Cytostatic Bacterial Metabolites Interfere with 5-Fluorouracil, Doxorubicin and Paclitaxel Efficiency in 4T1 Breast Cancer Cells
Abstract
1. Introduction
2. Results
2.1. General Consideratons
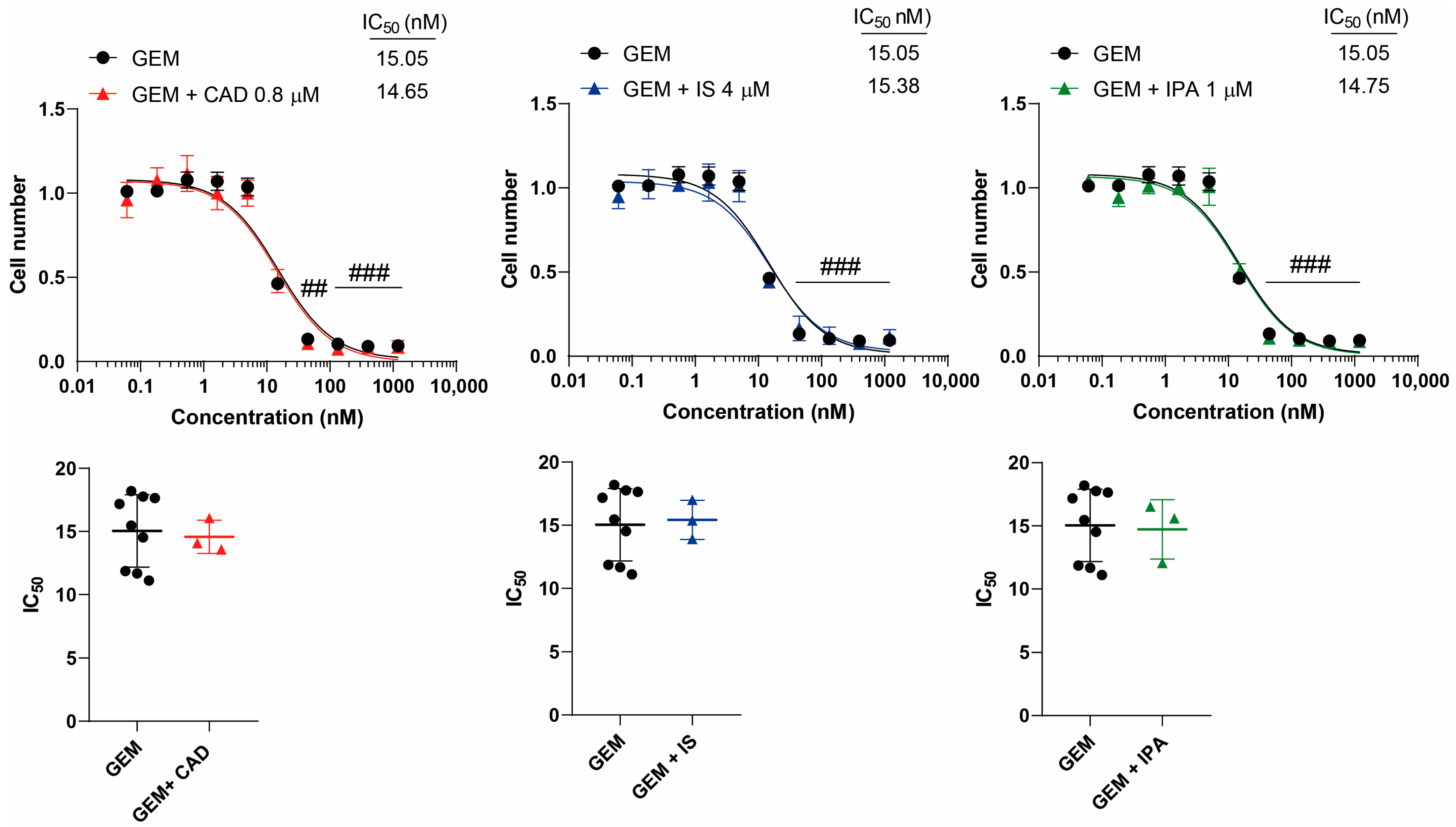
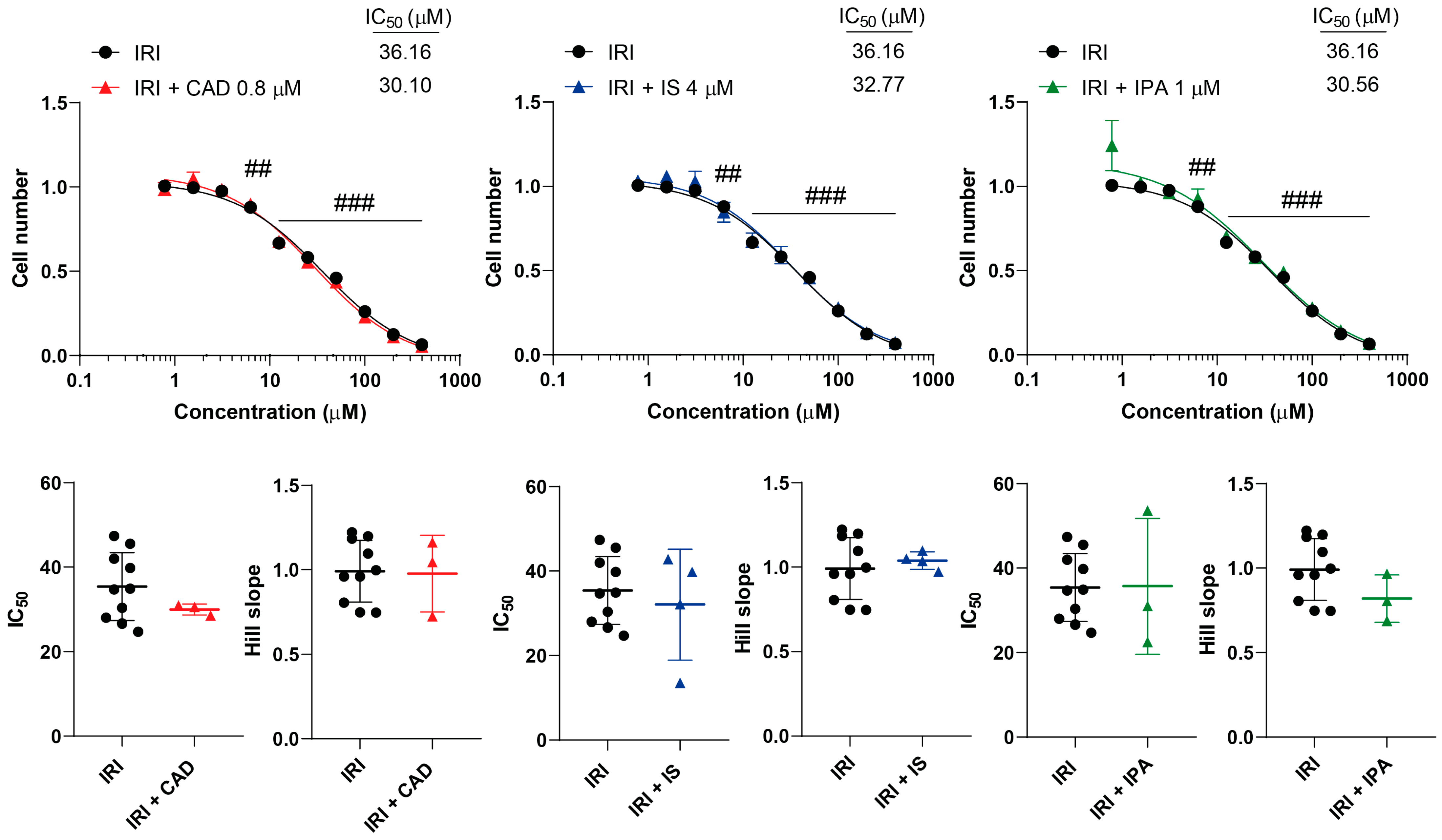
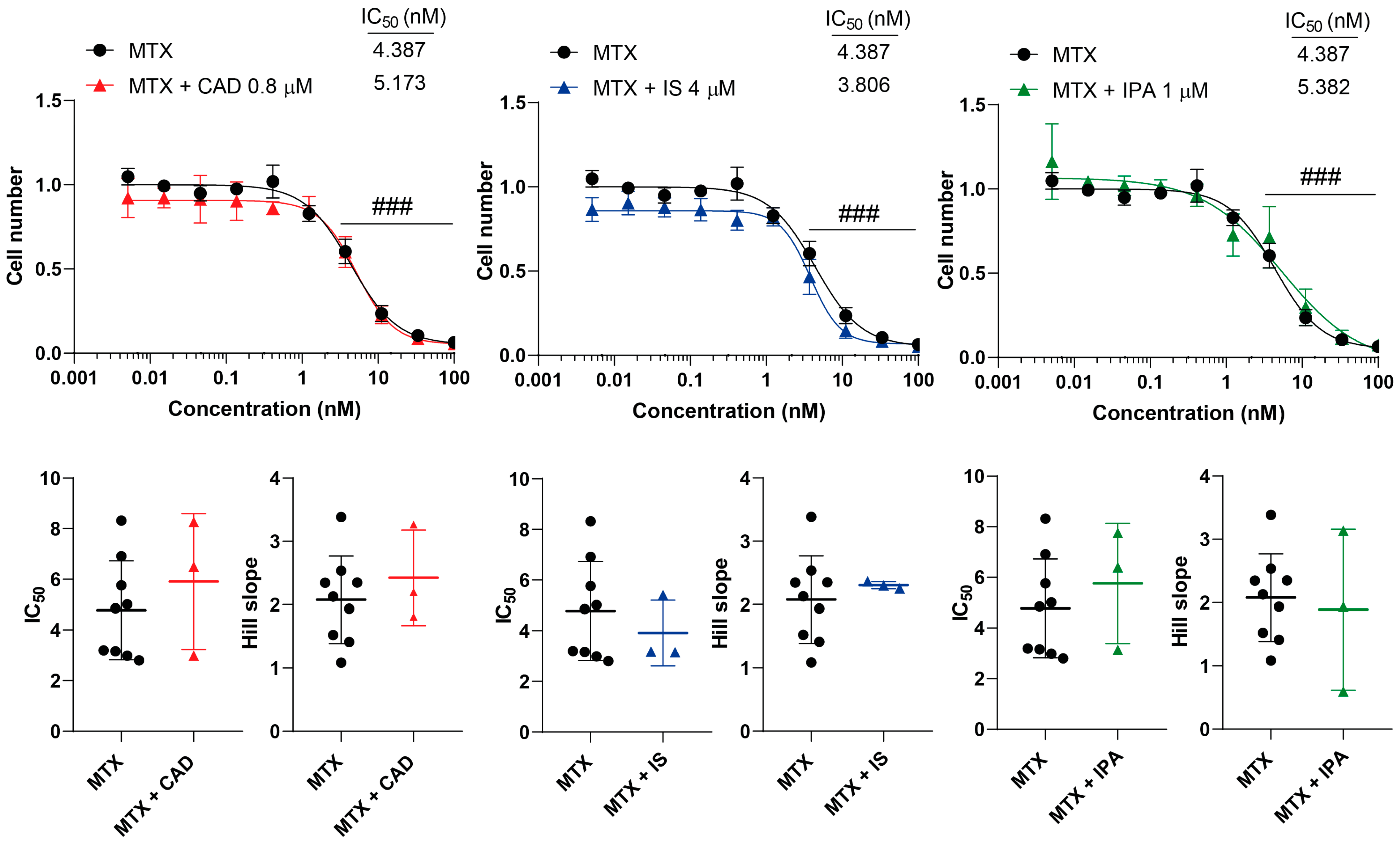
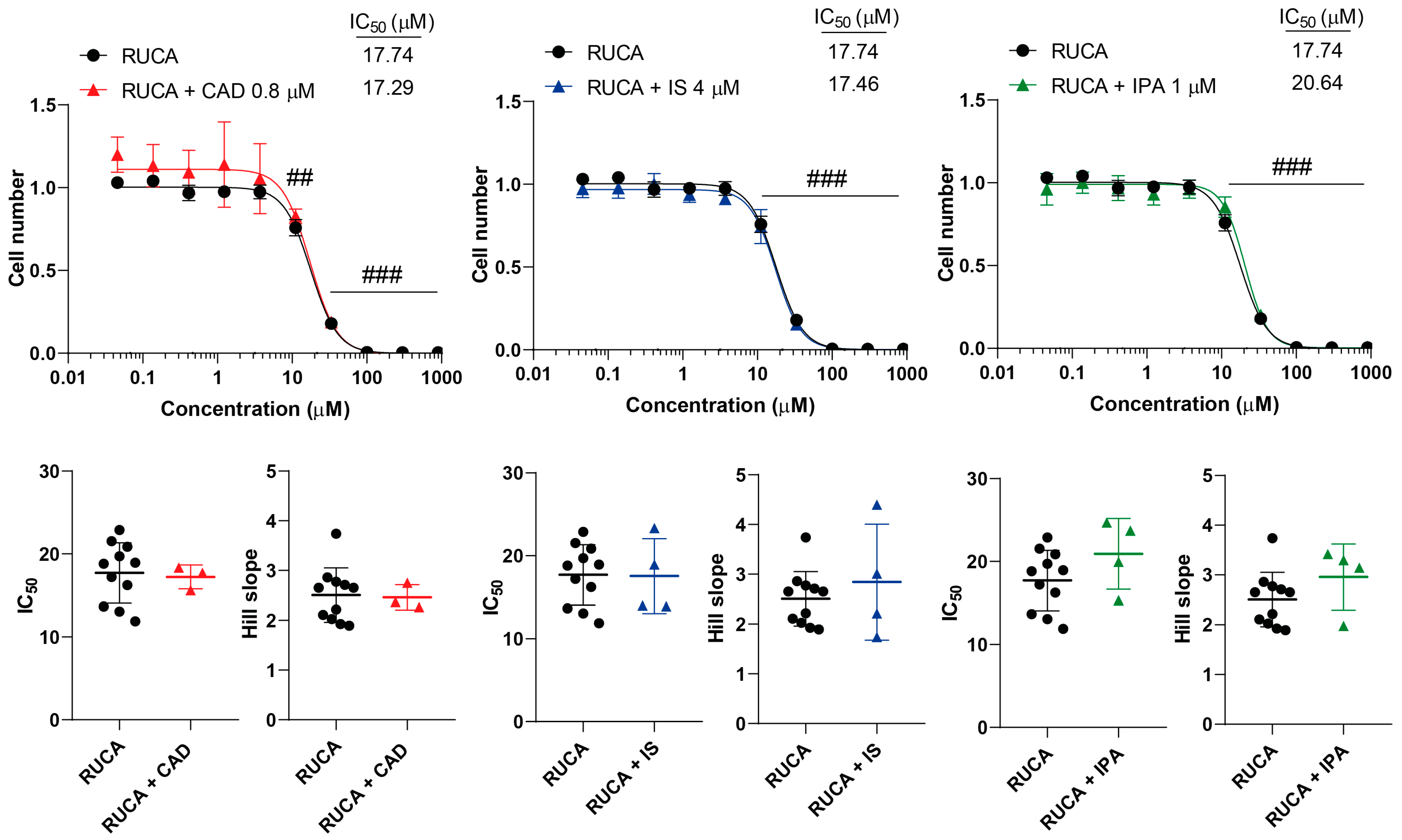
2.2. Bacterial Metabolites Do Not Interfere with Gemcitabine, Irinotecan, Methotrexate and Rucaparib Activity
2.3. Bacterial Metabolites Interfere with 5-Fluorouracil
2.4. Bacterial Metabolites Interfere with Paclitaxel
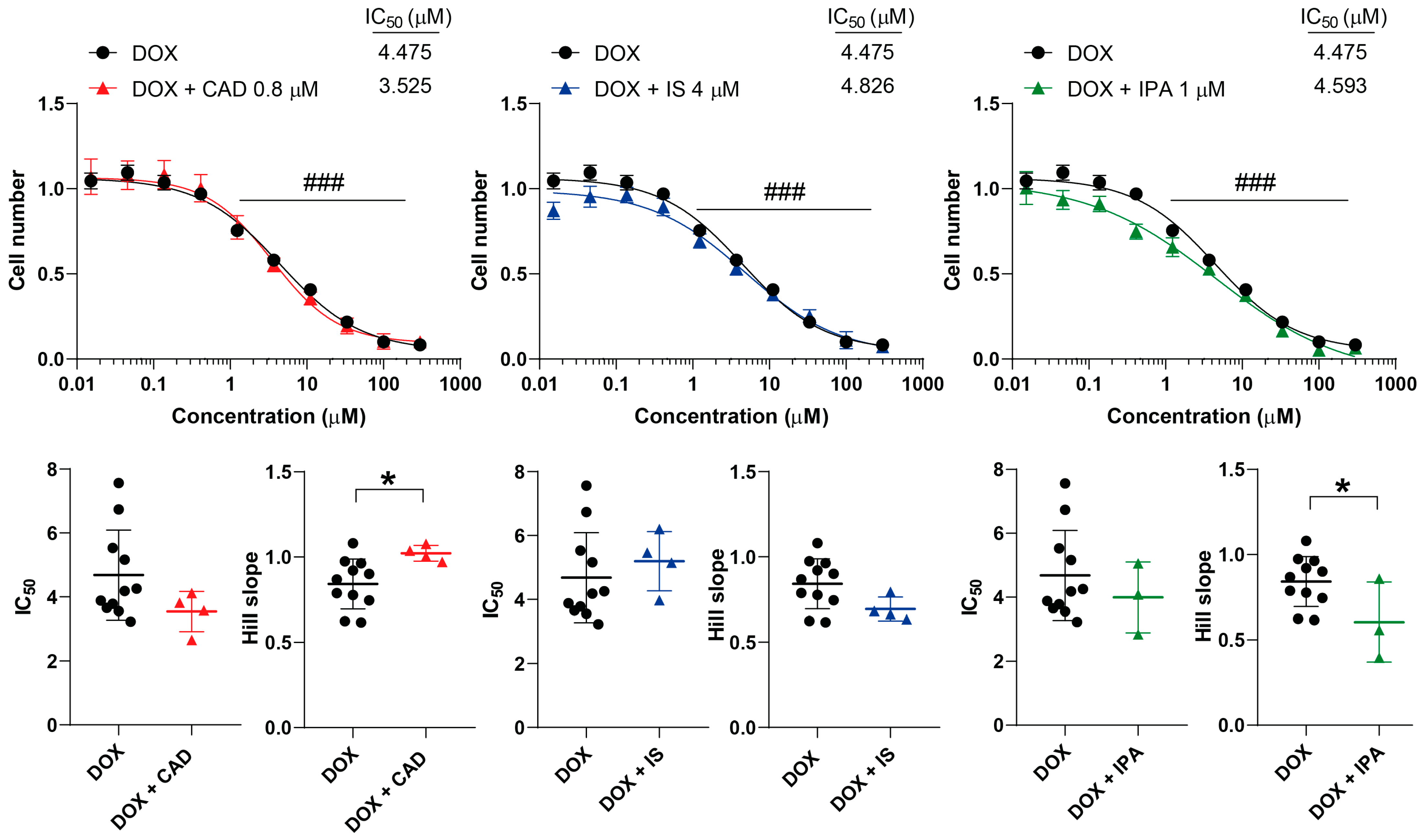
2.5. Bacterial Metabolites Interfere with Doxorubicin
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Line
4.3. MTT Assay
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cullin, N.; Azevedo Antunes, C.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef] [PubMed]
- Kovács, T.; Mikó, E.; Ujlaki, G.; Yousef, H.; Csontos, V.; Uray, K.; Bai, P. The involvement of oncobiosis and bacterial metabolite signaling in metastasis formation in breast cancer. Cancer Metastasis Rev. 2021, 40, 1223–1249. [Google Scholar] [CrossRef] [PubMed]
- Miko, E.; Vida, A.; Kovacs, T.; Ujlaki, G.; Trencsenyi, G.; Marton, J.; Sari, Z.; Kovacs, P.; Boratko, A.; Hujber, Z.; et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim. Biophys. Acta—Bioenerg. 2018, 1859, 958–974. [Google Scholar] [CrossRef] [PubMed]
- Kovács, T.; Mikó, E.; Vida, A.; Sebő, É.; Toth, J.; Csonka, T.; Boratkó, A.; Ujlaki, G.; Lente, G.; Kovács, P.; et al. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci. Rep. 2019, 9, 1300. [Google Scholar] [CrossRef] [PubMed]
- Sári, Z.; Mikó, E.; Kovács, T.; Jankó, L.; Csonka, T.; Sebő, E.; Toth, J.; Tóth, D.; Árkosy, P.; Boratkó, A.; et al. Indolepropionic acid, a metabolite of the microbiome, has cytostatic properties in breast cancer by activating AHR and PXR receptors and inducing oxidative stress. Cancers 2020, 12, 2411. [Google Scholar] [CrossRef] [PubMed]
- Sári, Z.; Mikó, E.; Kovács, T.; Boratkó, A.; Ujlaki, G.; Jankó, L.; Kiss, B.; Uray, K.; Bai, P. Indoxylsulfate, a Metabolite of the Microbiome, Has Cytostatic Effects in Breast Cancer via Activation of AHR and PXR Receptors and Induction of Oxidative Stress. Cancers 2020, 12, 2915. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Rong, X.; Zhao, G.; Zhou, Y.; Xiao, Y.; Ma, D.; Jin, X.; Wu, Y.; Yan, Y.; Yang, H.; et al. The microbial metabolite trimethylamine N-oxide promotes antitumor immunity in triple-negative breast cancer. Cell Metab. 2022, 34, 581–594.e588. