DPPA as a Potential Cell Membrane Component Responsible for Binding Amyloidogenic Protein Human Cystatin C
Abstract
1. Introduction
2. Results and Discussion
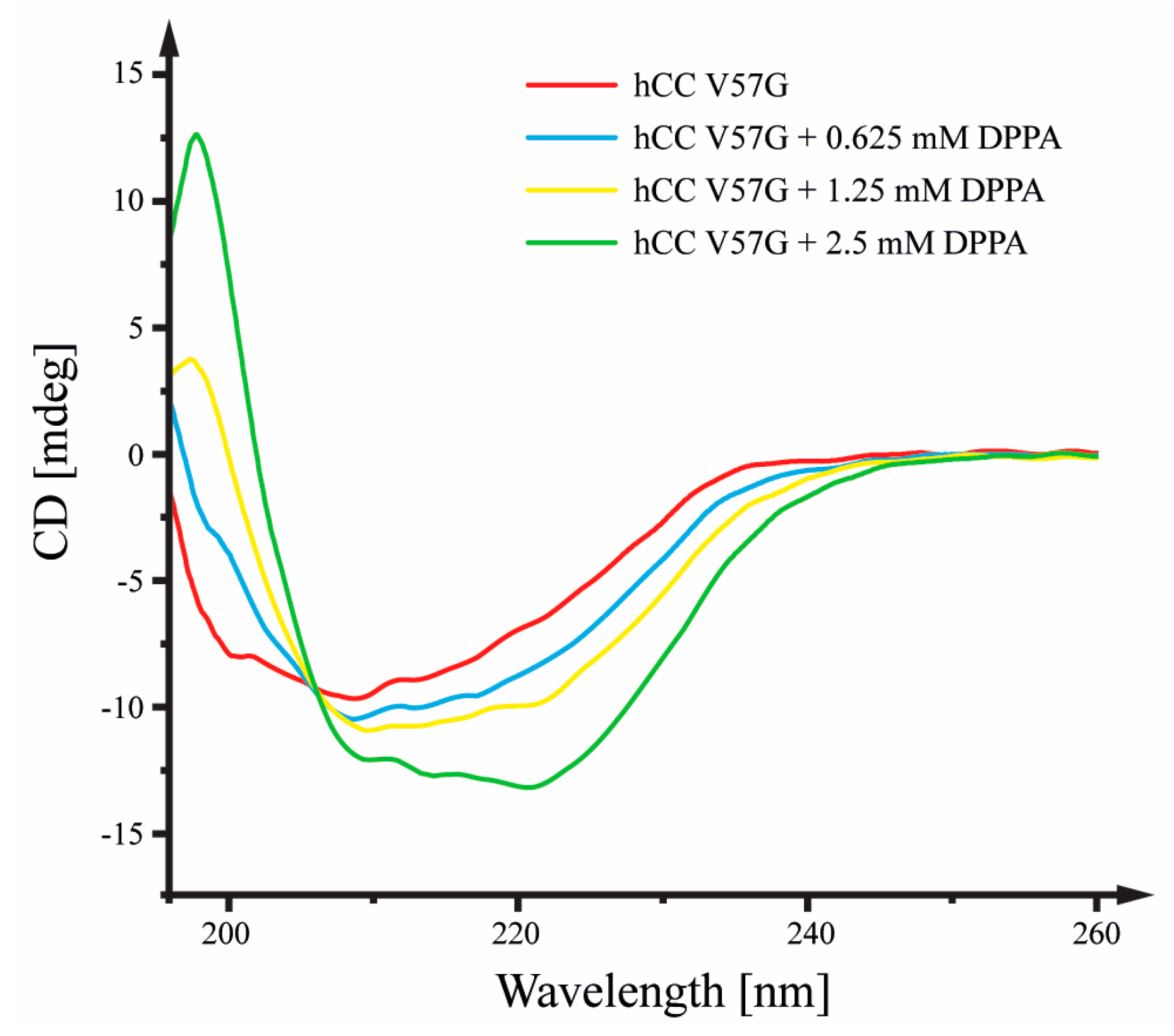
2.1. Circular Dichroism
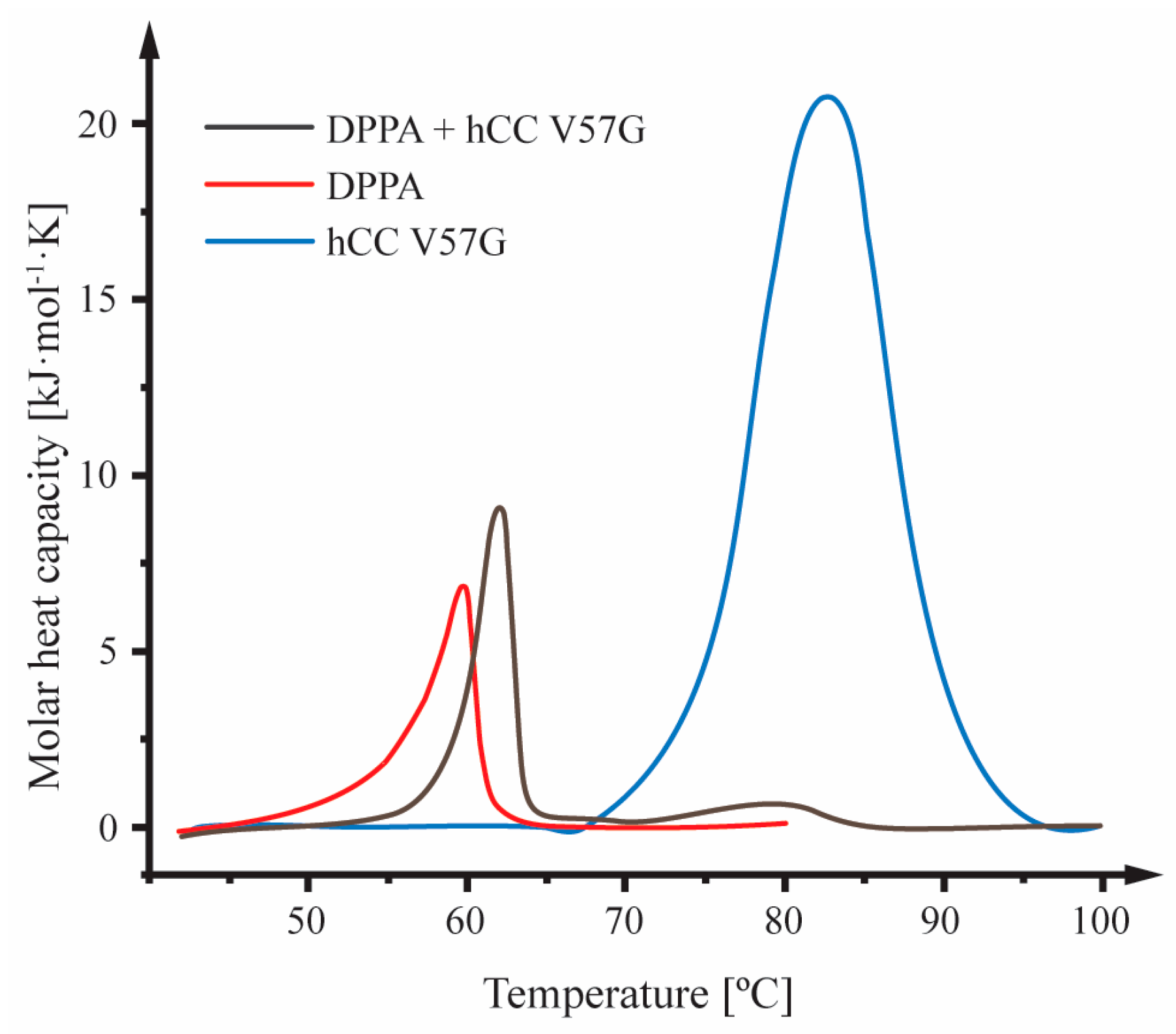
2.2. Differential Scanning Calorimetry
2.3. Molecular Dynamic Processes Occurring in the hCC V57G Protein in the Presence of DPPA Phospholipid Bilayer
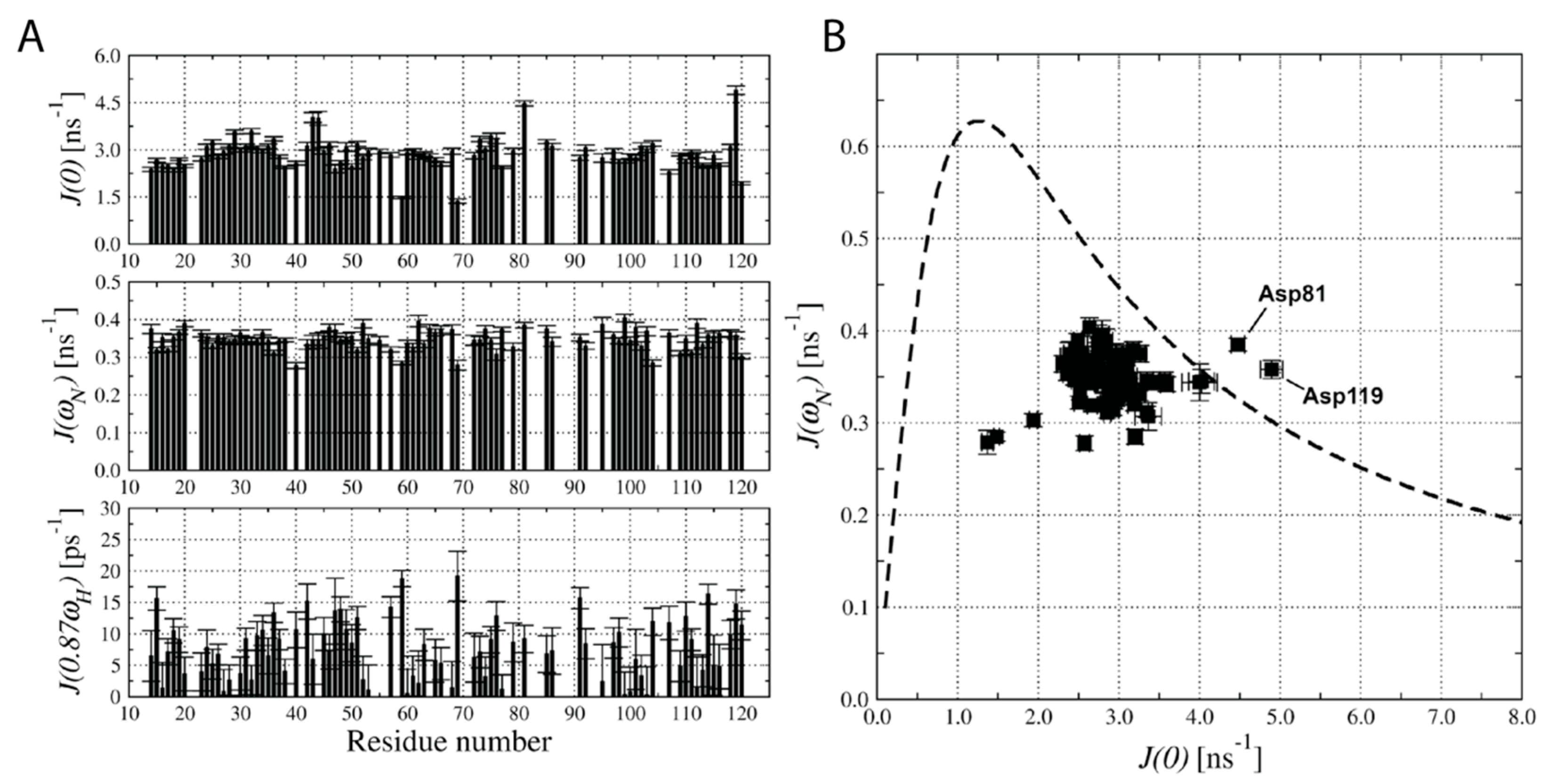
2.4. Analysis of 15N Relaxation Data with Spectral Density Mapping Approach
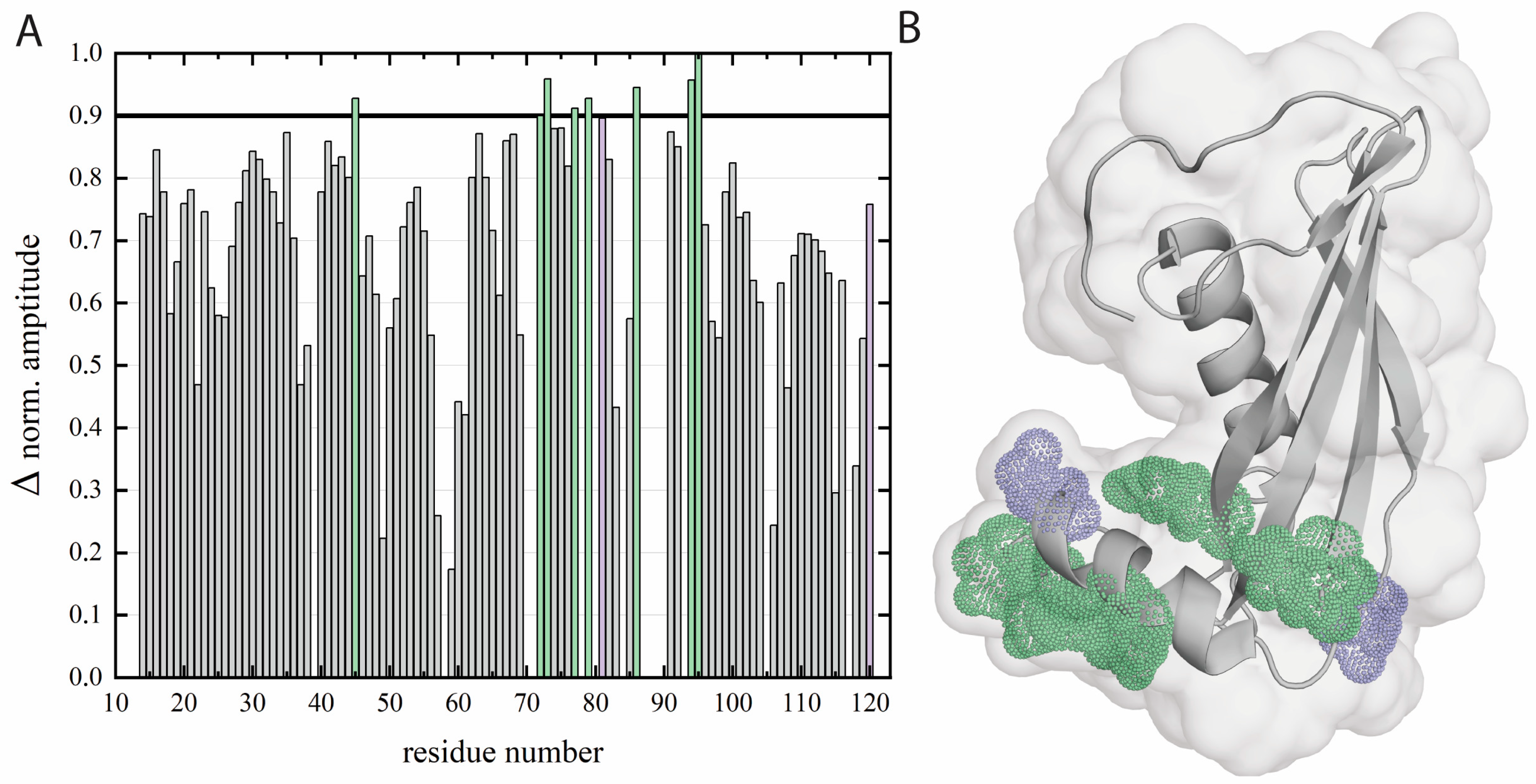
2.5. Determination of Amino Acid Residues Buried in the DPPA Phospholipid Bilayer—Paramagnetic Relaxation Enhancement Approach
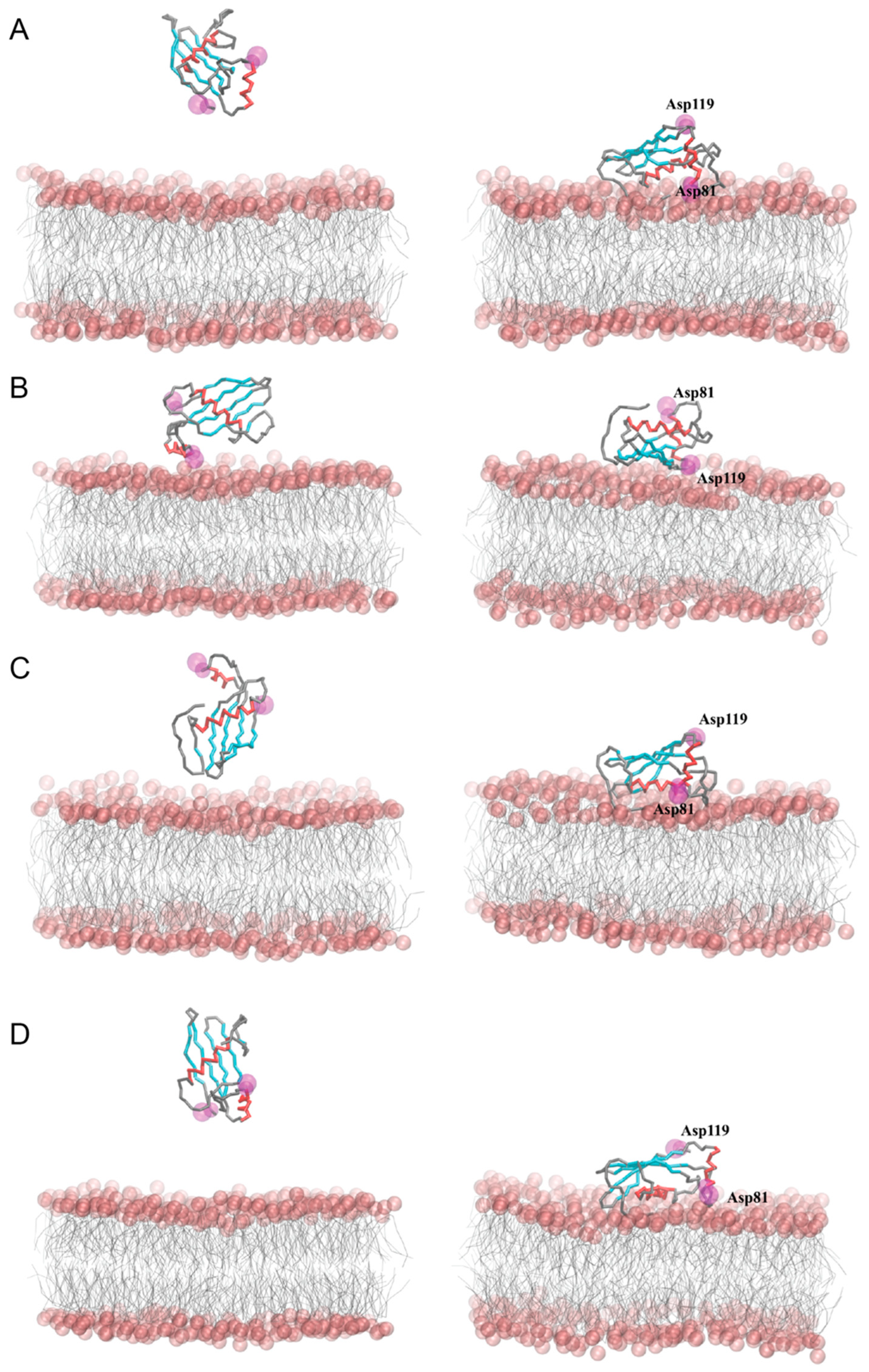
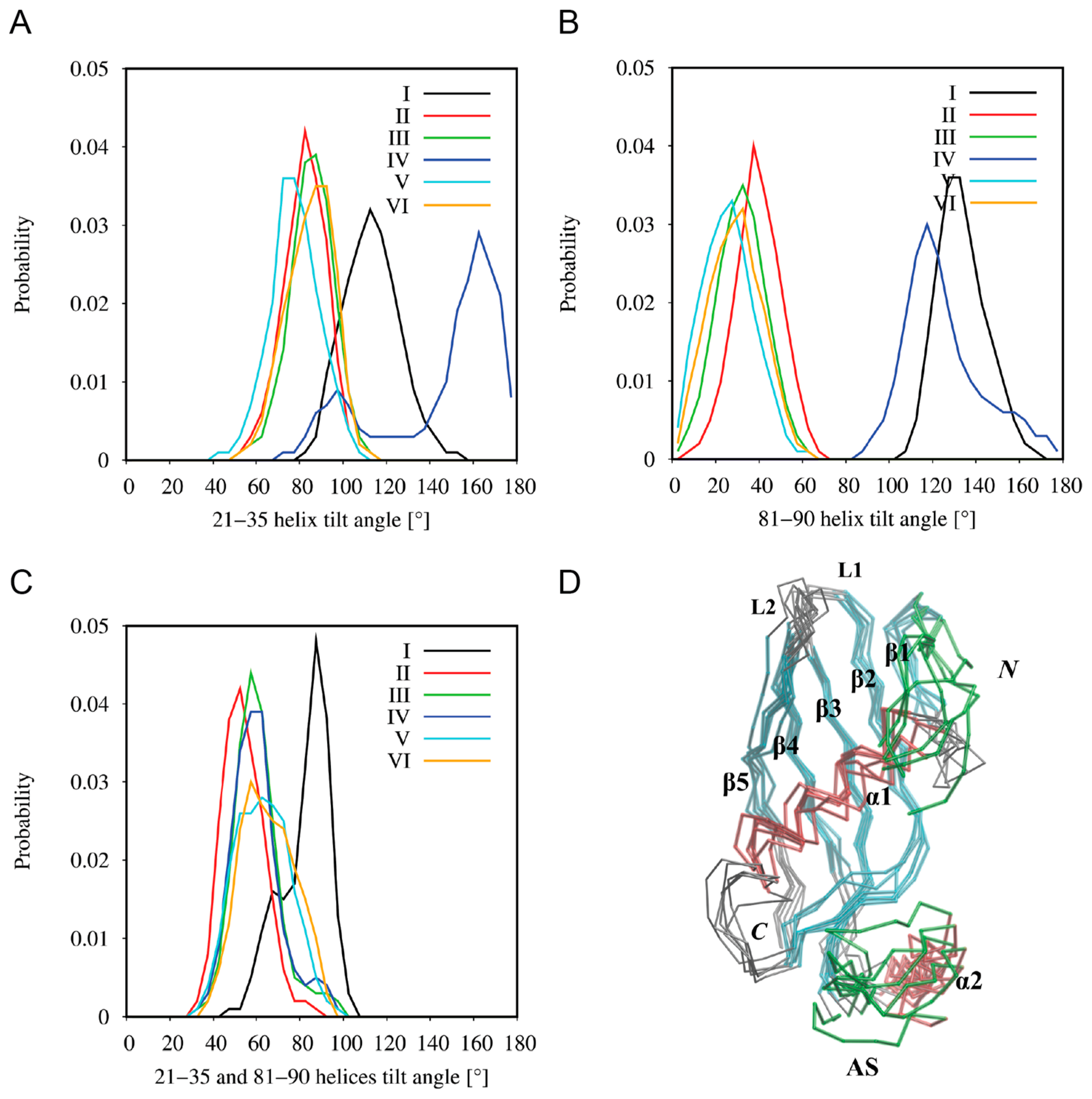
2.6. Coarse-Grained Molecular Dynamics Simulations
2.7. Summary
3. Materials and Methods
3.1. Protein Expression
3.2. Circular Dichroism Measurements
3.3. Differential Scanning Calorimetry Measurements
3.4. NMR Measurements
3.5. 15N Relaxation Measurements
3.6. 15N Relaxation Data Analysis
3.7. Determination of Amino Acid Residues Buried in the DPPA Phospholipid Bilayer
3.8. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296, Correction in Nat. Rev. Mol. Cell Biol. 2019, 20, 715. [Google Scholar] [CrossRef] [PubMed]
- Gaspard, G.J.; McMaster, C.R. Cardiolipin metabolism and its causal role in the etiology of the inherited cardiomyopathy Barth syndrome. Chem. Phys. Lipids 2015, 193, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bissig, C.; Gruenberg, J. Lipid sorting and multivesicular endosome biogenesis. Cold Spring Harb. Perspect. Biol. 2013, 5, a016816. