Characterization of the Components and Metabolites of Achyranthes Bidentata in the Plasma and Brain Tissue of Rats Based on Ultrahigh Performance Liquid Chromatography–High-Resolution Mass Spectrometry (UHPLC–HR-MS)
Abstract
1. Introduction
2. Results
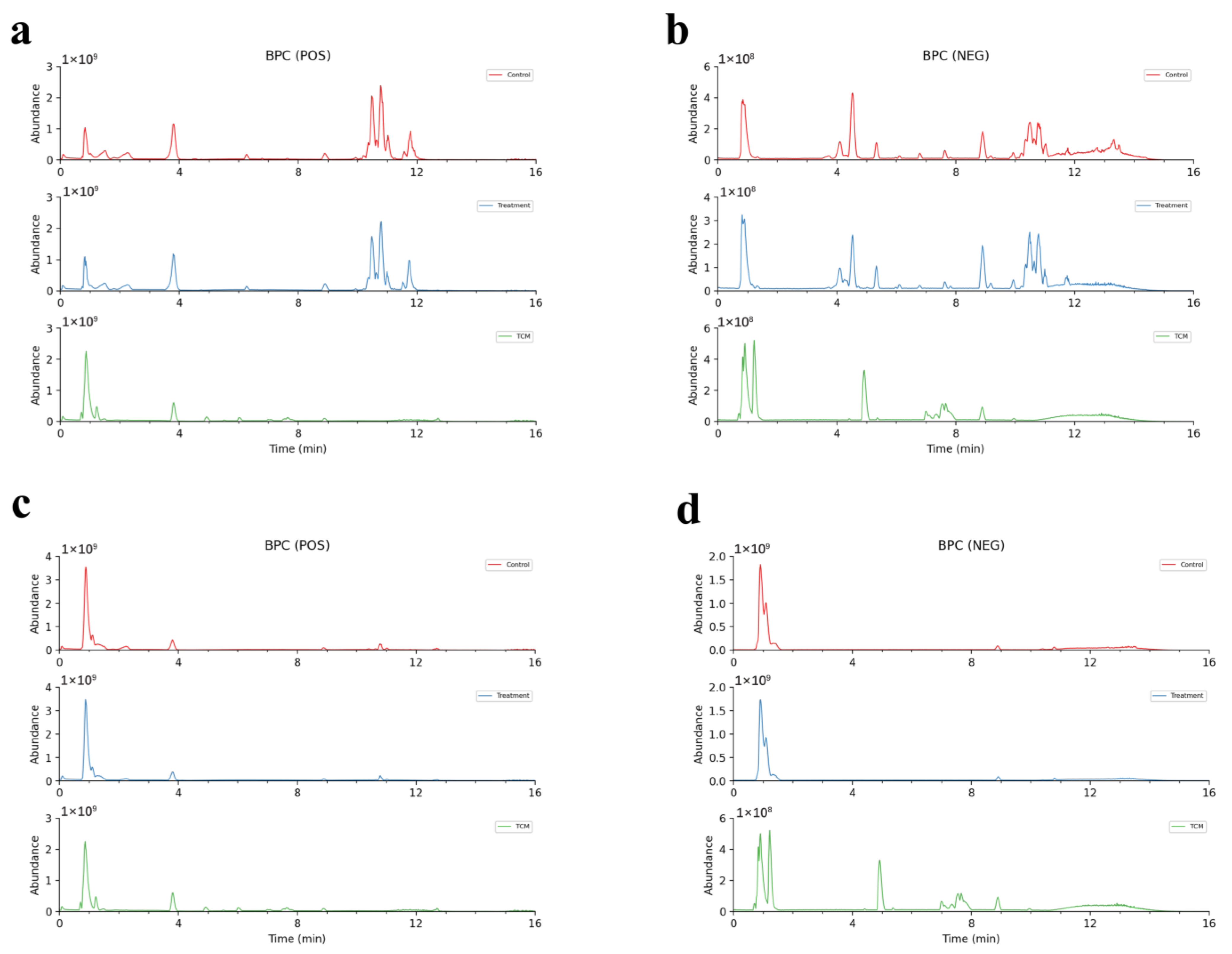
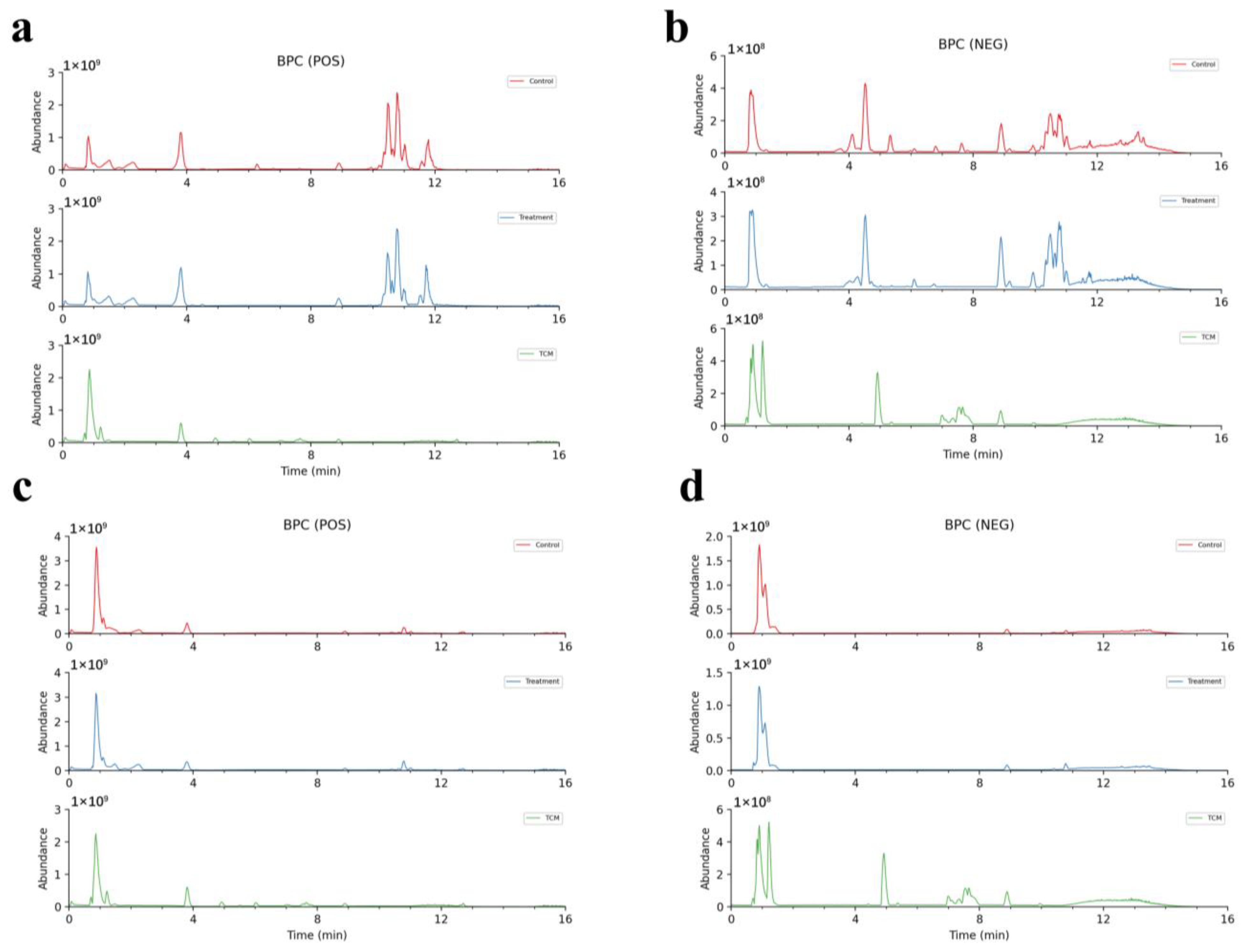
2.1. Model Verification
2.2. Component Identification and Analyses of TCM-AR
2.2.1. Sugars and Glycosides
2.2.2. Amino Acids and Peptides
2.2.3. Alkaloids
2.2.4. Terpenoids
2.2.5. Phenylpropanoids
2.2.6. Flavonoids
2.3. Analyses of AR Components Passing into Plasma and Brain after Administration
2.3.1. Analyses of the Prototype Components of AR into Plasma and Brain
2.3.2. Analyses of the Metabolites of AR in the Plasma and Brain
3. Discussion
4. Materials and Methods
4.1. Reagents and Materials
4.2. Preparation of Samples Achyranthes Bidentata (TCM-AR)
4.3. Preparation of Reference Substances
4.4. Animals
4.5. Preparation of Rat Plasma and Brain Tissue
4.6. Staining with 2,3,5-Triphenyltetrazolium Chloride (TTC)
4.7. Instruments and Experimental Conditions
4.8. Data Processing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AR | Achyranthes bidentata |
| BBB | Blood–brain barrier |
| BPC | Base peak chromatogram |
| CIRI | Cerebral ischemia-reperfusion injury |
| DDA | Data-dependent acquisition |
| EDTA | Ethylenediaminetetraacetic acid |
| EIC | Extracted ion chromatogram |
| HR–MS | High resolution-mass spectrometry |
| LC–MS | Liquid chromatography–mass spectrometry |
| MCAO | Middle cerebral artery occlusion |
| SD | Sprague Dawley |
| TTC | 2,3, 5-Triphenyltetrazolium chloride staining |
| TCM | Traditional Chinese medicine |