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Rodrigues, M.F.; Carvalho, E.; Pezzuto, P.; Rumjanek, F.D.; Amoedo, N.D. Reciprocal modulation of histone deacetylase inhibitors sodium butyrate and trichostatin A on the energy metabolism of breast cancer cells. J. Cell Biochem. 2015, 116, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain Fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [PubMed]
- Schulthess, J.; Pandey, S.; Capitani, M.; Rue-Albrecht, K.C.; Arnold, I.; Franchini, F.; Chomka, A.; Ilott, N.E.; Johnston, D.G.W.; Pires, E.; et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity 2019, 50, 432–445.e7. [Google Scholar] [CrossRef] [PubMed]
- Salimi, V.; Shahsavari, Z.; Safizadeh, B.; Hosseini, A.; Khademian, N.; Tavakoli-Yaraki, M. Sodium butyrate promotes apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment. Lipids Health Dis. 2017, 16, 208. [Google Scholar] [CrossRef]
- Ujlaki, G.; Kovács, T.; Vida, A.; Kókai, E.; Rauch, B.; Schwarcz, S.; Mikó, E.; Janka, E.; Sipos, A.; Hegedűs, C.; et al. Identification of Bacterial Metabolites Modulating Breast Cancer Cell Proliferation and Epithelial-Mesenchymal Transition. Molecules 2023, 28, 5898. [Google Scholar] [CrossRef] [PubMed]
- Radde, B.N.; Ivanova, M.M.; Mai, H.X.; Salabei, J.K.; Hill, B.G.; Klinge, C.M. Bioenergetic differences between MCF-7 and T47D breast cancer cells and their regulation by oestradiol and tamoxifen. Biochem. J. 2015, 465, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Radde, B.N.; Ivanova, M.M.; Mai, H.X.; Alizadeh-Rad, N.; Piell, K.; Van Hoose, P.; Cole, M.P.; Muluhngwi, P.; Kalbfleisch, T.S.; Rouchka, E.C.; et al. Nuclear respiratory factor-1 and bioenergetics in tamoxifen-resistant breast cancer cells. Exp. Cell Res. 2016, 347, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Sotgia, F.; Lisanti, M.P. Mitochondrial mRNA transcripts predict overall survival, tumor recurrence and progression in serous ovarian cancer: Companion diagnostics for cancer therapy. Oncotarget 2017, 8, 66925–66939. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, N.; Das, G.M. Metabolic Reprogramming in Breast Cancer and Its Therapeutic Implications. Cells 2019, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Zacksenhaus, E.; Shrestha, M.; Liu, J.C.; Vorobieva, I.; Chung, P.E.D.; Ju, Y.; Nir, U.; Jiang, Z. Mitochondrial OXPHOS Induced by RB1 Deficiency in Breast Cancer: Implications for Anabolic Metabolism, Stemness, and Metastasis. Trends Cancer 2017, 3, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Maximov, P.Y.; Abderrahman, B.; Curpan, R.F.; Hawsawi, Y.M.; Fan, P.; Jordan, V.C. A unifying biology of sex steroid-induced apoptosis in prostate and breast cancers. Endocr. Relat. Cancer 2018, 25, R83–R113. [Google Scholar] [CrossRef] [PubMed]