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, J.; Umeda, M.; Sims, P.J.; Nagata, S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature 2010, 468, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, J.; Denning, D.P.; Imanishi, E.; Horvitz, H.R.; Nagata, S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 2013, 341, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.; Adams, K. The Cell: A Molecular Approach, 9th ed.; Oxford University Press: New York, NY, USA, 2023. [Google Scholar]
- Butterfield, S.M.; Lashuel, H.A. Amyloidogenic protein–membrane interactions: Mechanistic insight from model systems. Angew. Chem. Int. Ed. 2010, 49, 5628–5654. [Google Scholar] [CrossRef] [PubMed]
- Sciacca, M.F.; Tempra, C.; Scollo, F.; Milardi, D.; La Rosa, C. Amyloid growth and membrane damage: Current themes and emerging perspectives from theory and experiments on Aβ and hIAPP. Biochim. Biophys. Acta (BBA) Biomembr. 2018, 1860, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Panahi, A.V.S.; Hultman, P.; Öllinger, K.; Westermark, G.T.; Lundmark, K. Lipid membranes accelerate amyloid formation in the mouse model of AA amyloidosis. Amyloid 2019, 26, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Terakawa, M.S.; Lin, Y.; Kinoshita, M.; Kanemura, S.; Itoh, D.; Sugiki, T.; Okumura, M.; Ramamoorthy, A.; Lee, Y.-H. Impact of membrane curvature on amyloid aggregation. Biochim. Biophys. Acta (BBA) Biomembr. 2018, 1860, 1741–1764. [Google Scholar] [CrossRef]
- Jurczak, P.; Zhukov, I.; Orlikowska, M.; Czaplewska, P.; Sikorska, E. Monitoring the interactions between POPG phospholipid bilayer and amyloid-forming protein human cystatin C. Does the bilayer influence the oligomeric state and structure of the protein? Biochim. Biophys. Acta (BBA) Biomembr. 2024, 1866, 184285. [Google Scholar] [CrossRef]
