Fluorescence Turn-Off Ligand for Parallel G-Quadruplexes
Abstract
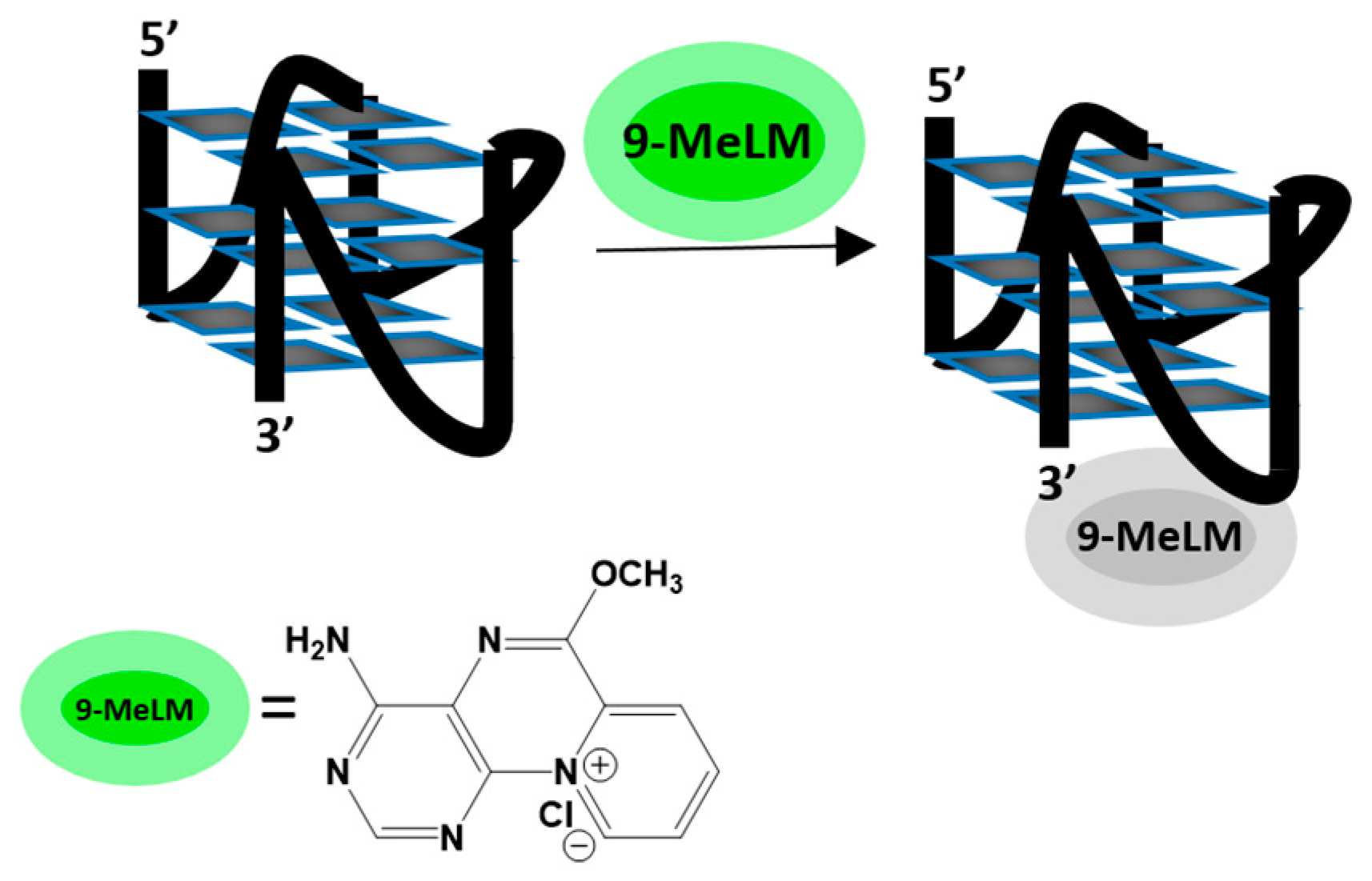
1. Introduction
2. Results and Discussion
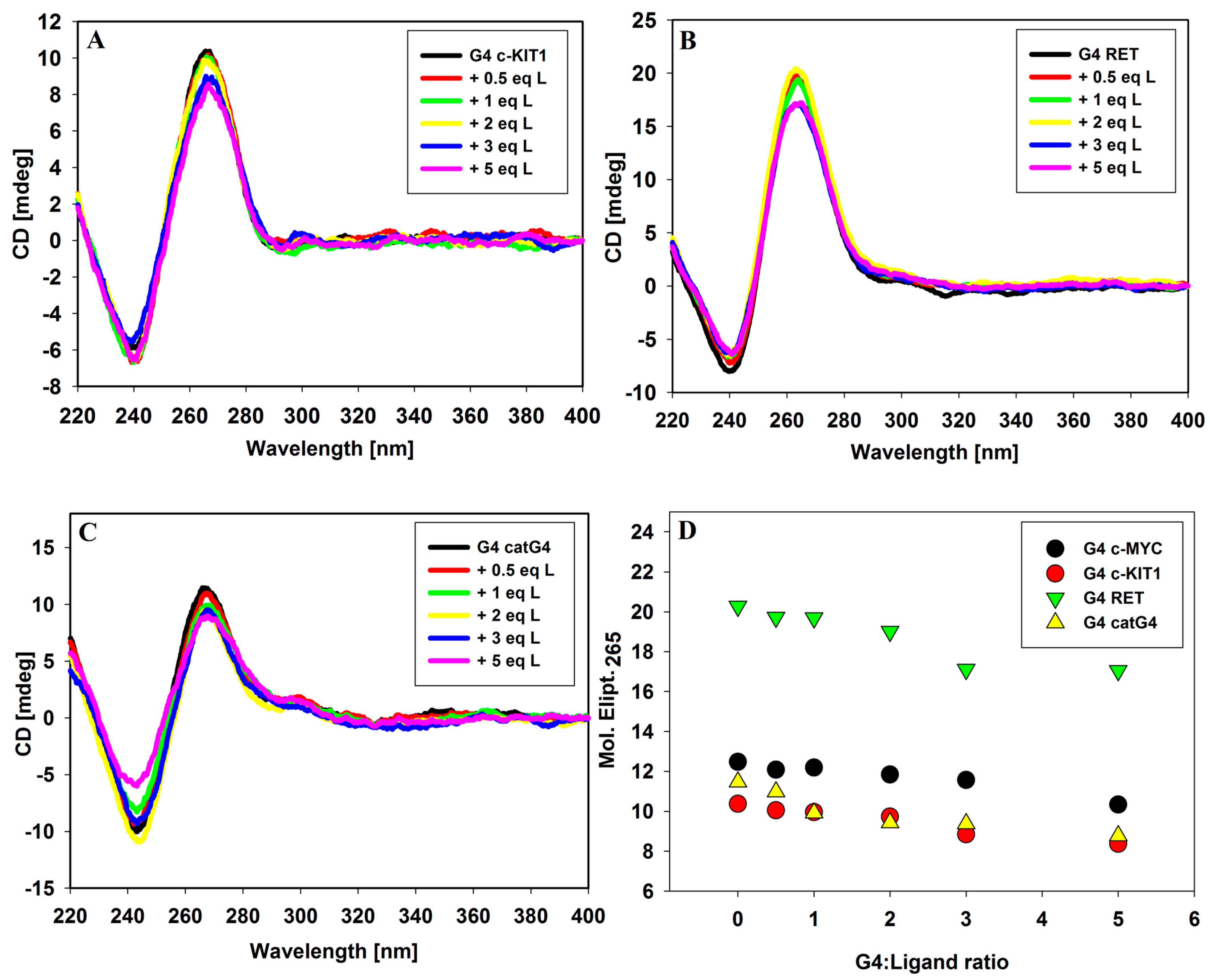
2.1. Circular Dichroism Spectroscopy
2.2. DNA Melting Studies
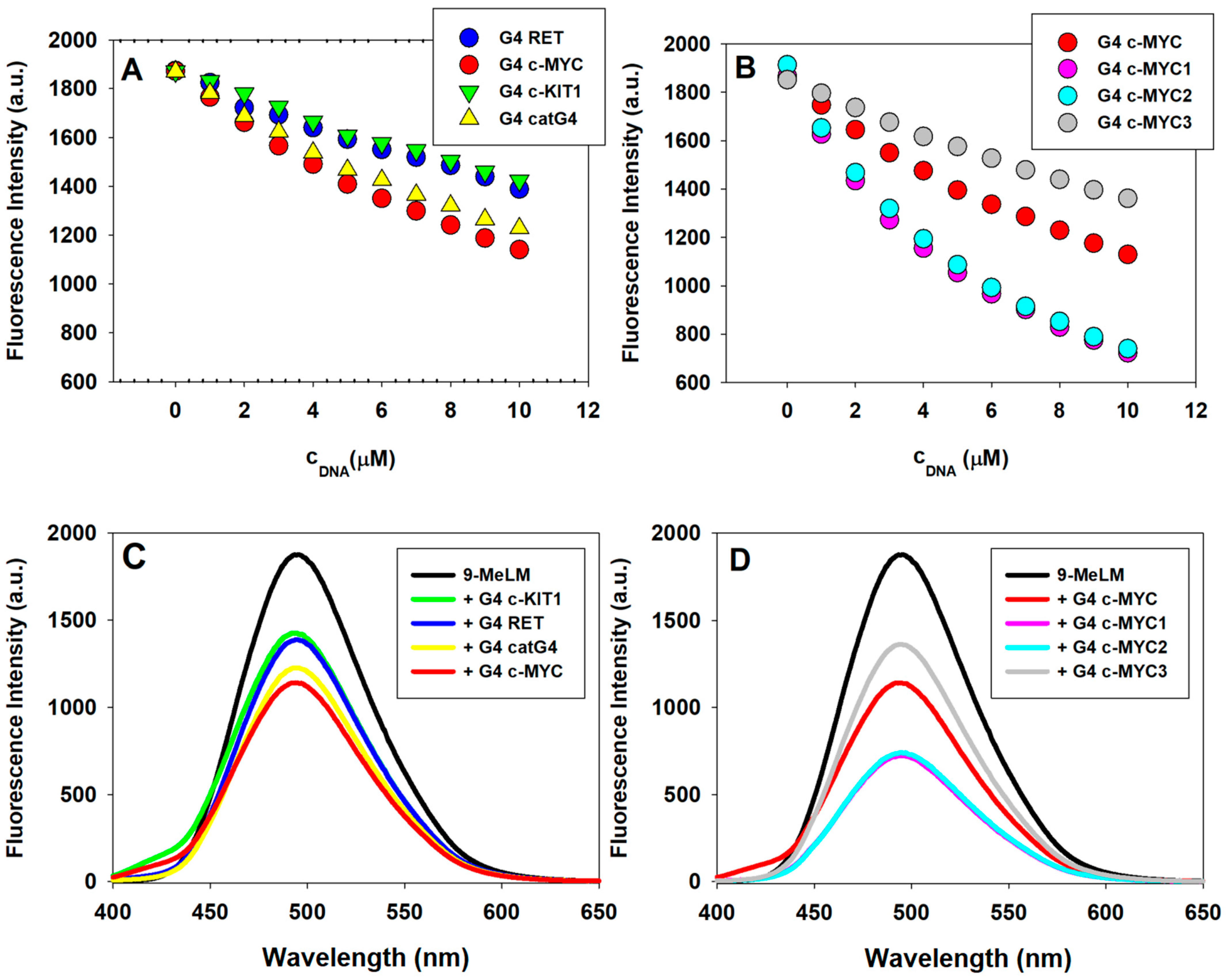
2.3. Binding Parameters via Fluorescence Spectroscopy
3. Material and Methods
3.1. Materials
3.2. Oligonucleotides and Sample Preparation
3.3. Circular Dichroism Spectroscopy (CD)
3.4. UV Melting Profiles
3.5. Fluorescence Spectroscopy
4. Summary and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [PubMed]
- Kosiol, N.; Juranek, S.; Brossart, P.; Heine, A.; Paeschke, K. G-quadruplexes: A promising target for cancer therapy. Mol. Cancer 2021, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Yang, Q.-F.; Lin, X.; Chen, D.; Wang, Z.-Y.; Chen, B.; Han, H.-Y.; Chen, H.-D.; Cai, K.-C.; Li, Q.; et al. G4LDB 2.2: A database for discovering and studying G-quadruplex and i-Motif ligands. Nucleic Acids Res. 2022, 50, D150–D160. [Google Scholar] [CrossRef]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Chem. Eur. J. 2019, 25, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef] [PubMed]
- Alessandrini, I.; Recagni, M.; Zaffaroni, N.; Folini, M. On the road to fight cancer: The potential of G-quadruplex ligands as novel therapeutic agents. Int. J. Mol. Sci. 2021, 22, 5947. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Debnath, M.; Müller, D.; Paul, R.; Das, T.; Bessi, I.; Schwalbe, H.; Dash, J. Cell penetrating thiazole peptides inhibit c-MYC expression via site-specific targeting of c-MYC G-quadruplex. Nucleic Acids Res. 2018, 46, 5355–5365. [Google Scholar] [CrossRef]
