Isovaleryl Sucrose Esters from Atractylodes japonica and Their Cytotoxic Activity
Abstract
1. Introduction
2. Results
2.1. Identification of the Isolated Compounds
2.2. Cytotoxicity Assays
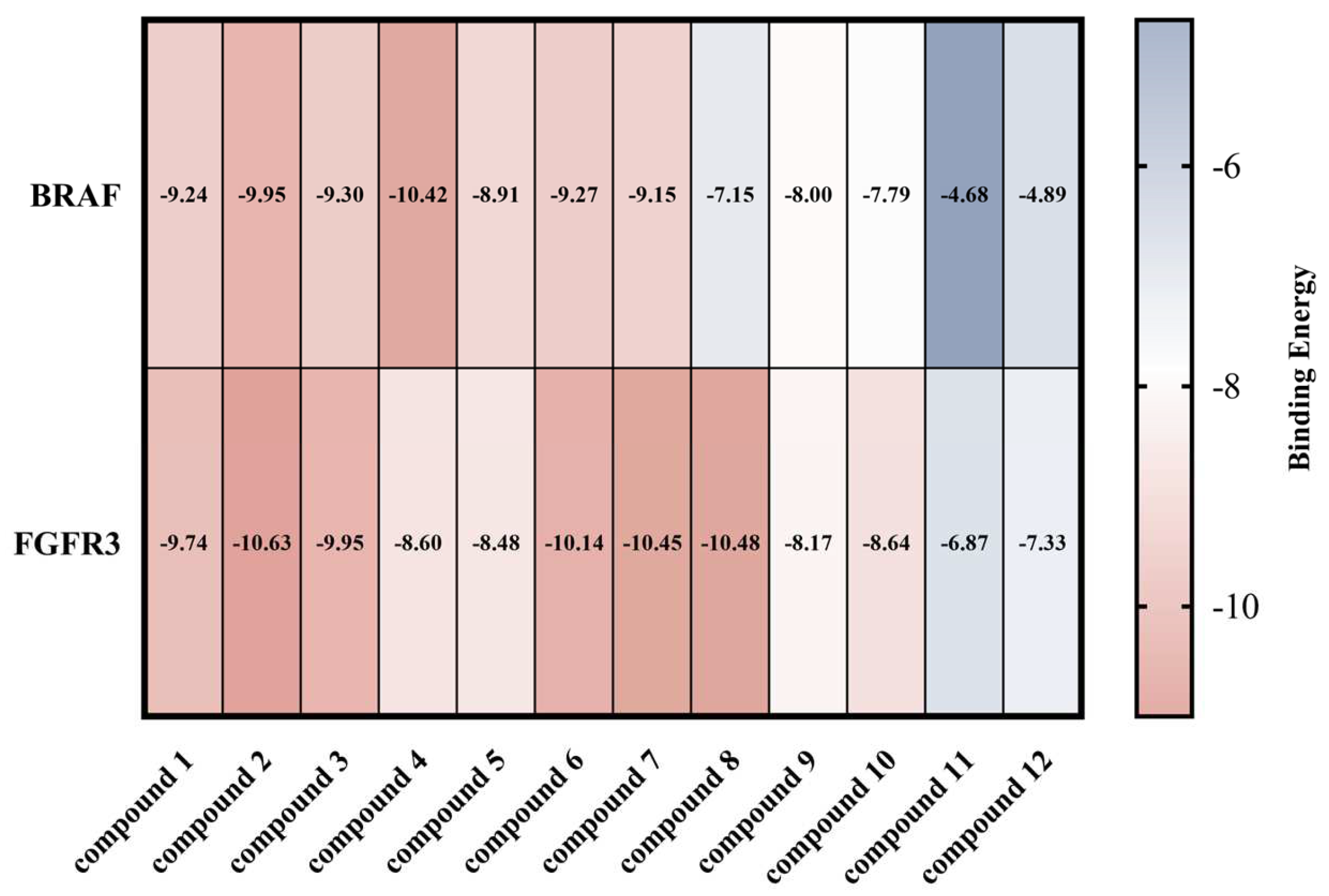
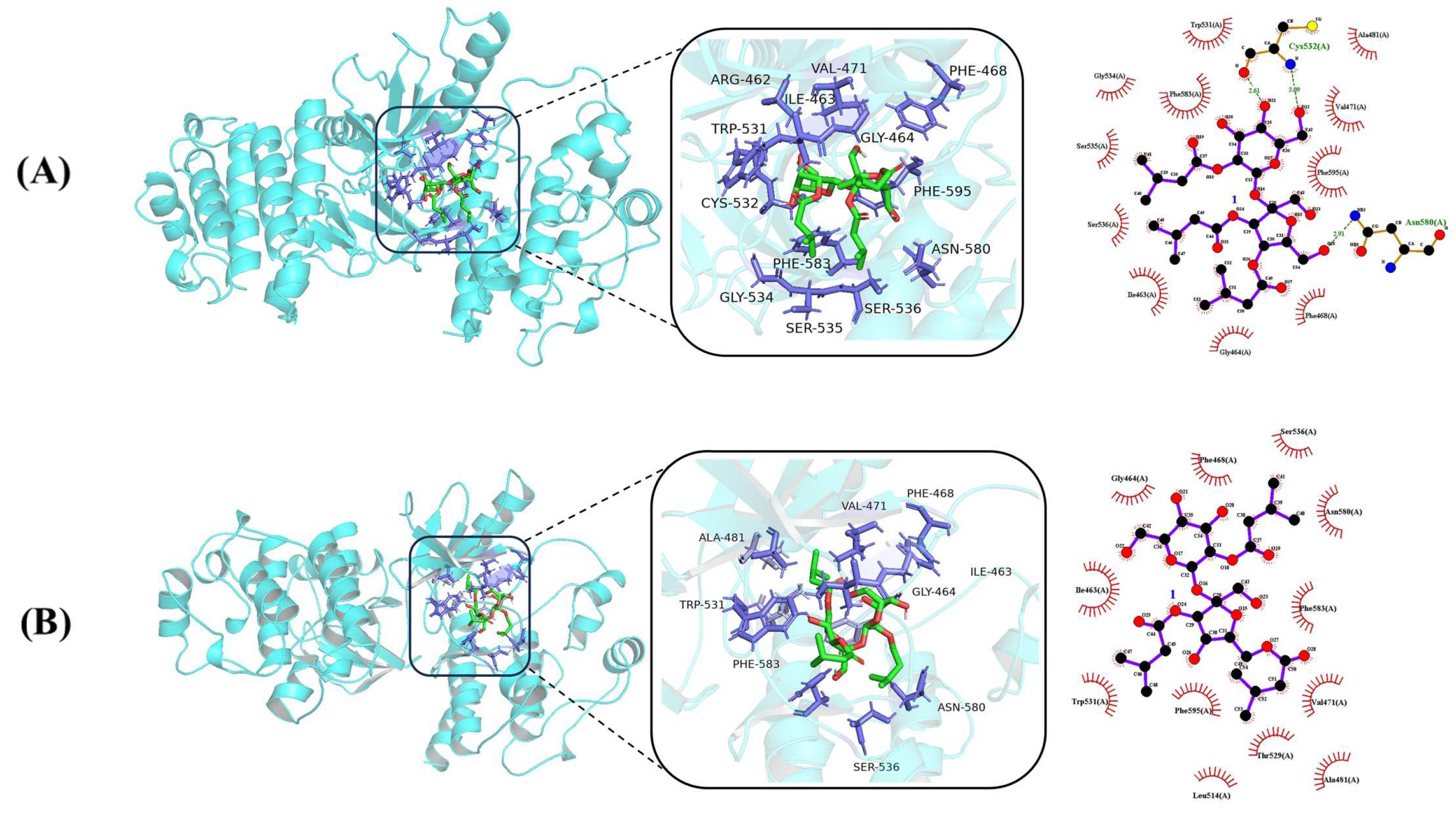
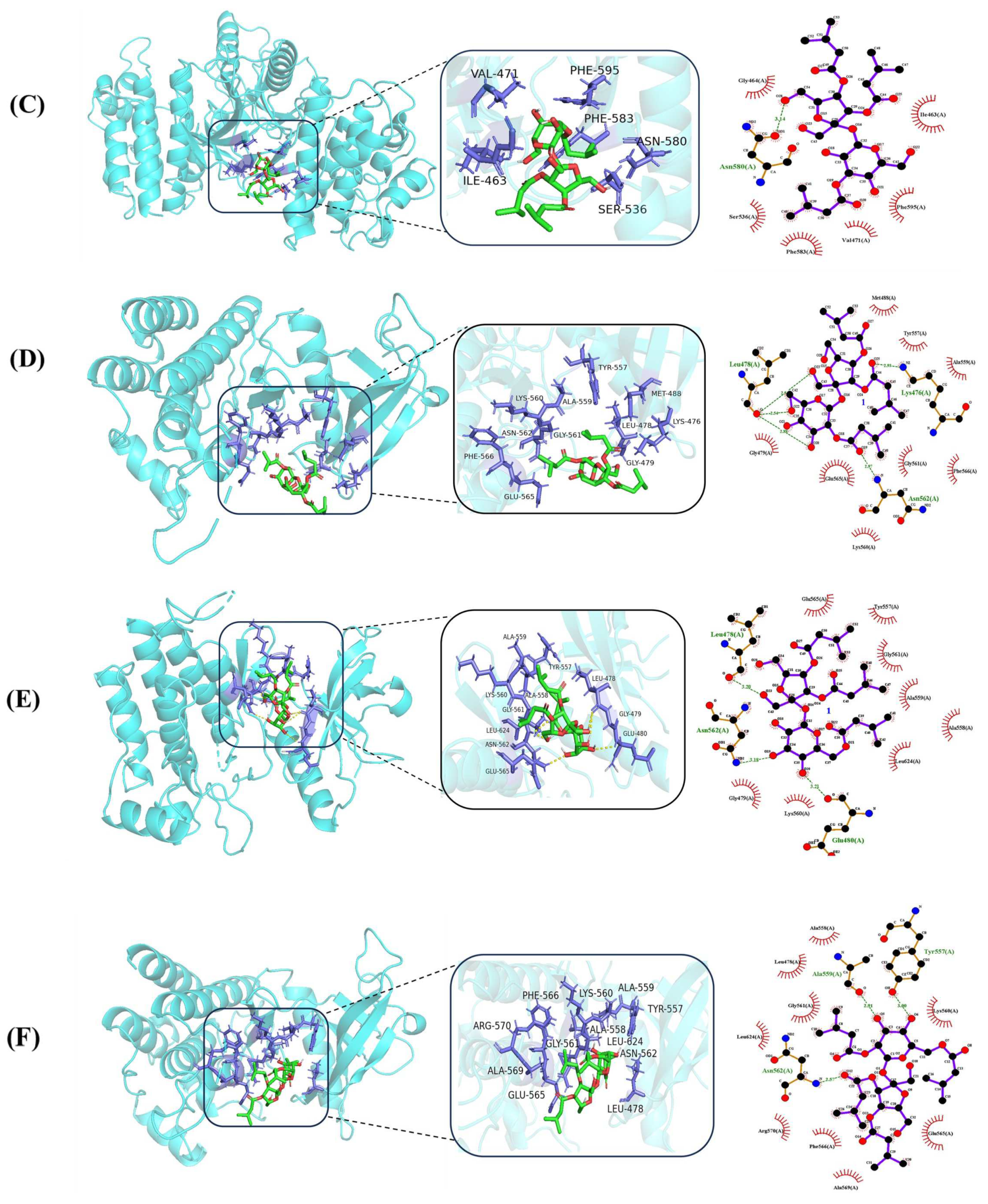
2.3. Docking Study
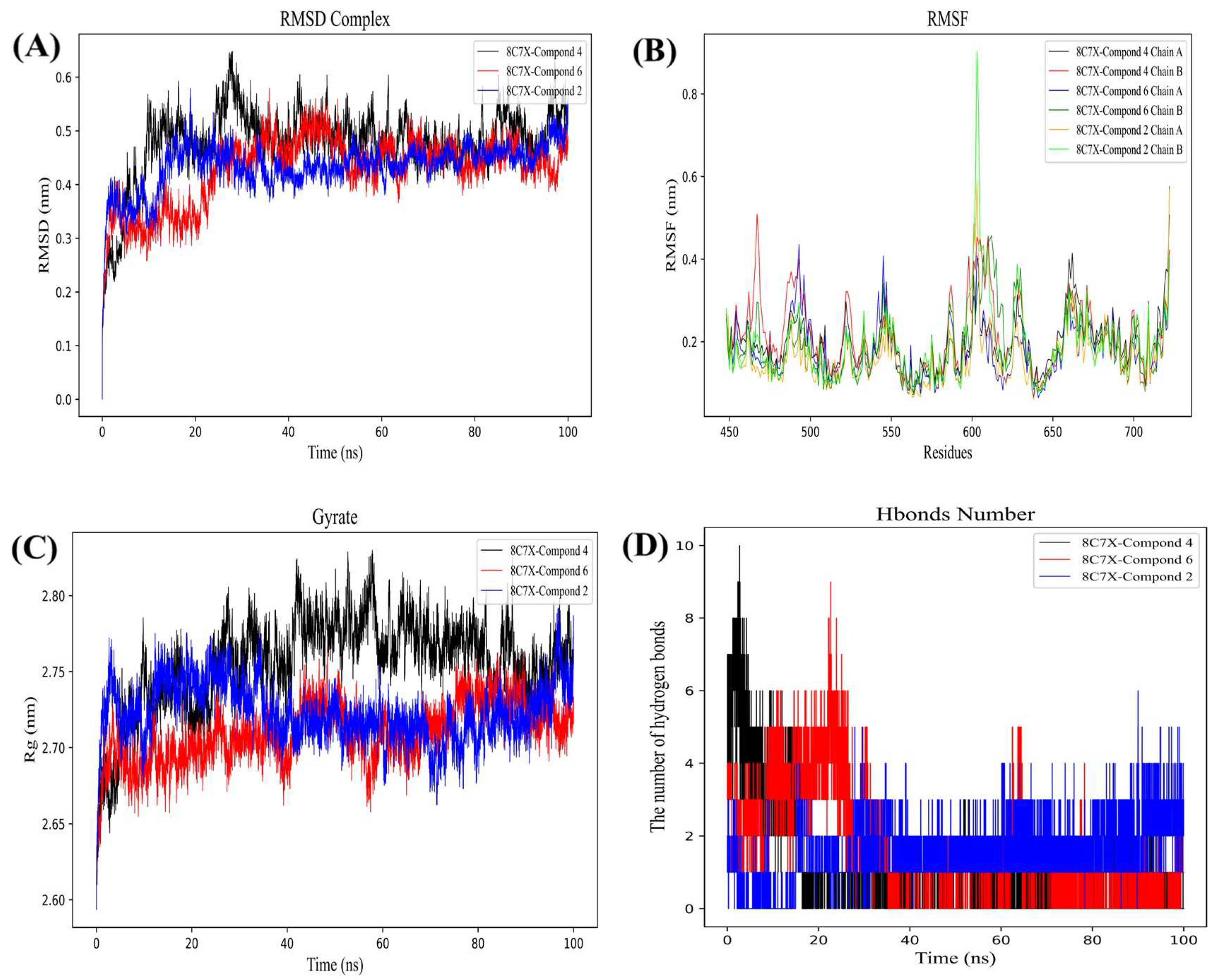
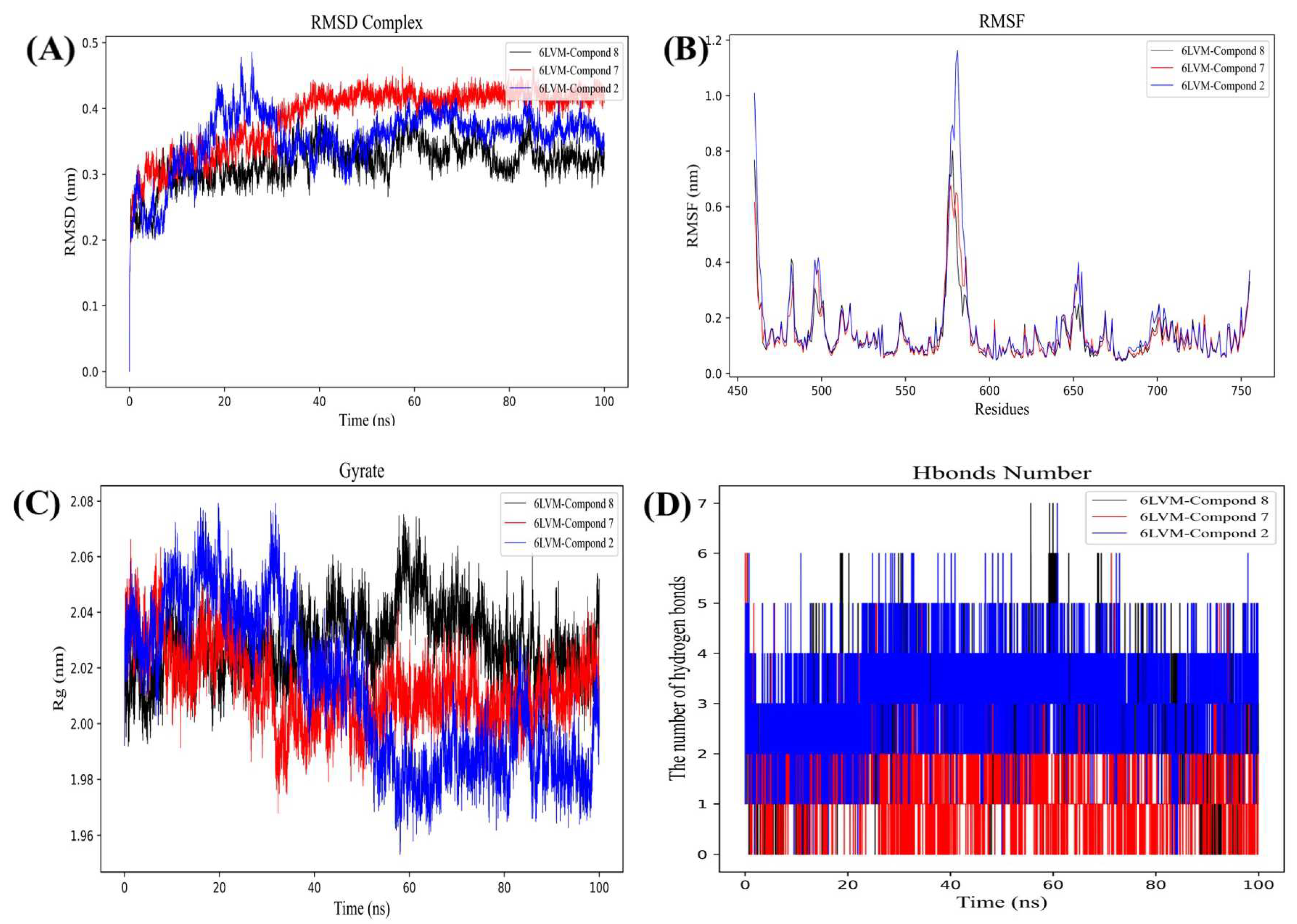
2.4. Molecular Dynamics Simulation
2.4.1. Stability Analysis of Small Molecule-Protein Receptor Complexes
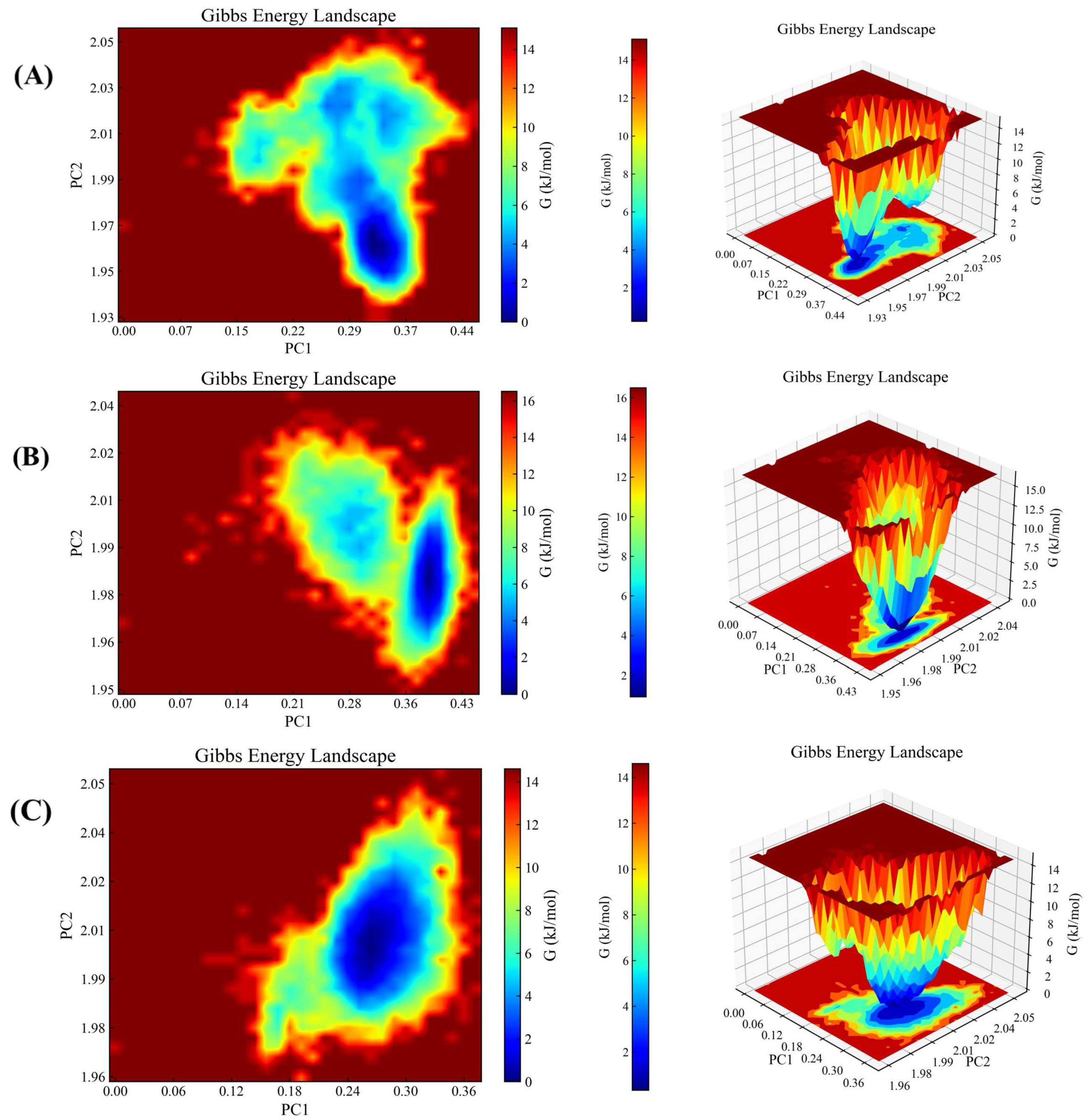
2.4.2. Gibbs Free Energy Analysis
2.4.3. MM/GBSA and Binding Free Energy Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. General Experimental Procedures
4.3. Extraction, Isolation, and Structure Identification
4.4. Cytotoxicity Assays
4.5. Molecular Docking Analysis
4.6. Molecular Dynamics Simulation Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, J.; Zhao, J.; Jiang, F.; Wang, L.; Xiao, Q.; Han, F.; Chen, J.; Yuan, S.; Wei, J.; Larsson, S.C.; et al. Identification of novel protein biomarkers and drug targets for colorectal cancer by integrating human plasma proteome with genome. Genome Med. 2023, 15, 75. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Sun, Y.; Bai, H.; Wang, Y.; Yang, B.; Wang, Q.; Kuang, H. Effects of saponins from Chinese herbal medicines on signal transduction pathways in cancer: A review. Front. Pharmacol. 2023, 14, 1159985. [Google Scholar] [CrossRef] [PubMed]
- Chrencik, J.E.; Staker, B.L.; Burgin, A.B.; Pourquier, P.; Pommier, Y.; Stewart, L.; Redinbo, M.R. Mechanisms of camptothecin resistance by human topoisomerase I mutations. J. Mol. Biol. 2004, 339, 773–784. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, A.; Zhang, S.; Kim, J.; Xia, J.; Zhang, F.; Wang, D.; Wang, Q.; Wang, J. Paclitaxel-loaded ginsenoside Rg3 liposomes for drug-resistant cancer therapy by dual targeting of the tumor microenvironment and cancer cells. J. Adv. Res. 2023, 49, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.Y.; Shi, L.P.; Zhu, J.L.; Bai, R.R.; Xie, T. Elemene Antitumor Drugs Development Based on “Molecular Compatibility Theory” and Clinical Application: A Retrospective and Prospective Outlook. Chin. J. Integr. Med. 2024, 30, 62–74. [Google Scholar] [CrossRef]