- Jurczak, P.; Szutkowski, K.; Lach, S.; Jurga, S.; Czaplewska, P.; Szymanska, A.; Zhukov, I. DMPC Phospholipid Bilayer as a Potential Interface for Human Cystatin C Oligomerization: Analysis of Protein-Liposome Interactions Using NMR Spectroscopy. Membranes 2020, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.D.; Wong, S.S.; Lieber, C.M.; Lansbury, P.T. Observation of metastable Aβ amyloid protofibrils by atomic force microscopy. Chem. Biol. 1997, 4, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Ionescu-Zanetti, C.; Khurana, R.; Gillespie, J.R.; Petrick, J.S.; Trabachino, L.C.; Minert, L.J.; Carter, S.A.; Fink, A.L. Monitoring the assembly of Ig light-chain amyloid fibrils by atomic force microscopy. Proc. Natl. Acad. Sci. USA 1999, 96, 13175–13179. [Google Scholar] [CrossRef] [PubMed]
- Wahlbom, M.; Wang, X.; Lindström, V.; Carlemalm, E.; Jaskolski, M.; Grubb, A. Fibrillogenic oligomers of human cystatin C are formed by propagated domain swapping. J. Biol. Chem. 2007, 282, 18318–18326. [Google Scholar] [CrossRef] [PubMed]
- Chrabąszczewska, M.; Sieradzan, A.K.; Rodziewicz-Motowidło, S.; Grubb, A.; Dobson, C.M.; Kumita, J.R.; Kozak, M. Structural Characterization of Covalently Stabilized Human Cystatin C Oligomers. Int. J. Mol. Sci. 2020, 21, 5860. [Google Scholar] [CrossRef] [PubMed]
- Abrahamson, M.; Barrett, A.J.; Salvesen, G.; Grubb, A. Isolation of six cysteine proteinase inhibitors from human urine. Their physicochemical and enzyme kinetic properties and concentrations in biological fluids. J. Biol. Chem. 1986, 261, 11282–11289. [Google Scholar] [CrossRef] [PubMed]
- Szymańska, A.; Radulska, A.; Czaplewska, P.; Grubb, A.; Grzonka, Z.; Rodziewicz-Motowidło, S. Governing the monomer-dimer ratio of human cystatin c by single amino acid substitution in the hinge region. Acta Biochim. Pol. 2009, 56, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Kaeser, S.A.; Herzig, M.C.; Coomaraswamy, J.; Kilger, E.; Selenica, M.-L.; Winkler, D.T.; Staufenbiel, M.; Levy, E.; Grubb, A.; Jucker, M. Cystatin C modulates cerebral β-amyloidosis. Nat. Genet. 2007, 39, 1437–1439. [Google Scholar] [CrossRef] [PubMed]