- Pandith, A.; Nagarajachari, U.; Siddappa, R.K.G.; Lee, S.; Park, C.-J.; Sannathammegowda, K.; Seo, Y.J. Loop-mediated fluorescent probes for selective discrimination of parallel and antiparallel G-Quadruplexes. Bioorg. Med. Chem. 2021, 35, 116077. [Google Scholar] [CrossRef]
- Deiana, M.; Chand, K.; Jamroskovic, J.; Obi, I.; Chorell, E.; Sabouri, N. A Light-up Logic Platform for Selective Recognition of Parallel G-Quadruplex Structures via Disaggregation-Induced Emission. Angew. Chem. Int. Ed. 2020, 59, 896–902. [Google Scholar] [CrossRef]
- Deiana, M.; Chand, K.; Jamroskovic, J.; Das, R.N.; Obi, I.; Chorell, E.; Sabouri, N. A site-specific self-assembled light-up rotor probe for selective recognition and stabilization of c-MYC G-quadruplex DNA. Nanoscale 2020, 12, 12950–12957. [Google Scholar] [CrossRef] [PubMed]
- Zuffo, M.; Guédin, A.; Leriche, E.D.; Doria, F.; Pirota, V.; Gabelica, V.; Mergny, J.L.; Freccero, M. More is not always better: Finding the right trade-off between affinity and selectivity of a G-quadruplex ligand. Nucleic Acids Res. 2018, 46, e115. [Google Scholar] [CrossRef]
- Grande, V.; Shen, C.A.; Deiana, M.; Dudek, M.; Olesiak-Banska, J.; Matczyszyn, K.; Würthner, F. Selective parallel G-quadruplex recognition by a NIR-to-NIR two-photon squaraine. Chem. Sci. 2018, 9, 8375–8381. [Google Scholar] [CrossRef] [PubMed]
- Grande, V.; Doria, F.; Freccero, M.; Würthner, F. An Aggregating Amphiphilic Squaraine: A Light-up Probe That Discriminates Parallel G-Quadruplexes. Angew. Chem. Int. Ed. 2017, 56, 7520–7524. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Gao, C.; Ding, J.; Zhang, Y.; Islam, B.; Lan, W.; Hou, H.; Deng, H.; Li, J.; Hu, Z.; et al. Selective recognition of c-MYC Pu22 G-quadruplex by a fluorescent probe. Nucleic Acids Res. 2019, 47, 2190–2204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Er, J.C.; Ghosh, K.K.; Chung, W.J.; Yoo, J.; Xu, W.; Zhao, W.; Phan, A.T.; Chang, Y.T. Discovery of a structural-element specific G-quadruplex “light-up” probe. Sci. Rep. 2015, 4, 3776. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.B.; Wu, W.B.; Hu, M.H.; Ou, T.M.; Gu, L.Q.; Tan, J.H.; Huang, Z.S. Discovery of a new fluorescent light-up probe specific to parallel G-quadruplexes. Chem. Commun. 2014, 50, 12173–12176. [Google Scholar] [CrossRef]
- Wang, L.-X.; Zhang, J.-T.; Sun, X.; Yang, D.-W.; Tang, Y.-L. Highly selective recognition of intramolecular parallel G-quadruplex using a chiral supramolecular probe. Dye Pigment. 2021, 185, 108882. [Google Scholar] [CrossRef]
- Xie, X.; Reznichenko, O.; Chaput, L.; Martin, P.; Teulade-Fichou, M.P.; Granzhan, A. Topology-Selective, Fluorescent “Light-Up” Probes for G-Quadruplex DNA Based on Photoinduced Electron Transfer. Chem.-Eur. J. 2018, 24, 12638–12651. [Google Scholar] [CrossRef]
- Largy, E.; Granzhan, A.; Hamon, F.; Verga, D.; Teulade-Fichou, M.-P. Visualizing the Quadruplex: From Fluorescent Ligands to Light-Up Probes. Quadruplex Nucleic Acids 2012, 330, 111–177. [Google Scholar] [CrossRef]
- Wenska, G.; Skalski, B.; Tomska-Foralewska, I.; Paszyc, S. Synthesis and fluorescence quenching study of the novel cationic probe derived from luminarosine. Helv. Chim. Acta 2001, 84, 3726–3734. [Google Scholar] [CrossRef]
