scholar] [CrossRef]
- Mergaert, P. Role of Antimicrobial Peptides in Controlling Symbiotic Bacterial Populations. Nat. Prod. Rep. 2018, 35, 336–356. [Google Scholar] [CrossRef]
- Sambhy, V.; Peterson, B.R.; Sen, A. Antibacterial and Hemolytic Activities of Pyridinium Polymers as a Function of the Spatial Relationship between the Positive Charge and the Pendant Alkyl Tail. Angew. Chem. Int. Ed. 2008, 47, 1250–1254. [Google Scholar] [CrossRef]
- El Kenawy, R.; Worley, S.D.; Broughton, R. The Chemistry and Applications of Antimicrobial Polymers: A State-of-the-Art Review. Biomacromolecules 2007, 8, 1359–1384. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.J.; Wong, E.H.H.; Boyer, C.; Qiao, G.G. Antimicrobial Polymeric Nanoparticles. Prog. Polym. Sci. 2018, 76, 40–64. [Google Scholar] [CrossRef]
- Yan, S.; Chen, S.; Gou, X.; Yang, J.; An, J.; Jin, X.; Yang, Y.W.; Chen, L.; Gao, H. Biodegradable Supramolecular Materials Based on Cationic Polyaspartamides and Pillar[5]Arene for Targeting Gram-Positive Bacteria and Mitigating Antimicrobial Resistance. Adv. Funct. Mater. 2019, 29, 1904683. [Google Scholar] [CrossRef]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological Activity of Ionic Liquids and Their Application in Pharmaceutics and Medicine. Chem. Rev. 2017, 117, 7132–7189. [Google Scholar] [CrossRef] [PubMed]
- Misra, A.; Franco Castillo, I.; Muller, D.P.; Gonzalez, C.; Eyssautier-Chuine, S.; Ziegler, A.; de la Fuente, J.M.; Mitchell, S.G.; Streb, C. Polyoxometalate-Ionic Liquids (Pom-Ils) as Anticorrosion and Antibacterial Coatings for Natural Stones. Angew. Chem. Int. Ed. 2018, 57, 14926–14931. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Li, J.; Yu, M.; Jia, W.; Duan, S.; Cao, D.; Ding, X.; Yu, B.; Zhang, X.; Xu, F.J. Molecular Sizes and Antibacterial Performance Relationships of Flexible Ionic Liquid Derivatives. J. Am. Chem. Soc. 2020, 142, 20257–20269. [Google Scholar] [CrossRef]
- Baptista, P.V.; McCusker, M.P.; Carvalho, A.; Ferreira, D.A.; Mohan, N.M.; Martins, M.; Fernandes, A.R. Nano-Strategies to Fight Multidrug Resistant Bacteria—“A Battle of the Titans”. Front. Microbiol. 2018, 9, 1441. [Google Scholar] [CrossRef]
- Cheeseman, S.; Christofferson, A.J.; Kariuki, R.; Cozzolino, D.; Daeneke, T.; Crawford, R.J.; Truong, V.K.; Chapman, J.; Elbourne, A. Antimicrobial Metal Nanomaterials: From Passive to Stimuli-Activated Applications. Adv. Sci. 2020, 7, 1902913. [Google Scholar] [CrossRef]
- Howson, S.E.; Bolhuis, A.; Brabec, V.; Clarkson, G.J.; Malina, J.; Rodger, A.; Scott, P. Optically Pure, Water-Stable Metallo-Helical ‘Flexicate’ Assemblies with Antibiotic Activity. Nat. Chem. 2012, 4, 31–36. [Google Scholar] [CrossRef]
- Sepehrpour, H.; Fu, W.; Sun, Y.; Stang, P.J. Biomedically Relevant Self-Assembled Metallacycles and Metallacages. J. Am. Chem. Soc. 2019, 141, 14005–14020. [Google Scholar] [CrossRef]
- Gao, L.; Wang, H.; Zheng, B.; Huang, F. Combating Antibiotic Resistance: Current Strategies for the Discovery of Novel Antibacterial Materials Based on Macrocycle Supramolecular Chemistry. Giant 2021, 7, 100066. [Google Scholar] [CrossRef]