- Al-Howail, H.A.; Hakami, H.A.; Al-Otaibi, B.; Al-Mazrou, A.; Daghestani, M.H.; Al-Jammaz, I.; Al-Khalaf, H.H.; Aboussekhra, A. PAC down-regulates estrogen receptor alpha and suppresses epithelial-to-mesenchymal transition in breast cancer cells. BMC Cancer 2016, 16, 540. [Google Scholar] [CrossRef]
- Bouris, P.; Skandalis, S.S.; Piperigkou, Z.; Afratis, N.; Karamanou, K.; Aletras, A.J.; Moustakas, A.; Theocharis, A.D.; Karamanos, N.K. Estrogen receptor alpha mediates epithelial to mesenchymal transition, expression of specific matrix effectors and functional properties of breast cancer cells. Matrix Biol. 2015, 43, 42–60. [Google Scholar] [CrossRef] [PubMed]
- Kulkoyluoglu-Cotul, E.; Arca, A.; Madak-Erdogan, Z. Crosstalk between Estrogen Signaling and Breast Cancer Metabolism. Trends Endocrinol. Metab. 2019, 30, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Hatse, S.; Kenis, C.; Fernández-García, J.; Altea-Manzano, P.; Billen, J.; Planque, M.; Vandekeere, A.; Lambrechts, Y.; Richard, F.; et al. Serum methylmalonic acid concentrations at breast cancer diagnosis significantly correlate with clinical frailty. GeroScience 2024, 46, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Koteva, K.; Wright, G.D. An unusual class of anthracyclines potentiate Gram-positive antibiotics in intrinsically resistant Gram-negative bacteria. J. Antimicrob. Chemother. 2014, 69, 1844–1855. [Google Scholar] [CrossRef] [PubMed]
- Westman, E.L.; Canova, M.J.; Radhi, I.J.; Koteva, K.; Kireeva, I.; Waglechner, N.; Wright, G.D. Bacterial inactivation of the anticancer drug doxorubicin. Chem. Biol. 2012, 19, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, P.; Pandey, R.P.; Nguyen, T.H.T.; Dhakal, D.; Sohng, J.K. Substrate Scope of O-Methyltransferase from Streptomyces peucetius for Biosynthesis of Diverse Natural Products Methoxides. Appl. Biochem. Biotechnol. 2018, 184, 1404–1420. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, D.; Lim, S.K.; Kim, D.H.; Kim, B.G.; Yamaguchi, T.; Sohng, J.K. Complete genome sequence of Streptomyces peucetius ATCC 27952, the producer of anticancer anthracyclines and diverse secondary metabolites. J. Biotechnol. 