| UHPLC | Ultrahigh performance liquid chromatography |
| UHPLC–HR-MS | Ultrahigh performance liquid chromatography–high-resolution mass spectrometry |
| UPLC–MS/MS | ultra performance liquid chromatography/tandem mass spectrometry |
| UHPLC-Q Exactive Orbitrap-HRMS | Ultrahigh-performance liquid chromatography-Q Exactive Orbitrap-High resolution-mass spectrometry |
References
- Poustchi, F.; Amani, H.; Ahmadian, Z.; Niknezhad, S.V.; Mehrabi, S.; Santos, H.A.; Shahbazi, M.A. Combination Therapy of Killing Diseases by Injectable Hydrogels: From Concept to Medical Applications. Adv. Healthc. Mater. 2021, 10, e2001571. [Google Scholar] [CrossRef]
- Niu, L.; Jiang, S.W.; Wang, Y.; Peng, Y.; Fei, A.H.; Wang, H.R.; Li, Y.; Zhang, J.C.; Meaney, C.; Gao, C.J.; et al. Total cholesterol affects the outcome of patients with anterior cerebral artery-occluded acute ischemic stroke treated with thrombolysis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1504–1514. [Google Scholar] [CrossRef]
- Alawneh, K.Z.; Al Qawasmeh, M.; Raffee, L.A.; Abuzayed, B.; Bani Hani, D.A.; Abdalla, K.M.; Al-Mnayyis, A.M.; Fataftah, J. A snapshot of Ischemic stroke risk factors, sub-types, and its epidemiology: Cohort study. Ann. Med. Surg. 2020, 59, 101–105. [Google Scholar] [CrossRef]
- Lee, M.; Saver, J.L.; Liao, H.W.; Lin, C.H.; Ovbiagele, B. Pioglitazone for Secondary Stroke Prevention: A Systematic Review and Meta-Analysis. Stroke 2017, 48, 388–393. [Google Scholar] [CrossRef]
- Charidimou, A.; Turc, G.; Oppenheim, C.; Yan, S.; Scheitz, J.F.; Erdur, H.; Klinger-Gratz, P.P.; El-Koussy, M.; Takahashi, W.; Moriya, Y.; et al. Microbleeds, Cerebral Hemorrhage, and Functional Outcome After Stroke Thrombolysis. Stroke 2017, 48, 2084–2090. [Google Scholar] [CrossRef]