- Hirsch, A.K.; Fischer, F.R.; Diederich, F. Phosphate Recognition in Structural Biology. Angew. Chem. Int. Ed. 2007, 46, 338–352. [Google Scholar] [CrossRef]
- Violette, A.; Fournel, S.; Lamour, K.; Chaloin, O.; Frisch, B.; Briand, J.P.; Monteil, H.; Guichard, G. Mimicking Helical Antibacterial Peptides with Nonpeptidic Folding Oligomers. Chem. Biol. 2006, 13, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Claudon, P.; Violette, A.; Lamour, K.; Decossas, M.; Fournel, S.; Heurtault, B.; Godet, J.; Mely, Y.; Jamart-Gregoire, B.; Averlant-Petit, M.C.; et al. Consequences of Isostructural Main-Chain Modifications for the Design of Antimicrobial Foldamers: Helical Mimics of Host-Defense Peptides Based on a Heterogeneous Amide/Urea Backbone. Angew. Chem. Int. Ed. 2010, 49, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Grillot, A.L.; Le Tiran, A.; Shannon, D.; Krueger, E.; Liao, Y.; O’Dowd, H.; Tang, Q.; Ronkin, S.; Wang, T.; Waal, N.; et al. Second-Generation Antibacterial Benzimidazole Ureas: Discovery of a Preclinical Candidate with Reduced Metabolic Liability. J. Med. Chem. 2014, 57, 8792–8816. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, K.; Kitchen, J.A.; Blasco, S.; Paradisi, F.; Gunnlaugsson, T. Supramolecular Pyridyl Urea Gels as Soft Matter with Antibacterial Properties against Mrsa and/or E. Coli. Chem. Commun. 2014, 50, 10819–10822. [Google Scholar] [CrossRef]
- Antunes, S.; Corre, J.P.; Mikaty, G.; Douat, C.; Goossens, P.L.; Guichard, G. Effect of Replacing Main-Chain Ureas with Thiourea and Guanidinium Surrogates on the Bactericidal Activity of Membrane Active Oligourea Foldamers. Bioorg. Med. Chem. 2017, 25, 4245–4252. [Google Scholar] [CrossRef]
- Tyuleva, S.N.; Allen, N.; White, L.J.; Pepes, A.; Shepherd, H.J.; Saines, P.J.; Ellaby, R.J.; Mulvihill, D.P.; Hiscock, J.R. A Symbiotic Supramolecular Approach to the Design of Novel Amphiphiles with Antibacterial Properties against Msra. Chem. Commun. 2018, 55, 95–98. [Google Scholar] [CrossRef]
- Carreira-Barral, I.; Rumbo, C.; Mielczarek, M.; Alonso-Carrillo, D.; Herran, E.; Pastor, M.; Del Pozo, A.; Garcia-Valverde, M.; Quesada, R. Small Molecule Anion Transporters Display in Vitro Antimicrobial Activity against Clinically Relevant Bacterial Strains. Chem. Commun. 2019, 55, 10080–10083. [Google Scholar] [CrossRef]
- Davis, J.T.; Gale, P.A.; Quesada, R. Advances in Anion Transport and Supramolecular Medicinal Chemistry. Chem. Soc. Rev. 2020, 49, 6056–6086. [Google Scholar] [CrossRef]
- Shen, J.; Ye, R.; Liu, Z.; Zeng, H. Hybrid Pyridine-Pyridone Foldamer Channels as M2-Like Artificial Proton Channels. Angew. Chem. Int. Ed. 2022, 61, e202200259. [Google Scholar] [CrossRef] [PubMed]
- Kral, V.; Lang, K.; Kralova, J.; Dvorak, M.; Martasek, P.; Chin, A.O.; Andrievsky, A.; Lynch, V.; Sessler, J.L. Polyhydroxylated Sapphyrins: Multisite Non-Metallic Catalysts for Activated Phosphodiester Hydrolysis. J. Am. Chem. Soc. 2006, 128, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Iverson, B.L.; Shreder, K.; Kral, V.; Sessler, J.L. Phosphate Recognition by Sapphyrin. A New Approach to DNA Binding. J. Am. Chem. Soc. 1993, 115, 11022–11023. [Google Scholar] [CrossRef]