- Zhao, Q.L.; Wang, M.J.; Zhao, M.; Zheng, B.J. Research progress on Atractylodes japonica. Chin. Tradit. Herb. Drugs 2018, 49, 3797–3803. [Google Scholar] [CrossRef]
- Choi, E.M.; Kim, G.H.; Lee, Y.S. Atractylodes japonica root extract protects osteoblastic MC3T3-E1 cells against hydrogen peroxide-induced inhibition of osteoblastic differentiation. Phytother. Res. 2009, 23, 1537–1542. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Yamashita, K.; Hitomi, N.; Suzuki, A.; Oneda, K. Studies on the constituents of Atractylodes rhizome, constituents in the rhizome of Atractylodes japonica and TLC analysis of Jutsu. Jpn. J. Pharmacogn. 1993, 47, 12–16. [Google Scholar]
- Daudé, D.; Remaud-Siméon, M.; André, I. Sucrose analogs: An attractive (bio)source for glycodiversification. Nat. Prod. Rep. 2012, 29, 945–960. [Google Scholar] [CrossRef]
- Teng, Y.; Stewart, S.G.; Hai, Y.W.; Li, X.; Banwell, M.G.; Lan, P. Sucrose fatty acid esters: Synthesis, emulsifying capacities, biological activities and structure-property profiles. Crit. Rev. Food. Sci. Nutr. 2021, 61, 3297–3317. [Google Scholar] [CrossRef]
- Murakami, N.; Iwata, E.; Tamura, S.; Akiyama, S.; Kobayashi, M. New multidrug resistance modulators from Atractylodis lanceae rhizoma. Bioorg. Med. Chem. Lett. 2000, 10, 2629–2632. [Google Scholar] [CrossRef]
- Tanaka, K.; Ina, A. Structure elucidation of acylsucrose derivatives from Atractylodes lanceae rhizome and Atractylodes rhizome. Nat. Prod. Commun. 2009, 4, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Zhuo, Z.G.; Xu, X.K.; Ye, J.; Li, H.L.; Shen, Y.H.; Zhang, W.D. Cytotoxic isovaleryl sucrose esters from Ainsliaea yunnanensis: Reduction of mitochondrial membrane potential and increase of reactive oxygen species levels in A549 cells. RSC Adv. 2017, 7, 20865–20873. [Google Scholar] [CrossRef]