- Żyła, A.; Martel, A.; Jurczak, P.; Moliński, A.; Szymańska, A.; Kozak, M. Human cystatin C induces the disaggregation process of selected amyloid beta peptides: A structural and kinetic view. Sci. Rep. 2023, 13, 20833. [Google Scholar] [CrossRef]
- Pérez-González, R.; Sahoo, S.; Gauthier, S.A.; Kim, Y.; Li, M.; Kumar, A.; Pawlik, M.; Benussi, L.; Ghidoni, R.; Levy, E. Neuroprotection mediated by cystatin C-loaded extracellular vesicles. Sci. Rep. 2019, 9, 11104. [Google Scholar] [CrossRef]
- Taube, M.; Pietralik, Z.; Szymanska, A.; Szutkowski, K.; Clemens, D.; Grubb, A.; Kozak, M. The domain swapping of human cystatin C induced by synchrotron radiation. Sci. Rep. 2019, 9, 8548. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowska, D.; Taube, M.; Rucińska, K.; Maksim, J.; Kozak, M. Oligomerization of Human Cystatin C—An Amyloidogenic Protein: An Analysis of Small Oligomeric Subspecies. Int. J. Mol. Sci. 2022, 23, 13441. [Google Scholar] [CrossRef] [PubMed]
- Żygowska, J.; Orlikowska, M.; Zhukov, I.; Bal, W.; Szymańska, A. Copper interaction with cystatin C: Effects on protein structure and oligomerization. FEBS J. 2024, 291, 1974–1991. [Google Scholar] [CrossRef] [PubMed]
- Ekiel, I.; Abrahamson, M. Folding-related dimerization of human cystatin C. J. Biol. Chem. 1996, 271, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Ayscough, S.E.; Clifton, L.A.; Skoda, M.W.; Titmuss, S. Suspended phospholipid bilayers: A new biological membrane mimetic. J. Colloid Interface Sci. 2023, 633, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Mukundan, H.; Price, D.N.; Goertz, M.; Parthasarathi, R.; Montaño, G.A.; Kumar, S.; Scholfield, M.R.; Anderson, A.S.; Gnanakaran, S.; Iyer, S.; et al. Understanding the interaction of Lipoarabinomannan with membrane mimetic architectures. Tuberculosis 2012, 92, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Trewby, W.; Faraudo, J.; Voïtchovsky, K. Long-lived ionic nano-domains can modulate the stiffness of soft interfaces. Nanoscale 2019, 11, 4376–4384. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.J.; Loewen, C.J. Putting the pH into phosphatidic acid signaling. BMC Biol. 2011, 9, 85. [Google Scholar] [CrossRef]
- Young, B.P.; Shin, J.J.H.; Orij, R.; Chao, J.T.; Li, S.C.; Guan, X.L.; Khong, A.; Jan, E.; Wenk, M.R.; Prinz, W.A.; et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science 2010, 329, 1085–1088. [Google Scholar] [CrossRef]
- Kulig, W.; Korolainen, H.; Zatorska, M.; Kwolek, U.; Wydro, P.; Kepczynski, M.; Róg, T. Complex Behavior of Phosphatidylcholine–Phosphatidic Acid Bilayers and Monolayers: Effect of Acyl Chain Unsaturation. Langmuir 2019, 35, 5944–5956. [Google Scholar] [CrossRef]
- Szymańska, A.; Jankowska, E.; Orlikowska, M.; Behrendt, I.; Czaplewska, P.; Rodziewicz-Motowidło, S. Influence of point mutations on the stability, dimerization, and oligomerization of human cystatin C and its L68Q variant. Front. Mol. Neurosci. 2012, 5, 27530. [Google Scholar] [CrossRef]
- Orlikowska, M.; Szymańska, A.; Borek, D.; Otwinowski, Z.; Skowron, P.; Jankowska, E. Structural characterization of V57D and V57P mutants of human cystatin C, an amyloidogenic protein. Acta Crystallogr. Sect. D Struct. Biol. 2013, 69 Pt 4, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Orlikowska, M.; Jankowska, E.; Kołodziejczyk, R.; Jaskólski, M.; Szymańska, A. Hinge-loop mutation can be used to control 3D domain swapping and amyloidogenesis of human cystatin C. J. Struct. Biol. 2011, 173, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Maszota-Zieleniak, M.; Jurczak, P.; Orlikowska, M.; Zhukov, I.; Borek, D.; Otwinowski, Z.; Skowron, P.; Pietralik, Z.; Kozak, M.; Szymańska, A.; et al. NMR and crystallographic structural studies of the extremely stable monomeric variant of human cystatin C with single amino acid substitution. FEBS J. 2020, 287, 361–376. [Google Scholar] [CrossRef] [PubMed]