- He, Q.; Vargas-Zúñiga, G.I.; Kim, S.H.; Kim, S.K.; Sessler, J.L. Macrocycles as Ion Pair Receptors. Chem. Rev. 2019, 119, 9753–9835. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Berry, S.N.; Wu, X.; Howe, E.N.W.; Gale, P.A. Advances in Anion Receptor Chemistry. Chem 2020, 6, 61–141. [Google Scholar] [CrossRef]
- Manna, U.; Das, G. An Overview of Anion Coordination by Hydroxyl, Amine and Amide Based Rigid and Symmetric Neutral Dipodal Receptors. Coord. Chem. Rev. 2021, 427, 213547. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, W.; Chen, C.-H.; Flood, A.H. Chloride Capture Using a C–H Hydrogen-Bonding Cage. Science 2019, 365, 159–161. [Google Scholar] [CrossRef]
- Bowman-James, K. Supramolecular Cages Trap Pesky Anions. Science 2019, 365, 124–125. [Google Scholar] [CrossRef]
- Sommer, F.; Marcus, Y.; Kubik, S. Effects of Solvent Properties on the Anion Binding of Neutral Water-Soluble Bis(cyclopeptides) in Water and Aqueous Solvent Mixtures. ACS Omega 2017, 2, 3669–3680. [Google Scholar] [CrossRef]
- Langton, M.J.; Serpell, C.J.; Beer, P.D. Anion Recognition in Water: Recent Advances from a Supramolecular and Macromolecular Perspective. Angew. Chem. Int. Ed. 2016, 55, 1974–1987. [Google Scholar] [CrossRef]
- Kataev, E.A.; Müller, C. Recent Advances in Molecular Recognition in Water: Artificial Receptors and Supramolecular Catalysis. Tetrahedron 2014, 70, 137–167. [Google Scholar] [CrossRef]
- Langton, M.J.; Robinson, S.W.; Marques, I.; Félix, V.; Beer, P.D. Halogen Bonding in Water Results in Enhanced Anion Recognition in Acyclic and Rotaxane Hosts. Nat. Chem. 2014, 6, 1039–1043. [Google Scholar] [CrossRef]
- Zhou, H.; Zhao, Y.; Gao, G.; Li, S.; Lan, J.; You, J. Highly Selective Fluorescent Recognition of Sulfate in Water by Two Rigid Tetrakisimidazolium Macrocycles with Peripheral Chains. J. Am. Chem. Soc. 2013, 135, 14908–14911. [Google Scholar] [CrossRef]
- Kubik, S. Anion Recognition in Water. Chem. Soc. Rev. 2010, 39, 3648–3663. [Google Scholar] [CrossRef] [PubMed]
- Oshovsky, G.V.; Reinhoudt, D.N.; Verboom, W. Supramolecular Chemistry in Water. Angew. Chem. Int. Ed. 2007, 46, 2366–2393. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Huang, Z.; Li, A.; Zhao, Y.; Zuo, W.; Li, Y.; Miao, H.; Ma, J.; Sun, W.; Wang, X.; et al. Crown Ether Functionalized Potassium-Responsive Anionocages for Cascaded Guest Delivery. Angew. Chem. Int. Ed. 2021, 60, 9573–9579. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Jia, C.; Wu, B.; Jansone-Popova, S.; Seipp, C.A.; Custelcean, R. Selective Binding of (Thio)sulfate and Phosphate in Water by Quaternary Ammonium Functionalized Oligo-ureas. Chem. Commun. 2019, 55, 1714–1717. [Google Scholar] [CrossRef]
- Zuo, W.; Huang, Z.; Zhao, Y.; Xu, W.; Liu, Z.; Yang, X.-J.; Jia, C.; Wu, B. Chirality Sensing of Choline Derivatives by a Triple Anion Helicate Cage through Induced Circular Dichroism. Chem. Commun. 2018, 54, 7378–7381. [Google Scholar] [CrossRef]