- Tchinda, A.T.; Tane, P.; Ayafor, J.F.; Connolly, J.D. Stigmastane derivatives and isovaleryl sucrose esters from Vernonia guineensis (Asteraceae). Phytochemistry 2003, 63, 841–846. [Google Scholar] [CrossRef]
- Wu, Q.; Cho, J.G.; Lee, D.S.; Lee, D.Y.; Song, N.Y.; Kim, Y.C.; Lee, K.T.; Chung, H.G.; Choi, M.S.; Jeong, T.S. Carbohydrate derivatives from the roots of Brassica rapa ssp. campestris and their effects on ROS production and glutamate-induced cell death in HT-22 cells. Carbohydr. Res. 2013, 372, 9–14. [Google Scholar] [CrossRef]
- Kim, C.T.; Jung, M.H.; Kim, H.S.; Kim, H.J.; Kang, S.H. Inhibitors of melanogenesis from Euphorbieae lathyridis Semen. Korean J. Pharmacogn. 2000, 31, 168–173. [Google Scholar]
- Ullah, R.; Yin, Q.; Snell, A.H.; Wan, L. RAF-MEK-ERK pathway in cancer evolution and treatment. Semin. Cancer Biol. 2022, 85, 123–154. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chen, T.; Ding, Z.; Wang, Y.; Wei, Y.; Wei, X. Inhibition of FGF-FGFR and VEGF-VEGFR signalling in cancer treatment. Cell Prolif. 2021, 54, e13009. [Google Scholar] [CrossRef] [PubMed]
- Molina-Cerrillo, J.; San Román, M.; Pozas, J.; Alonso-Gordoa, T.; Pozas, M.; Conde, E.; Rosas, M.; Grande, E.; García-Bermejo, M.L.; Carrato, A. BRAF Mutated Colorectal Cancer: New Treatment Approaches. Cancers 2020, 12, 1571. [Google Scholar] [CrossRef]
- Johansson, C.H.; Brage, S.E. BRAF inhibitors in cancer therapy. Pharmacol. Ther. 2014, 142, 176–182. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, D.C.; Shi, L.; Zhu, B.; Min, Z.; Jin, J. Genome analyses identify the genetic modification of lung cancer subtypes. Semin. Cancer Biol. 2017, 42, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, M.; Wu, A.; Yao, X.; Wang, Q. Structure-Based Discovery and Biological Assays of a Novel PRMT5 Inhibitor for Non-Small Cell Lung Cancer. Molecules 2022, 27, 7436. [Google Scholar] [CrossRef] [PubMed]
- Saif, M.W.; Elfiky, A.; Salem, R.R. Gastrointestinal perforation due to bevacizumab in colorectal cancer. Ann. Surg. Oncol. 2007, 14, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