- Nowak-Karnowska, J.; Głuszyńska, A.; Kosman, J.; Neunert, G.; Dembska, A. Interaction of 9-Methoxyluminarine with Different G-Quadruplex Topologies: Fluorescence and Circular Dichroism Studies. Int. J. Mol. Sci. 2021, 22, 10399. [Google Scholar] [CrossRef] [PubMed]
- Biver, T. Discriminating between Parallel, Anti-Parallel and Hybrid G-Quadruplexes: Mechanistic Details on Their Binding to Small Molecules. Molecules 2022, 27, 4165. [Google Scholar] [CrossRef]
- Deiana, M.; Obi, I.; Andreasson, M.; Tamilselvi, S.; Chand, K.; Chorell, E.; Sabouri, N. A Minimalistic Coumarin Turn-On Probe for Selective Recognition of Parallel G-Quadruplex DNA Structures. ACS Chem. Biol. 2021, 16, 1365–1376. [Google Scholar] [CrossRef] [PubMed]
- Gai, W.; Yang, Q.; Xiang, J.; Sun, H.; Shang, Q.; Li, Q.; Jiang, W.; Guan, A.; Zhang, H.; Tang, Y.; et al. Roles of flanking sequences in the binding between unimolecular parallel-stranded G-quadruplexes and ligands. Chin. Sci. Bull. 2013, 58, 731–740. [Google Scholar] [CrossRef][Green Version]
- Lennartsson, J.; Rönnstrand, L. Stem cell factor receptor/c-kit: From basic science to clinical implications. Physiol. Rev. 2012, 92, 1619–1649. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wu, Y.-L.; Chen, B.-J.; Zhang, W.; Tanaka, Y.; Sugiyama, H. The C-Kit Receptor-Mediated Signal Transduction and Tumor-Related Diseases. Int. J. Biol. Sci. 2013, 9, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, E.; Tran, T.; Vranic, S.; Levy, A.; Bonfil, R.D. Role and significance of c-KIT receptor tyrosine kinase in cancer: A review. Bosn. J. Basic. Med. Sci. 2022, 22, 683–698. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.V.; Burge, S.; Neidle, S.; Patel, D.J. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007, 129, 4386–4392. [Google Scholar] [CrossRef]