2018, 267, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Zabala, D.; Brana, A.F.; Florez, A.B.; Salas, J.A.; Mendez, C. Engineering precursor metabolite pools for increasing production of antitumor mithramycins in Streptomyces argillaceus. Metab. Eng. 2013, 20, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Dou, T.Y.; Luan, H.W.; Liu, X.B.; Li, S.Y.; Du, X.F.; Yang, L. Enzymatic hydrolysis of 7-xylosyltaxanes by an extracellular xylosidase from Cellulosimicrobium cellulans. Biotechnol. Lett. 2015, 37, 1905–1910. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.J.; Pan, J.; Yu, H.L.; Zheng, G.W.; Xu, J.H. Target-oriented discovery of a new esterase-producing strain Enterobacter sp. ECU1107 for whole cell-catalyzed production of (2S,3R)-3-phenylglycidate as a chiral synthon of Taxol. Appl. Microbiol. Biotechnol. 2013, 97, 6293–6300. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gonzalez, A.P.; Ritter, A.D.; Shrestha, S.; Andersen, E.C.; Yilmaz, L.S.; Walhout, A.J.M. Bacterial Metabolism Affects the C. elegans Response to Cancer Chemotherapeutics. Cell 2017, 169, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.A.; Quintaneiro, L.M.; Norvaisas, P.; Lui, P.P.; Wilson, M.P.; Leung, K.Y.; Herrera-Dominguez, L.; Sudiwala, S.; Pessia, A.; Clayton, P.T.; et al. Host-Microbe Co-metabolism Dictates Cancer Drug Efficacy in C. elegans. Cell 2017, 169, 442–456.e18. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Straussman, R. Intratumoral bacteria may elicit chemoresistance by metabolizing anticancer agents. Mol. Cell Oncol. 2018, 5, e1405139. [Google Scholar] [CrossRef] [PubMed]
- Sandrini, M.P.; Shannon, O.; Clausen, A.R.; Bjorck, L.; Piskur, J. Deoxyribonucleoside kinases activate nucleoside antibiotics in severely pathogenic bacteria. Antimicrob. Agents Chemother. 2007, 51, 2726–2732. [Google Scholar] [CrossRef] [PubMed]
- Sandrini, M.P.; Clausen, A.R.; On, S.L.; Aarestrup, F.M.; Munch-Petersen, B.; Piskur, J. Nucleoside analogues are activated by bacterial deoxyribonucleoside kinases in a species-specific manner. J. Antimicrob. Chemother. 2007, 60, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Vande Voorde, J.; Sabuncuoglu, S.; Noppen, S.; Hofer, A.; Ranjbarian, F.; Fieuws, S.; Balzarini, J.; Liekens, S. Nucleoside-catabolizing enzymes in mycoplasma-infected tumor cell cultures compromise the cytostatic activity of the anticancer drug gemcitabine. J. Biol. Chem. 2014, 289, 13054–13065. [Google Scholar] [CrossRef]
- Lehouritis, P.; Cummins, J.; Stanton, M.; Murphy, C.T.; McCarthy, F.O.; Reid, G.; Urbaniak, C.; Byrne, W.L.; Tangney, M. Local bacteria affect the efficacy of chemotherapeutic drugs. Sci. Rep. 2015, 5, 14554. [Google Scholar] [CrossRef]