- Bai, X.; Jia, C.; Zhao, Y.; Yang, D.; Wang, S.-C.; Li, A.; Chan, Y.-T.; Wang, Y.-Y.; Yang, X.-J.; Wu, B. Peripheral Templation-Modulated Interconversion between an A4L6 Tetrahedral Anion Cage and A2L3 Triple Helicate with Guest Capture/Release. Angew. Chem. Int. Ed. 2018, 57, 1851–1855. [Google Scholar] [CrossRef]
- Wu, G.-Y.; Shi, X.; Phan, H.; Qu, H.; Hu, Y.-X.; Yin, G.-Q.; Zhao, X.-L.; Li, X.; Xu, L.; Yu, Q.; et al. Efficient Self-Assembly of Heterometallic Triangular Necklace with Strong Antibacterial Activity. Nat. Commun. 2020, 11, 3178. [Google Scholar] [CrossRef]
- Jia, C.; Zuo, W.; Zhang, D.; Yang, X.-J.; Wu, B. Anion Recognition by Oligo-(Thio)Urea-Based Receptors. Chem. Commun. 2016, 52, 9614–9627. [Google Scholar] [CrossRef] [PubMed]
- Hargrove, A.E.; Nieto, S.; Zhang, T.; Sessler, J.L.; Anslyn, E.V. Artificial Receptors for the Recognition of Phosphorylated Molecules. Chem. Rev. 2011, 111, 6603–6782. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Zhao, W.; Yang, X.J.; Wu, B. Anion-Coordination-Driven Assembly. Acc. Chem. Res. 2022, 55, 3218–3229. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Tao, Y.; Luo, Z.; Li, A.; Wang, S.; Qiao, X.; Ma, F.; Jia, C. Stereoselective Assembly of Hydrogen-Bonded Anionic Cages Dictated by Organophosphate-Based Chiral Nodes. Angew. Chem. Int. Ed. 2023, 62, e202300470. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Qiao, X.; Zuo, W.; Tao, Y.; Li, A.; Luo, Z.; Liu, Y.; Liu, X.; Wang, X.; Sun, W.; et al. Less Is More: A Shortcut for Anionocages Design Based on (RPO32−)-Monourea Coordination. Angew. Chem. Int. Ed. 2022, 61, e202210478. [Google Scholar] [CrossRef]
- Katayev, E.A.; Boev, N.V.; Myshkovskaya, E.; Khrustalev, V.N.; Ustynyuk, Y.A. Expanding Sapphyrin: Towards Selective Phosphate Binding. Chem.-Eur. J. 2008, 14, 9065–9073. [Google Scholar] [CrossRef]
- Fasting, C.; Schalley, C.A.; Weber, M.; Seitz, O.; Hecht, S.; Koksch, B.; Dernedde, J.; Graf, C.; Knapp, E.W.; Haag, R. Multivalency as a Chemical Organization and Action Principle. Angew. Chem. Int. Ed. 2012, 51, 10472–10498. [Google Scholar] [CrossRef]
- Bhatia, S.; Camacho, L.C.; Haag, R. Pathogen Inhibition by Multivalent Ligand Architectures. J. Am. Chem. Soc. 2016, 138, 8654–8666. [Google Scholar] [CrossRef]
- Boulos, L.; Prevost, M.; Barbeau, B.; Coallier, J.; Desjardins, R. Live/Dead Baclight: Application of a New Rapid Staining Method for Direct Enumeration of Viable and Total Bacteria in Drinking Water. J. Microbiol. Methods 1999, 37, 77–86. [Google Scholar] [CrossRef]
- Jin, Y.; Cowan, J.A. DNA Cleavage by Copper-Atcun Complexes. Factors Influencing Cleavage Mechanism and Linearization of Dsdna. J. Am. Chem. Soc. 2005, 127, 8408–8415. [Google Scholar] [CrossRef]