- Jhiang, S.M. The RET proto-oncogene in human cancers. Oncogene 2000, 19, 5590–5597. [Google Scholar] [CrossRef]
- Romei, C.; Ciampi, R.; Elisei, R. A comprehensive overview of the role of the RET proto-oncogene in thyroid carcinoma. Nat. Rev. Endocrinol. 2016, 12, 192–202. [Google Scholar] [CrossRef]
- Tong, X.; Lan, W.; Zhang, X.; Wu, H.; Liu, M.; Cao, C. Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence. Nucleic Acids Res. 2011, 39, 6753. [Google Scholar] [CrossRef]
- Kosman, J.; Juskowiak, B. Hemin/G-quadruplex structure and activity alteration induced by magnesium cations. Int. J. Biol. Macromol. 2016, 85, 555–564. [Google Scholar] [CrossRef]
- Shumayrikh, N.; Sen, D. Heme G-quadruplex DNAzymes: Conditions for maximizing their peroxidase activity. Methods Mol. Biol. 2019, 2035, 357–368. [Google Scholar] [CrossRef]
- Balagurumoorthy, P.; Brahmachari, S.K.; Mohanty, D.; Bansal, M.; Sasisekharan, V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992, 20, 4061–4067. [Google Scholar] [CrossRef]
- Masiero, S.; Trotta, R.; Pieraccini, S.; De Tito, S.; Perone, R.; Randazzo, A.; Spada, G.P. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 2010, 8, 2683–2692. [Google Scholar] [CrossRef] [PubMed]
- White, E.W.; Tanious, F.; Ismail, M.A.; Reszka, A.P.; Neidle, S.; Boykin, D.W.; Wilson, W.D. Structure-specific recognition of quadruplex DNA by organic cations: Influence of shape, substituents and charge. Biophys. Chem. 2007, 126, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Jana, J.; Mondal, S.; Bhattacharjee, P.; Sengupta, P.; Roychowdhury, T.; Saha, P.; Kundu, P.; Chatterjee, S. Chelerythrine down regulates expression of VEGFA, BCL2 and KRAS by arresting G-Quadruplex structures at their promoter regions. Sci. Rep. 2017, 7, 40706. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Pradhan, S.K.; Kar, A.; Chowdhury, S.; Dasgupta, D. Molecular basis of recognition of quadruplexes human telomere and c-myc promoter by the putative anticancer agent sanguinarine. Biochim. Biophys. Acta 2013, 1830, 4189–4201. [Google Scholar] [CrossRef]
- Bhadra, K.; Kumar, G.S. Interaction of berberine, palmatine, coralyne, and sanguinarine to quadruplex DNA: A comparative spectroscopic and calorimetric study. Biochim. Biophys. Acta 2011, 1810, 485–496. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef]
- Rachwal, P.A.; Fox, K.R. Quadruplex melting. Methods 2007, 43, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: Boston, MA, USA, 2006; pp. 1–954. [Google Scholar]
- Lavrik, N.L.; Bazhin, N.M. On the Question of Defining the Association Constants by the Method of Fluorescence Quenching. Am. J. Anal. Chem. 2014, 5, 1065–1068. [Google Scholar] [CrossRef]
- Tataurov, A.V.; You, Y.; Owczarzy, R. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 2008, 133, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.-L.; Phan, A.T.; Lacroix, L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef]
- Viglasky, V.; Bauer, L.; Tluckova, K. Structural features of intra-and intermolecular G-quadruplexes derived from telomeric repeats. Biochemistry 2010, 49, 2110–2120. [Google Scholar] [CrossRef] [PubMed]

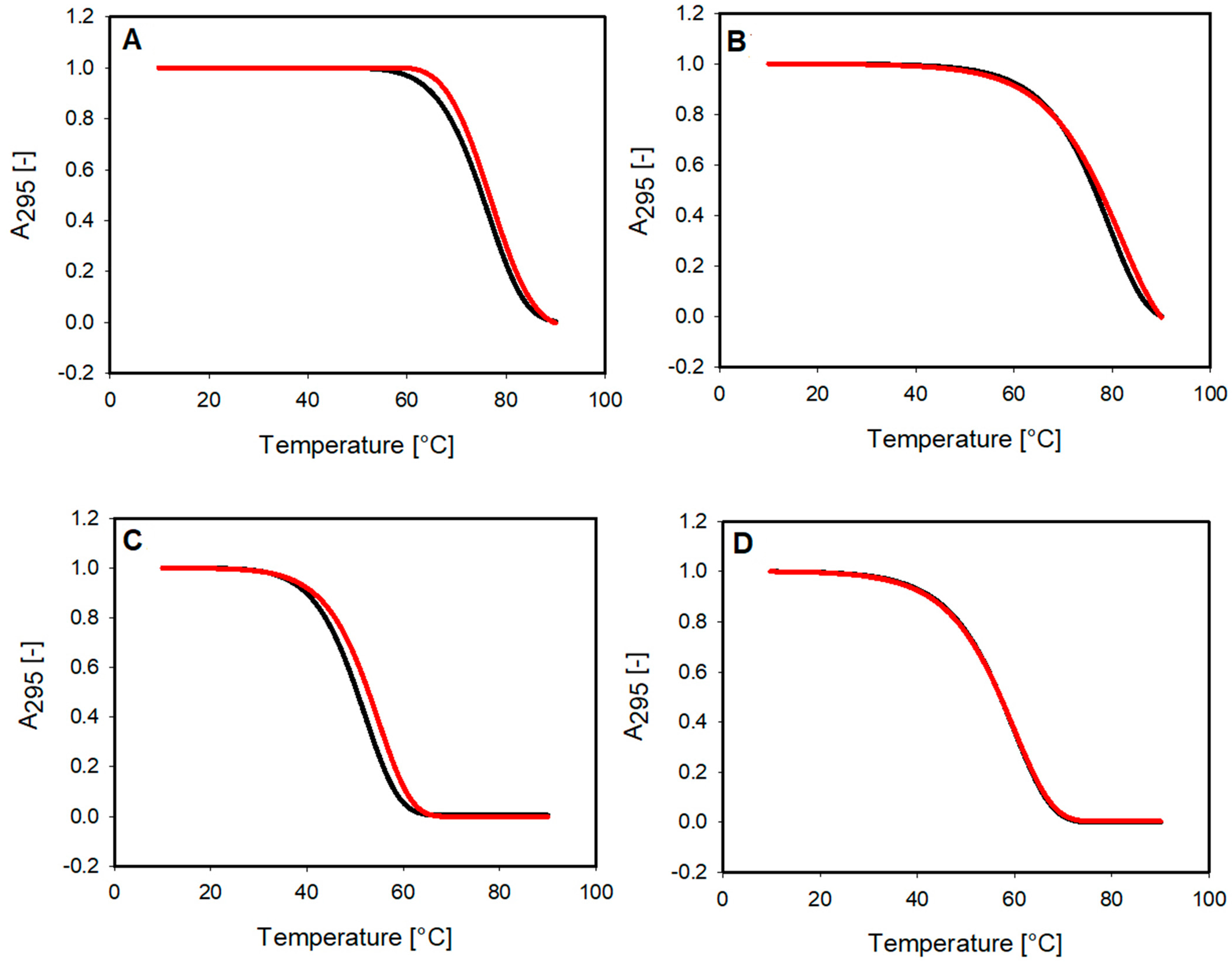
| G4 DNA | Tm [°C] a | Tm G4 + L [°C] b |