- Ghosh, S.; Singh, R.; Vanwinkle, Z.M.; Guo, H.; Vemula, P.K.; Goel, A.; Haribabu, B.; Jala, V.R. Microbial metabolite restricts 5-fluorouracil-resistant colonic tumor progression by sensitizing drug transporters via regulation of FOXO3-FOXM1 axis. Theranostics 2022, 12, 5574–5595. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, B.; Zheng, Q.; Li, H.; Meng, X.; Zhou, F.; Zhang, L. A Review of Gut Microbiota-Derived Metabolites in Tumor Progression and Cancer Therapy. Adv. Sci. 2023, 10, e2207366. [Google Scholar] [CrossRef] [PubMed]
- Farhana, L.; Banerjee, H.N.; Verma, M.; Majumdar, A.P.N. Role of Microbiome in Carcinogenesis Process and Epigenetic Regulation of Colorectal Cancer. Methods Mol. Biol. 2018, 1856, 35–55. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lau, H.C.; Yu, J. Microbial metabolites in colorectal tumorigenesis and cancer therapy. Gut Microbes 2023, 15, 2203968. [Google Scholar] [CrossRef] [PubMed]
- Al-Khazaleh, A.K.; Chang, D.; Münch, G.W.; Bhuyan, D.J. The Gut Connection: Exploring the Possibility of Implementing Gut Microbial Metabolites in Lymphoma Treatment. Cancers 2024, 16, 1464. [Google Scholar] [CrossRef] [PubMed]
- Giurini, E.F.; Godla, A.; Gupta, K.H. Redefining bioactive small molecules from microbial metabolites as revolutionary anticancer agents. Cancer Gene Ther. 2024, 31, 187–206. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Diaz, J.; Álvarez-Mercado, A.I. The Interplay between Microbiota and Chemotherapy-Derived Metabolites in Breast Cancer. Metabolites 2023, 13, 703. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Curry, K.P.; Bultman, S.J. Microbial oncotarget: Bacterial-produced butyrate, chemoprevention and Warburg effect. Oncotarget 2013, 4, 182–183. [Google Scholar] [CrossRef]
- Loser, C.; Folsch, U.R.; Paprotny, C.; Creutzfeldt, W. Polyamine concentrations in pancreatic tissue, serum, and urine of patients with pancreatic cancer. Pancreas 1990, 5, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Loser, C.; Folsch, U.R.; Paprotny, C.; Creutzfeldt, W. Polyamines in colorectal cancer. Evaluation of polyamine concentrations in the colon tissue, serum, and urine of 50 patients with colorectal cancer. Cancer 1990, 65, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Danaceau, J.P.; Anderson, G.M.; McMahon, W.M.; Crouch, D.J. A liquid chromatographic-tandem mass spectrometric method for the analysis of serotonin and related indoles in human whole blood. J. Anal. Toxicol. 2003, 27, 440–444. [Google Scholar] [CrossRef]