- Prahallad, A.; Sun, C.; Huang, S.; Di Nicolantonio, F.; Salazar, R.; Zecchin, D.; Beijersbergen, R.L.; Bardelli, A. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012, 483, 7387. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Adjei, A.A. FGFR Signaling as a Target for Lung Cancer Therapy. J. Thorac. Oncol. 2016, 11, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Subbiah, V.; Iannotti, N.O.; Gutierrez, M.; Smith, D.C.; Féliz, L.; Lihou, C.F.; Tian, C.; Silverman, I.M.; Ji, T.; Saleh, M. FIGHT-101, a first-in-human study of potent and selective FGFR 1–3 inhibitor pemigatinib in pan-cancer patients with FGF/FGFR alterations and advanced malignancies. Ann. Oncol. 2022, 33, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Helsten, T.; Elkin, S.; Arthur, E.; Tomson, B.N.; Carter, J.; Kurzrock, R. The FGFR Landscape in Cancer: Analysis of 4853 Tumors by Next-Generation Sequencing. Clin. Cancer Res. 2016, 22, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Bock, K.; Pedersen, C.; Pedersen, H. Carbon-13 nuclear magnetic resonance data for oligosaccharides. Adv. Carbohydr. Chem. Biochem. 1984, 42, 193–225. [Google Scholar] [CrossRef]
- Tabopda, T.K.; Mitaine-Offer, A.C.; Paululat, T.; Delemasure, S.; Dutartre, P.; Ngadjui, B.T.; Lacaille-Dubois, M.A. Steroidal saponins from Chlorophytum deistelianum. Phytochemistry 2016, 126, 34–40. [Google Scholar] [CrossRef]
- Nana, H.; Penglonh, W.; Hongshan, C. Basic research on antioxidant substances of gentiana macrophylla based on composition analysis-activity screening-network pharmacology. Chin. J. Tradit. Chin. Med. 2022, 37, 5883–5890. [Google Scholar]
- Arora, R.; Linders, J.T.M.; Aci-Sèche, S.; Verheyen, T.; Van Heerde, E.; Brehmer, D.; Chaikuad, A.; Knapp, S.; Bonnet, P. Design, synthesis and characterisation of a novel type II B-RAF paradox breaker inhibitor. Eur. J. Med. Chem. 2023, 250, 115231. [Google Scholar] [CrossRef]
- Kuriwaki, I.; Kameda, M.; Hisamichi, H.; Kikuchi, S.; Iikubo, K.; Kawamoto, Y.; Moritomo, H.; Kondoh, Y.; Amano, Y.; Tateishi, Y.; et al. Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2. Bioorg. Med. Chem. 2020, 28, 115453. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.Z.; Ventikos, Y. Molecular dynamics simulation: A new way to understand the functionality of the endothelial glycocalyx. Curr. Opin. Struct. Biol. 2022, 73, 102330. [Google Scholar] [CrossRef]