- Chen, J.; Wang, X.; Shao, Y.; Zhu, J.; Zhu, Y.; Li, Y.; Xu, Q.; Guo, Z. A Trinuclear Copper(Ii) Complex of 2,4,6-Tris(Di-2-Pyridylamine)-1,3,5-Triazine Shows Prominent DNA Cleavage Activity. Inorg. Chem. 2007, 46, 3306–3312. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Rhodes, A.L.; Wyatt, M.D.; Forrow, S.; Hartley, J.A. Gc Base Sequence Recognition by Oligo(Imidazolecarboxamide) and C-Terminus-Modified Analogues of Distamycin Deduced from Circular Dichroism, Proton Nuclear Magnetic Resonance, and Methidiumpropylethylenediaminetetraacetate-Iron(Ii) Footprinting Studies. Biochemistry 1993, 32, 4237–4245. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Wang, X.; Lin, M.; Sun, H.; Yang, X.; Guo, Z. Promotive Effect of the Platinum Moiety on the DNA Cleavage Activity of Copper-Based Artificial Nucleases. Inorg. Chem. 2010, 49, 2541–2549. [Google Scholar] [CrossRef]
- Kuzmic, P. Program Dynafit for the Analysis of Enzyme Kinetic Data: Application to Hiv Proteinase. Anal. Biochem. 1996, 237, 260–273. [Google Scholar] [CrossRef] [PubMed]

| Bacterial Strains | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | P. aeruginosa | S. aureus | S. epidermidis | E. faecalis | ||||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| D1 | 120 | 240 | 60 | 120 | 60 | 120 | 1.88 | 3.75 | 1.88 | 3.75 |
| D2 | 30 | 60 | 30 | 60 | 30 | 60 | 1.88 | 3.75 | 0.94 | 1.88 |
| D3 | 15 | 30 | 15 | 30 | 15 | 30 | 1.88 | 3.75 | 0.47 | 0.94 |
| Polymyxin B | 1.88 | 3.75 | - | - | 60 | 120 | - | - | - | - |
| Compound | Ka/M−1 | Compound | Ka1/M−1 | Ka2/M−1 |
|---|---|---|---|---|
| M1 | (7.08 ± 0.64) × 103 | D1 | (8.90 ± 1.1) × 104 | (1.43 ± 0.11) × 102 |
| M2 | (1.88 ± 0.1) × 104 | D2 | (2.80 ± 0.36) × 105 | (1.21 ± 0.14) × 104 |
| M3 | (2.71 ± 0.19) × 104 | D3 | (5.14 ± 0.63) × 105 | (1.49 ± 0.13) × 104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, X.; Yang, F.; Lv, G.; Qiu, Y.; Jia, X.; Hu, Q.; Zhang, J.; Yang, J.; Ouyang, X.; Gao, L.; et al. Multi-Hydrogen Bonding on Quaternized-Oligourea Receptor Facilitated Its Interaction with Bacterial Cell Membranes and DNA for Broad-Spectrum Bacteria Killing. Molecules 2024, 29, 3937. https://doi.org/10.3390/molecules29163937
Yan X, Yang F, Lv G, Qiu Y, Jia X, Hu Q, Zhang J, Yang J, Ouyang X, Gao L, et al. Multi-Hydrogen Bonding on Quaternized-Oligourea Receptor Facilitated Its Interaction with Bacterial Cell Membranes and DNA for Broad-Spectrum Bacteria Killing. Molecules. 2024; 29(16):3937. https://doi.org/10.3390/molecules29163937
Chicago/Turabian StyleYan, Xiaojin, Fan Yang, Guanghao Lv, Yuping Qiu, Xiaoying Jia, Qirong Hu, Jia Zhang, Jing Yang, Xiangyuan Ouyang, Lingyan Gao, and et al. 2024. "Multi-Hydrogen Bonding on Quaternized-Oligourea Receptor Facilitated Its Interaction with Bacterial Cell Membranes and DNA for Broad-Spectrum Bacteria Killing" Molecules 29, no. 16: 3937. https://doi.org/10.3390/molecules29163937
APA StyleYan, X., Yang, F., Lv, G., Qiu, Y., Jia, X., Hu, Q., Zhang, J., Yang, J., Ouyang, X., Gao, L., & Jia, C. (2024). Multi-Hydrogen Bonding on Quaternized-Oligourea Receptor Facilitated Its Interaction with Bacterial Cell Membranes and DNA for Broad-Spectrum Bacteria Killing. Molecules, 29(16), 3937. https://doi.org/10.3390/molecules29163937