- Rosas, H.D.; Doros, G.; Bhasin, S.; Thomas, B.; Gevorkian, S.; Malarick, K.; Matson, W.; Hersch, S.M. A systems-level “misunderstanding”: The plasma metabolome in Huntington’s disease. Ann. Clin. Transl. Neurol. 2015, 2, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-N.; Wu, I.W.; Huang, Y.-F.; Peng, S.-Y.; Huang, Y.-C.; Ning, H.-C. Measuring serum total and free indoxyl sulfate and p-cresyl sulfate in chronic kidney disease using UPLC-MS/MS. J. Food Drug Anal. 2019, 27, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Kacsir, I.; Sipos, A.; Kiss, T.; Major, E.; Bajusz, N.; Tóth, E.; Buglyo, P.; Somsak, L.; Kardos, G.; Bai, P.; et al. Half Sandwich-Type Osmium, Ruthenium, Iridium and Rhodium Complexes with Bidentate Glycosyl Heterocyclic Ligands Induce Cytostasis in Platinum-Resistant Ovarian Cancer Cells and Bacteriostasis in Gram-Positive Multiresistant Bacteria. Front. Chem. 2023, 11, 1086267. [Google Scholar] [CrossRef] [PubMed]
- Senkus, E.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rutgers, E.; Zackrisson, S.; Cardoso, F. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26, v8–v30. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.; Szabo, C. Therapeutic Applications of PARP Inhibitors: Anticancer Therapy and Beyond. Mol. Asp. Med. 2013, 34, 1217–1256. [Google Scholar] [CrossRef] [PubMed]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O’Connor, M.J.; et al. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. N. Engl. J. Med. 2009, 361, 123–134. [Google Scholar] [CrossRef]
- Kwapisz, D. Cyclin-dependent kinase 4/6 inhibitors in breast cancer: Palbociclib, ribociclib, and abemaciclib. Breast Cancer Res. Treat. 2017, 166, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Liang, L.; Tang, X.; Zhu, J.; Mu, L.; Wang, M.; Huang, X.; Gong, S.; Xu, J.; Liu, T.; et al. Changes in the gut microbiota structure and function in rats with doxorubicin-induced heart failure. Front. Cell Infect. Microbiol. 2023, 13, 1135428. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Qiao, X.; Wang, Z.; Li, X.; Pan, Y.; Wei, X.; Lv, Z.; Li, P.; Du, Q.; Wei, W.; et al. Cisplatin and doxorubicin chemotherapy alters gut microbiota in a murine osteosarcoma model. Aging 2024, 16, 1336–1351. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Tintelnot, J.; Xu, Y.; Lesker, T.R.; Schönlein, M.; Konczalla, L.; Giannou, A.D.; Pelczar, P.; Kylies, D.; Puelles, V.G.; Bielecka, A.A.; et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature 2023, 615, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef]
- Lee, S.; Cho, Y.Y.; Cho, E.J.; Yu, S.J.; Lee, J.H.; Yoon, J.H.; Kim, Y.J. Synergistic effect of ursodeoxycholic acid on the antitumor activity of sorafenib in hepatocellular carcinoma cells via modulation of STAT3 and ERK. Int. J. Mol. Med. 2018, 42, 2551–2559. [Google Scholar] [CrossRef] [PubMed]