- Zhu, J.; Li, K.; Xu, L.; Cai, Y.; Chen, Y.; Zhao, X.; Li, H.; Huang, G.; Jin, J. Discovery of novel selective PI3Kγ inhibitors through combining machine learning-based virtual screening with multiple protein structures and bio-evaluation. J. Adv. Res. 2021, 36, 1–13. [Google Scholar] [CrossRef]
- Dorado, G.; Gálvez, S.; Rosales, T.E.; Vásquez, V.F.; Hernández, P. Analyzing Modern Biomolecules: The Revolution of Nucleic-Acid Sequencing—Review. Biomolecules 2021, 11, 1111. [Google Scholar] [CrossRef]
- Salike, S.; Bhatt, N. Thermodynamically consistent estimation of Gibbs free energy from data: Data reconciliation approach. Bioinformatics 2020, 36, 1219–1225. [Google Scholar] [CrossRef]
- Jia, Z.J.; Lan, X.W.; Lu, K.; Meng, X.; Jing, W.J.; Jia, S.R.; Zhao, K.; Dai, Y.J. Synthesis, molecular docking, and binding Gibbs free energy calculation of β-nitrostyrene derivatives: Potential inhibitors of SARS-CoV-2 3CL protease. J. Mol. Struct. 2023, 1284, 135409. [Google Scholar] [CrossRef]
- Dasmahapatra, U.; Kumar, C.K.; Das, S.; Subramanian, P.T.; Murali, P.; Isaac, A.E.; Ramanathan, K.; Mm, B.; Chanda, K. In-silico molecular modelling, MM/GBSA binding free energy and molecular dynamics simulation study of novel pyrido fused imidazo[4,5-c] quinolines as potential anti-tumor agents. Front. Chem. 2022, 10, 991369. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.F.; Sun, H.H.; Tan, J.B.; Huang, Q.; Cheng, F.; Xu, K.P.; Zou, Z.X.; Tan, G.S. New cytotoxic biflavones from Selaginella doederleinii. Nat. Prod. Res. 2021, 35, 930–936. [Google Scholar] [CrossRef]
- Ashiru, M.A.; Ogunyemi, S.O.; Temionu, O.R.; Ajibare, A.C.; Cicero-Mfon, N.C.; Ihekuna, O.A.; Jagun, M.O.; Abdulmumin, L.; Adisa, Q.K.; Asibor, Y.E.; et al. Identification of EGFR inhibitors as potential agents for cancer therapy: Pharmacophore-based modeling, molecular docking, and molecular dynamics investigations. J. Mol. Model. 2023, 29, 128. [Google Scholar] [CrossRef]