- Garidel, P.; Johann, C.; Blume, A. Non-ideal mixing and fluid–fluid immiscibility in phosphatidic acid–phosphatidylethanolamine mixed bilayers. Eur. Biophys. J. 2011, 40, 891–905. [Google Scholar] [CrossRef] [PubMed]
- Jankowska, E.; Wiczk, W.; Grzonka, Z. Thermal and guanidine hydrochloride-induced denaturation of human cystatin C. Eur. Biophys. J. 2004, 33, 454–461. [Google Scholar] [CrossRef]
- Heimburg, T. Thermal Biophysics of Membranes; Wiley-VCH Verlag: Weinheim, Germany, 2007. [Google Scholar]
- Pérez-Isidoro, R.; Ruiz-Suárez, J. Thermal behavior of a lipid-protein membrane model and the effects produced by anesthetics and neurotransmitters. Biochim. Biophys. Acta (BBA) Biomembr. 2020, 1862, 183099. [Google Scholar] [CrossRef] [PubMed]
- Volmer, R.; Ron, D. Lipid-dependent regulation of the unfolded protein response. Curr. Opin. Cell Biol. 2015, 33, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yoo, S.D.; Li, L.; Fang, L.; Wen, Z.; Li, T. Formulation and characterization of boanmycin-loaded liposomes prepared by pH gradient experimental design. Drug Deliv. 2012, 19, 90–101. [Google Scholar] [CrossRef]
- Palsdottir, H.; Hunte, C. Lipids in membrane protein structures. Biochim. Biophys. Acta (BBA) Biomembr. 2004, 1666, 2–18. [Google Scholar] [CrossRef]
- Neunert, G.; Tomaszewska-Gras, J.; Baj, A.; Gauza-Włodarczyk, M.; Witkowski, S.; Polewski, K. Phase Transitions and Structural Changes in DPPC Liposomes Induced by a 1-Carba-Alpha-Tocopherol Analogue. Molecules 2021, 26, 2851. [Google Scholar] [CrossRef] [PubMed]
- Watson, H. Biological membranes. Essays Biochem. 2015, 59, 43–69. [Google Scholar] [CrossRef] [PubMed]