- Kovács, P.; Csonka, T.; Kovács, T.; Sári, Z.; Ujlaki, G.; Sipos, A.; Karányi, Z.; Szeőcs, D.; Hegedűs, C.; Uray, K.; et al. Lithocholic acid, a metabolite of the microbiome, increases oxidative stress in breast cancer. Cancers 2019, 11, 1255. [Google Scholar] [CrossRef] [PubMed]
- Schwarcz, S.; Kovács, P.; Kovács, T.; Ujlaki, G.; Nyerges, P.; Uray, K.; Bai, P.; Mikó, E. The pro- and antineoplastic effects of deoxycholic acid in pancreatic adenocarcinoma cell models. Mol. Biol. Rep. 2023, 50, 5273–5282. [Google Scholar] [CrossRef] [PubMed]
- Schwarcz, S.; Kovács, P.; Nyerges, P.; Ujlaki, G.; Sipos, A.; Uray, K.; Bai, P.; Mikó, E. The bacterial metabolite, lithocholic acid, has antineoplastic effects in pancreatic adenocarcinoma. Cell Death Discov. 2024, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Chou, W.C.; Lai, Y.; Liang, K.; Tam, J.W.; Brickey, W.J.; Chen, L.; Montgomery, N.D.; Li, X.; Bohannon, L.M.; et al. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science 2020, 370, eaay9097. [Google Scholar] [CrossRef] [PubMed]
- Sigurdsson, V.; Haga, Y.; Takei, H.; Mansell, E.; Okamatsu-Haga, C.; Suzuki, M.; Radulovic, V.; van der Garde, M.; Koide, S.; Soboleva, S.; et al. Induction of blood-circulating bile acids supports recovery from myelosuppressive chemotherapy. Blood Adv. 2020, 4, 1833–1843. [Google Scholar] [CrossRef] [PubMed]
- Gesztelyi, R.; Zsuga, J.; Kemeny-Beke, A.; Varga, B.; Juhasz, B.; Tosaki, A. The Hill equation and the origin of quantitative pharmacology. Arch. Hist. Exact Sci. 2012, 66, 427–438. [Google Scholar] [CrossRef]
- Yokoyama, M.T.; Carlson, J.R. Microbial metabolites of tryptophan in the intestinal tract with special reference to skatole. Am. J. Clin. Nutr. 1979, 32, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Auslander, N.; Yizhak, K.; Weinstock, A.; Budhu, A.; Tang, W.; Wang, X.W.; Ambs, S.; Ruppin, E. A joint analysis of transcriptomic and metabolomic data uncovers enhanced enzyme-metabolite coupling in breast cancer. Sci. Rep. 2016, 6, 29662. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Lin, C.C.; Spasojevic, I.; Iversen, E.S.; Chi, J.T.; Marks, J.R. A joint analysis of metabolomics and genetics of breast cancer. Breast Cancer Res. 2014, 16, 415. [Google Scholar] [CrossRef] [PubMed]
- Box, G.E.P.; Cox, D.R. An analysis of transformations. J. R. Stat. Soc. B 1964, 26, 211–234. [Google Scholar] [CrossRef]



| Chemotherapeutic Agent | Metabolite | IC50 (±SD) | Hill Coefficient (±SD) |
|---|---|---|---|