- Huang, L.; Wang, Z.; Wang, F.; Wang, S.; Wang, D.; Gao, M.; Li, H.; Song, M.; Zhang, X. Triterpenoids from the Leaves of Diospyros digyna and Their PTP1B Inhibitory Activity. Molecules 2024, 29, 1640. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, Q. Simultaneous Extraction and Analysis of Seven Major Saikosaponins from Bupleuri Radix and the Exploration of Antioxidant Activity and Its Mechanism. Molecules 2023, 28, 5872. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 19, 22. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Lobato-Tapia, C.A.; Moreno-Hernández, Y.; Olivo-Vidal, Z.E. In Silico Studies of Four Compounds of Cecropia obtusifolia against Malaria Parasite. Molecules 2023, 28, 6912. [Google Scholar] [CrossRef]
- Filipe, H.A.L.; Loura, L.M.S. Molecular Dynamics Simulations: Advances and Applications. Molecules 2022, 27, 2105. [Google Scholar] [CrossRef]
- Hu, X.; Zeng, Z.; Zhang, J.; Wu, D.; Li, H.; Geng, F. Molecular dynamics simulation of the interaction of food proteins with small molecules. Food Chem. 2023, 405, 134824. [Google Scholar] [CrossRef]
- Ouyang, J.; Hu, N.; Wang, H. Isolation, Purification and Tyrosinase Inhibitory Activity of Anthocyanins and Their Novel Degradation Compounds from Solanum tuberosum L. Molecules 2024, 29, 1492. [Google Scholar] [CrossRef]
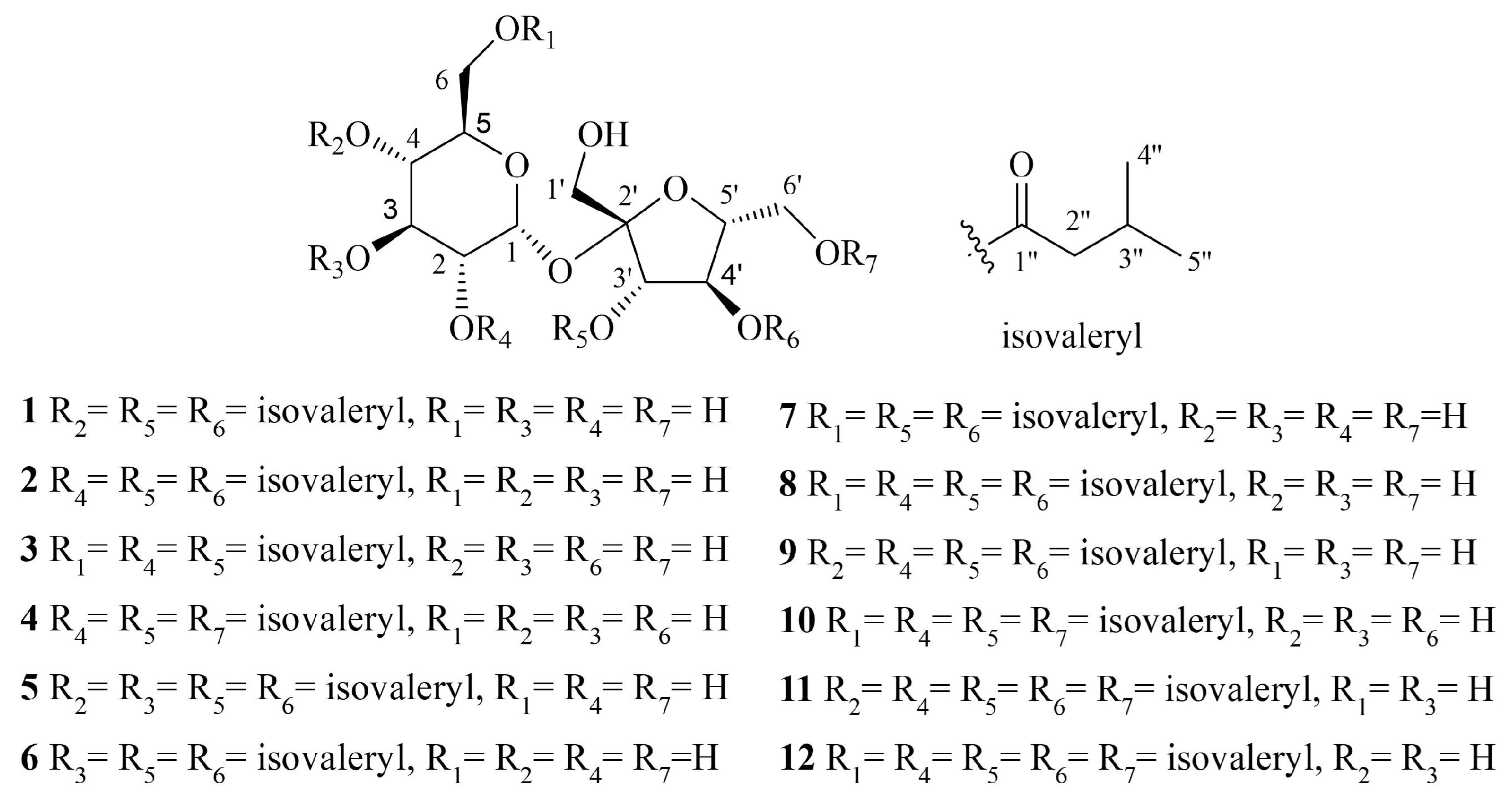
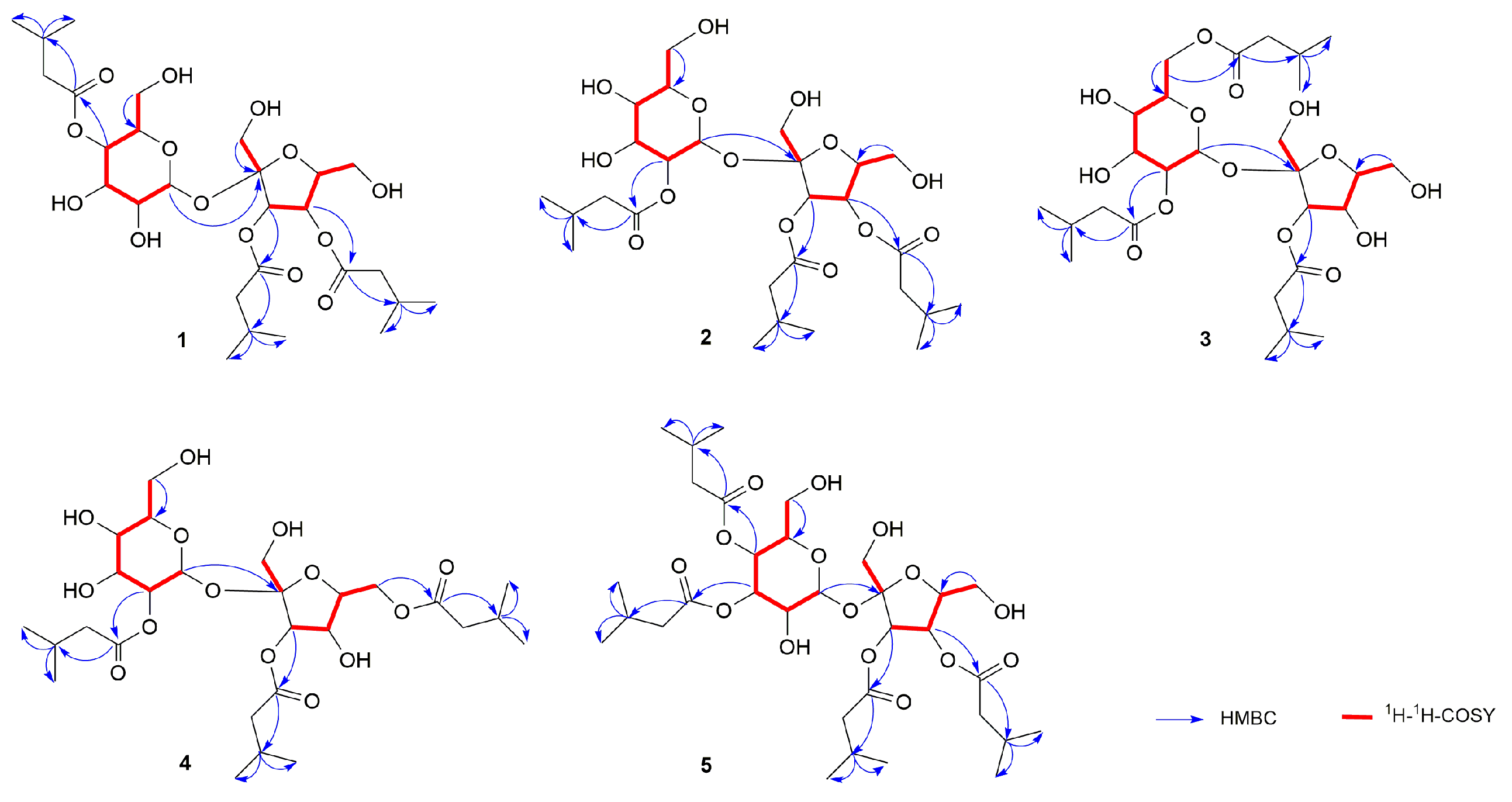
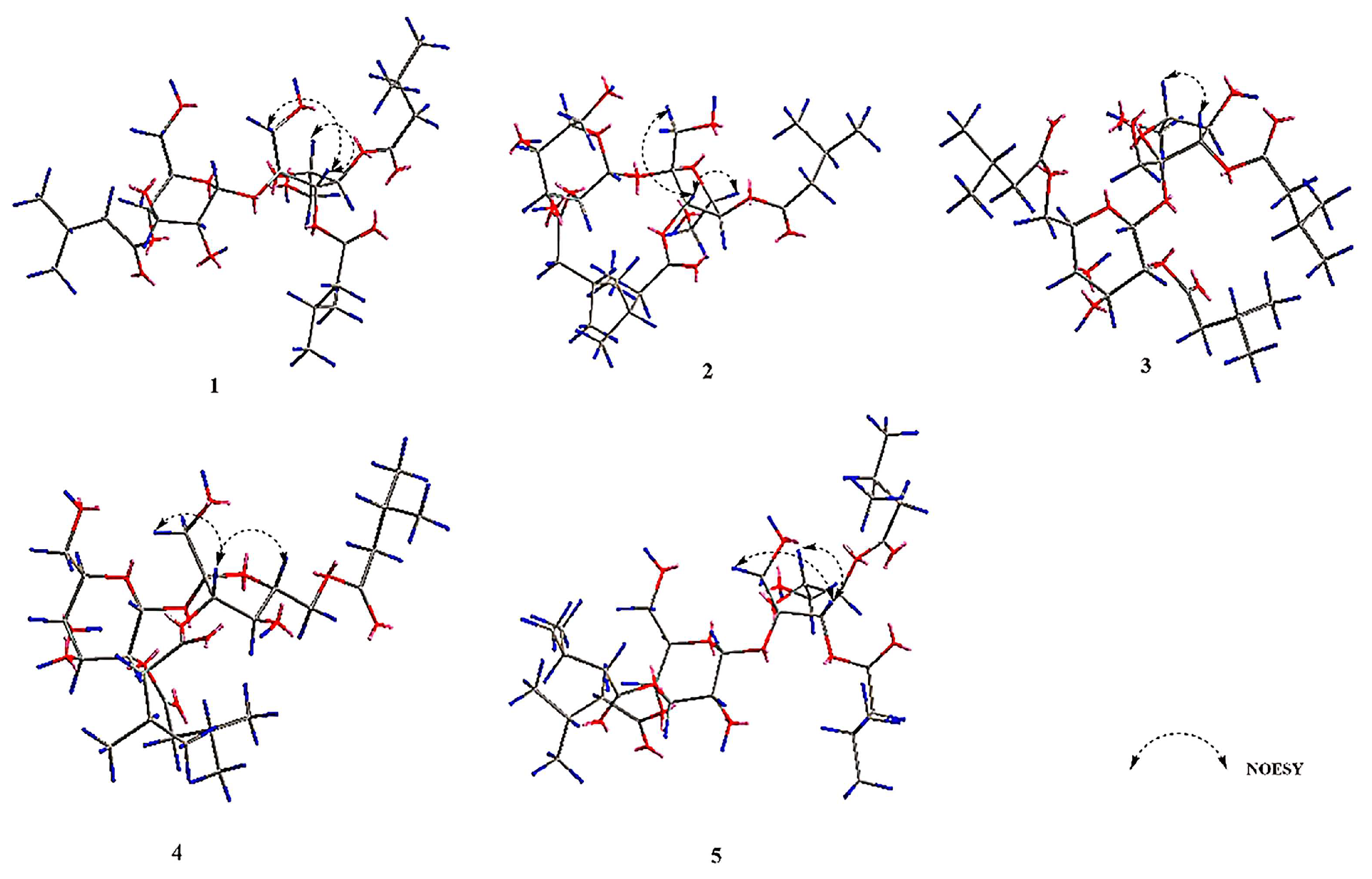

| No | 1 a | 2 a | 3 b | 4 b | 5 b |
|---|---|---|---|---|---|
| 1 | 5.53 (d, 3.5) | 5.54 (d, 3.7) | 5.50 (d, 3.8) | 5.50 (d, 3.8) | 5.53 (d, 3.5) |
| 2 | 3.73 m | 4.77 (dd, 10.1, 3.7) | 4.63 (dd, 10.1, 3.8) | 4.62 (dd, 10.2, 3.8) | 3.73 m |
| 3 | 3.81 m | 3.99 m | 3.73 m | 3.77 m | 5.32 (t, 9.8) |
| 4 | 4.86 (t, 9.6) | 3.59 m | 3.38 (t, 9.1) | 3.44 m | 5.08 (t, 9.8) |
| 5 | 4.00 m | 3.95 m | 4.01 m | 3.86 m | 4.16 m |
| 6 | 3.75 m | 3.88 m | 4. 45 (dd, 12.0, 2.0) | 3.84 m | 3.75 m |
| 3.58 m | 3.82 m | 4.16 (dd, 12.0, 5.3) | 3.79 m | 3.58 m | |
| 1′ | 3.65 m | 3.88 m | 3.35 (d, 11.8) | 3.50 m | 3.79 m |