- Escribá, P.V.; González-Ros, J.M.; Goñi, F.M.; Kinnunen, P.K.J.; Vigh, L.; Sánchez-Magraner, L.; Fernández, A.M.; Busquets, X.; Horváth, I.; Barceló-Coblijn, G. Membranes: A meeting point for lipids, proteins and therapies. J. Cell. Mol. Med. 2008, 12, 829–875. [Google Scholar] [CrossRef] [PubMed]
- Farrow, N.A.; Muhandiram, R.; Singer, A.U.; Pascal, S.M.; Kay, C.M.; Gish, G.; Shoelson, S.E.; Pawson, T.; Forman-Kay, J.D.; Kay, L.E. Backbone Dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR Relaxation. Biochemistry 1994, 33, 5984–6003. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Ng, W.J.; Bhattacharjya, S. NMR structure and localization of the host defense antimicrobial peptide thanatin in zwitterionic dodecylphosphocholine micelle: Implications in antimicrobial activity. Biochim. Biophys. Acta (BBA) Biomembr. 2020, 1862, 183432. [Google Scholar] [CrossRef] [PubMed]
- Khvedelidze, M.; Mdzinarashvili, T.; Shekiladze, E.; Schneider, M.; Moersdorf, D.; Bernhardt, I. Structure of drug delivery DPPA and DPPC liposomes with ligands and their permeability through cells. J. Liposome Res. 2015, 25, 20–31. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef]
- D’auvergne, E.J.; Gooley, P.R. Optimisation of NMR dynamic models I. Minimisation algorithms and their performance within the model-free and Brownian rotational diffusion spaces. J. Biomol. NMR 2008, 40, 107–119. [Google Scholar] [CrossRef]
- Mandel, A.M.; Akke, M.; Palmer, I.A.G. Backbone dynamics of ribonuclease HI: Correlations with structure and function in an active enzyme. J. Mol. Biol. 1995, 246, 144–163. [Google Scholar] [CrossRef]