|---|---|---|
| c-MYC | 79.10 ± 0.10 | 80.6 ± 1.7 |
| c-KIT1 | 52.9 ± 1.0 | 54.40 ± 0.60 |
| RET | 75.30 ± 0.80 | 76.4 ± 1.2 |
| catG4 | 63.30 ± 0.10 | 62.80 ± 0.30 |
| G4 | K (×104 M−1) | kq (×1012 M−1s−1) | R |
|---|---|---|---|
| RET | 3.0 ± 0.3 | 3 | 0.994 |
| c-KIT1 | 3.7 ± 0.6 | 4 | 0.990 |
| catG4 | 5.3 ± 0.1 | 5 | 0.999 |
| c-MYC | 5.9 ± 0.7 | 6 | 0.999 |
| c-MYC1 | 20 ± 4 | 20 | 0.997 |
| c-MYC2 | 17 ± 3 | 17 | 0.999 |
| c-MYC3 | 4.5 ± 0.5 | 5 | 0.990 |
| G4 | Sequence 5′-3′ | PDB ID |
|---|---|---|
| RET | GGGGCGGGGCGGGGCGGGGT | 2L88 |
| c-KIT1 | AGGGAGGGCGCTGGGAGGAGGG | 2O3M |
| catG4 | TGGGTAGGGCGGGTTGGGAAA | - |
| c-MYC | TGAGGGTGGGTAGGGTGGGTAA | 2L7V |
| c-MYC1 | GGGTGGGTAGGGTGGG | - |
| c-MYC2 | TAAGGGTGGGTAGGGTGGG | - |
| c-MYC3 | GGGTGGGTAGGGTGGGTAA | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowak-Karnowska, J.; Głuszyńska, A.; Kosman, J.; Dembska, A. Fluorescence Turn-Off Ligand for Parallel G-Quadruplexes. Molecules 2024, 29, 3907. https://doi.org/10.3390/molecules29163907
Nowak-Karnowska J, Głuszyńska A, Kosman J, Dembska A. Fluorescence Turn-Off Ligand for Parallel G-Quadruplexes. Molecules. 2024; 29(16):3907. https://doi.org/10.3390/molecules29163907
Chicago/Turabian StyleNowak-Karnowska, Joanna, Agata Głuszyńska, Joanna Kosman, and Anna Dembska. 2024. "Fluorescence Turn-Off Ligand for Parallel G-Quadruplexes" Molecules 29, no. 16: 3907. https://doi.org/10.3390/molecules29163907
APA StyleNowak-Karnowska, J., Głuszyńska, A., Kosman, J., & Dembska, A. (2024). Fluorescence Turn-Off Ligand for Parallel G-Quadruplexes. Molecules, 29(16), 3907. https://doi.org/10.3390/molecules29163907