| 3.70 m | 3.72 m | 3.46 (d, 11.8) | 3.37 m | 3.77 m | |
| 3′ | 5.37 (d, 5.9) | 5.48 (d, 7.4) | 5.43 (d, 8.6) | 5.45 (d, 8.6) | 5.64 (d, 7.2) |
| 4′ | 5.43 (t, 5.9) | 5.45 (t, 7.1) | 4.23 (t, 8.6) | 4.32 (t, 8.5) | 5.48 (t, 7.3) |
| 5′ | 4.08 m | 4.08 m | 3.88 m | 4.08 m | 4.13 m |
| 6′ | 3.78 m | 3.63 m | 3.77 m | 4.35m | 3.56 m |
| 3.67 m | 3.65 (d, 12.4) | ||||
| 2″ | 2.21–2.31 m | 2.15–2.42 m | 2.18–2.36 m | 2.19–2.32 m | 2.30–2.22 m |
| 3″ | 2.06–2.16 m | 1.98–2.13 m | 2.14–2.16 m | 2.04–2.18 m | 2.09–2.02 m |
| 4″/5″ | 0.89–1.02 m | 0.99–0.90 m | 0.96–1.02 m | 0.95–1.01 m | 1.02–0.96 m |
| No. | 1 a | 2 a | 3 b | 4 b | 5 b |
|---|---|---|---|---|---|
| 1 | 92.0 | 90.7 | 90.6 | 90.6 | 93.1 |
| 2 | 72.1 | 72.3 | 73.8 | 74.1 | 70.9 |
| 3 | 71.9 | 72.9 | 72.2 | 72.3 | 74.0 |
| 4 | 70.3 | 70.4 | 71.7 | 71.6 | 69.7 |
| 5 | 72.2 | 71.5 | 71.9 | 74.1 | 72.2 |
| 6 | 61.2 | 61.4 | 64.4 | 62.4 | 61.3 |
| 1′ | 61.1 | 62.0 | 64.4 | 64.3 | 63.2 |
| 2′ | 104.5 | 104.6 | 104.9 | 105.1 | 105.4 |
| 3′ | 77.7 | 76.1 | 78.1 | 77.8 | 77.1 |
| 4′ | 74.2 | 73.5 | 73.6 | 74.2 | 75.8 |
| 5′ | 82.0 | 81.4 | 84.3 | 80.9 | 82.5 |
| 6′ | 64.9 | 63.8 | 63.8 | 65.9 | 64.6 |
| 1″ | 173.7 | 173.4 | 174.7 | 174.7 | 174.0 |
| 172.6 | 172.6 | 174.2 | 174.1 | 173.8 | |
| 172.3 | 172.5 | 174.1 | 174.0 | 173.4 | |
| 173.3 | |||||
| 2″ | 43.5 | 43.2 | 44.1 × 3 | 44.1 | 44.2 × 2 |
| 43.1 × 2 | 43.1 | 44.0 × 2 | 43.9 × 2 | ||
| 42.8 | |||||
| 3″ | 25.9 | 25.7 × 2 | 27.0 | 27.0 | 26.8 |
| 25.7 × 2 | 25.5 | 26.9 | 26.8 | 26.6 × 2 | |
| 26.5 | 26.7 | 26.5 | |||
| 4″/5″ | 22.6 × 2 | 22.4 × 3 | 22.9 | 22.9 | 22.9 × 2 |
| 22.5 | 22.3 × 3 | 22.8 × 3 | 22.8 × 3 | 22.8 × 2 | |
| 22.4 × 3 | 22.7 × 2 | 22.7 × 2 | 22.7 × 2 | ||
| 22.6 × 2 |
| IC50 (μM) | |||||
|---|---|---|---|---|---|
| No. | Hct116 | A549 | No. | Hct116 | A549 |
| 1 | 18.77 ± 1.56 | 8.36 ± 0.77 | 8 | 20.10 ± 0.26 | 22.08 ± 2.19 |
| 2 | 7.49 ± 0.48 | 15.60 ± 0.53 | 9 | 24.75 ± 0.24 | 18.25 ± 1.98 |
| 3 | 21.75 ± 0.24 | 12.26 ± 1.17 | 10 | >50 | 40.50 ± 0.76 |
| 4 | 9.03 ± 0.21 | 21.95 ± 0.33 | 11 | >50 | >50 |
| 5 | 22.06 ± 0.68 | 30.58 ± 2.22 | 12 | >50 | 32.60 ± 0.69 |