- Souza, P.C.T.; Alessandri, R.; Barnoud, J.; Thallmair, S.; Faustino, I.; Grünewald, F.; Patmanidis, I.; Abdizadeh, H.; Bruininks, B.M.H.; Wassenaar, T.A.; et al. Martini 3: A general purpose force field for coarse-grained molecular dynamics. Nat. Methods 2021, 18, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks, C.L., III; MacKerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Gapsys, V.; de Groot, B.L.; Briones, R. Computational analysis of local membrane properties. J. Comput. Mol. Des. 2013, 27, 845–858. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]


| System | ΔH [kJ/mol] | ΔS [J/mol K] | Tm [°C] | Td [°C] | ΔT1/2 [°C] a |
|---|---|---|---|---|---|
| DPPA liposomes | 32.8 b | 98.5 b | 59.8 | - | 3.50 |
| DPPA liposomes | 25.8 b | 76.9 b | 62.1 | - | 2.68 |
| with hCC V57G | 4.11 c | 11.6 c | - | 80.5 | 7.00 |
| hCC V57G solution | 281.9 c | 792.5 c | - | 82.6 | 10.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhukov, I.; Sikorska, E.; Orlikowska, M.; Górniewicz-Lorens, M.; Kepczynski, M.; Jurczak, P. DPPA as a Potential Cell Membrane Component Responsible for Binding Amyloidogenic Protein Human Cystatin C. Molecules 2024, 29, 3446. https://doi.org/10.3390/molecules29153446
Zhukov I, Sikorska E, Orlikowska M, Górniewicz-Lorens M, Kepczynski M, Jurczak P. DPPA as a Potential Cell Membrane Component Responsible for Binding Amyloidogenic Protein Human Cystatin C. Molecules. 2024; 29(15):3446. https://doi.org/10.3390/molecules29153446
Chicago/Turabian StyleZhukov, Igor, Emilia Sikorska, Marta Orlikowska, Magdalena Górniewicz-Lorens, Mariusz Kepczynski, and Przemyslaw Jurczak. 2024. "DPPA as a Potential Cell Membrane Component Responsible for Binding Amyloidogenic Protein Human Cystatin C" Molecules 29, no. 15: 3446. https://doi.org/10.3390/molecules29153446
APA StyleZhukov, I., Sikorska, E., Orlikowska, M., Górniewicz-Lorens, M., Kepczynski, M., & Jurczak, P. (2024). DPPA as a Potential Cell Membrane Component Responsible for Binding Amyloidogenic Protein Human Cystatin C. Molecules, 29(15), 3446. https://doi.org/10.3390/molecules29153446