| Gemcitabine | - | 15.05 (±2.87) | - |
| Cadaverine | 14.65 (±1.31) | - | |
| Indoxylsulfate | 15.38 (±1.55) | - | |
| Indolepropionic acid | 14.75 (±2.35) | - | |
| Irinotecan | - | 36.16 (±8.03) | 0.93 (±0.18) |
| Cadaverine | 30.10 (±1.30) | 0.97 (±0.22) | |
| Indoxylsulfate | 32.77 (±13.13) | 1.03 (±0.05) | |
| Indolepropionic acid | 31.56 (±16.07) | 0.81(±0.14) | |
| Methotrexate | - | 4.38 (±1.94) | 2.07 (±0.69) |
| Cadaverine | 5.17 (±2.68) | 2.42 (±0.75) | |
| Indoxylsulfate | 3.80 (±1.29) | 2.30 (±0.05) | |
| Indolepropionic acid | 5.38 (±2.37) | 1.88 (±1.27) | |
| Rucaparib | - | 17.74 (±3.63) | 2.50 (±0.54) |
| Cadaverine | 17.29 (±1.43) | 2.46 (±0.25) | |
| Indoxylsulfate | 17.46 (±4.52) | 2.84 (±1.16) | |
| Indolepropionic acid | 20.64 (±4.26) | 2.95 (±0.66) | |
| 5-fluorouracil | - | 0.69 (±0.16) | 1.24 (±0.44) |
| Cadaverine | 0.81 (±0.17) | 2.12 (±0.64) * | |
| Indoxylsulfate | 0.67 (±0.30) | 1.50 (±0.65) | |
| Indolepropionic acid | 0.99 (±0.27) * | 1.04 (±0.32) | |
| Paclitaxel | - | 12.21 (±2.12) | 1.30 (±0.34) |
| Cadaverine | 11.54 (±3.38) | 1.61 (±0.34) | |
| Indoxylsulfate | 7.77 (±3.86) | 1.09 (±0.47) | |
| Indolepropionic acid | 6.03 (±2.76) ** | 1.18 (±0.45) | |
| Doxorubicin | - | 4.47 (±1.40) | 0.84 (±0.14) |
| Cadaverine | 3.52 (±0.62) | 1.02 (±0.04) * | |
| Indoxylsulfate | 4.82 (±0.92) | 0.69 (±0.06) | |
| Indolepropionic acid | 4.59 (±1.10) | 0.60 (±0.23) * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwarcz, S.; Nyerges, P.; Bíró, T.I.; Janka, E.; Bai, P.; Mikó, E. Cytostatic Bacterial Metabolites Interfere with 5-Fluorouracil, Doxorubicin and Paclitaxel Efficiency in 4T1 Breast Cancer Cells. Molecules 2024, 29, 3073. https://doi.org/10.3390/molecules29133073
Schwarcz S, Nyerges P, Bíró TI, Janka E, Bai P, Mikó E. Cytostatic Bacterial Metabolites Interfere with 5-Fluorouracil, Doxorubicin and Paclitaxel Efficiency in 4T1 Breast Cancer Cells. Molecules. 2024; 29(13):3073. https://doi.org/10.3390/molecules29133073
Chicago/Turabian StyleSchwarcz, Szandra, Petra Nyerges, Tímea Ingrid Bíró, Eszter Janka, Péter Bai, and Edit Mikó. 2024. "Cytostatic Bacterial Metabolites Interfere with 5-Fluorouracil, Doxorubicin and Paclitaxel Efficiency in 4T1 Breast Cancer Cells" Molecules 29, no. 13: 3073. https://doi.org/10.3390/molecules29133073
APA StyleSchwarcz, S., Nyerges, P., Bíró, T. I., Janka, E., Bai, P., & Mikó, E. (2024). Cytostatic Bacterial Metabolites Interfere with 5-Fluorouracil, Doxorubicin and Paclitaxel Efficiency in 4T1 Breast Cancer Cells. Molecules, 29(13), 3073. https://doi.org/10.3390/molecules29133073