| 6 | 13.49 ± 1.45 | 7.10 ± 0.52 | Doxorubicin | 2.14 ± 1.08 | 1.78 ± 0.56 |
| 7 | 21.65 ± 0.40 | 18.55 ± 1.56 | |||
| Compound 2-FGFR3 | Compound 7-FGFR3 | Compound 8-FGFR3 | |
|---|---|---|---|
| VDWAALS | −54.70 ± 0.04 | −46.64 ± 1.04 | −59.22 ± 1.40 |
| ΔEEl | −40.18 ± 5.61 | −25.16 ± 1.60 | −30.25 ± 2.92 |
| ΔEGB | 58.51 ± 2.13 | 41.21 ± 0.29 | 51.72 ± 1.43 |
| ΔEsurf | −8.06 ± 0.09 | −6.64 ± 0.06 | −8.12 ± 0.14 |
| ΔGgas | −94.88 ± 5.61 | −71.80 ± 1.91 | −89.47 ± 3.24 |
| ΔGsolvation | 50.45 ± 2.13 | 34.57 ± 0.30 | 43.60 ± 1.44 |
| ΔTotal | −44.43 ± 6.00 | −37.23 ± 1.93 | −45.87 ± 3.55 |
| Compound 2-BRAF | Compound 4-BRAF | Compound 6-BRAF | |
|---|---|---|---|
| VDWAALS | −59.33 ± 0.60 | −61.31 ± 1.73 | −57.46 ± 0.31 |
| ΔEEl | −29.46 ± 2.19 | −37.77 ± 3.24 | −36.74 ± 2.05 |
| ΔEGB | 53.19 ± 0.49 | 55.22 ± 2.02 | 51.97 ± 0.21 |
| ΔEsurf | −8.59 ± 0.01 | −8.07 ± 0.00 | −8.01 ± 0.07 |
| ΔGgas | −88.79 ± 2.27 | −99.08 ± 3.68 | −94.19 ± 2.08 |
| ΔGsolvation | 44.60 ± 0.49 | 47.15 ± 2.02 | 43.96 ± 0.23 |
| ΔTotal | −44.19 ± 2.33 | −51.92 ± 4.19 | −50.24 ± 2.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wang, Z.; Sun, Y.; Zhu, M.; Jiang, Y.; Bai, H.; Yang, B.; Kuang, H. Isovaleryl Sucrose Esters from Atractylodes japonica and Their Cytotoxic Activity. Molecules 2024, 29, 3069. https://doi.org/10.3390/molecules29133069
Wang Y, Wang Z, Sun Y, Zhu M, Jiang Y, Bai H, Yang B, Kuang H. Isovaleryl Sucrose Esters from Atractylodes japonica and Their Cytotoxic Activity. Molecules. 2024; 29(13):3069. https://doi.org/10.3390/molecules29133069
Chicago/Turabian StyleWang, Yimeng, Zhibin Wang, Yanping Sun, Mingtao Zhu, Yong Jiang, Haodong Bai, Bingyou Yang, and Haixue Kuang. 2024. "Isovaleryl Sucrose Esters from Atractylodes japonica and Their Cytotoxic Activity" Molecules 29, no. 13: 3069. https://doi.org/10.3390/molecules29133069
APA StyleWang, Y., Wang, Z., Sun, Y., Zhu, M., Jiang, Y., Bai, H., Yang, B., & Kuang, H. (2024). Isovaleryl Sucrose Esters from Atractylodes japonica and Their Cytotoxic Activity. Molecules, 29(13), 3069. https://doi.org/10.3390/molecules29133069