- Parikh, N.S.; Merkler, A.E.; Iadecola, C. Inflammation, Autoimmunity, Infection, and Stroke: Epidemiology and Lessons From Therapeutic Intervention. Stroke 2020, 51, 711–718. [Google Scholar] [CrossRef]
- Rufus, P.; Mohamed, N.; Shuid, A.N. Beneficial effects of traditional Chinese medicine on the treatment of osteoporosis on ovariectomised rat models. Curr. Drug Targets 2013, 14, 1689–1693. [Google Scholar] [CrossRef]
- Cai, Z.; Wang, L.; Qiu, S.L.; Tong, L.; Jin, M. Professor Jin Mei summed up the experience in the treatment of hypertension with the application of medicine differentiation. J. Chang. Univ. Chin. Med. 2023, 39, 732–736. [Google Scholar] [CrossRef]
- Sun, M.Z.; Zhao, M.L.; Zhang, L.Z. Progress of clinical application of Xuefu Zhuyu Decoction in traditional Chinese medicine. World’s Latest Med. Inf. Dig. 2019, 19, 81+83. [Google Scholar] [CrossRef]
- He, X.; Wang, X.; Fang, J.; Chang, Y.; Ning, N.; Guo, H.; Huang, L.; Huang, X. The genus Achyranthes: A review on traditional uses, phytochemistry, and pharmacological activities. J. Ethnopharmacol. 2017, 203, 260–278. [Google Scholar] [CrossRef]
- Li, J.; Qi, H.; Qi, L.W.; Yi, L.; Li, P. Simultaneous determination of main phytoecdysones and triterpenoids in radix achyranthis bidentatae by high-performance liquid chromatography with diode array-evaporative light scattering detectors and mass spectrometry. Anal. Chim. Acta 2007, 596, 264–272. [Google Scholar] [CrossRef]
- Ali, S.I.; Gopalakrishnan, B.; Venkatesalu, V. Pharmacognosy, Phytochemistry and Pharmacological Properties of Achillea millefolium L.: A Review. Phytother. Res. 2017, 31, 1140–1161. [Google Scholar] [CrossRef]
- Yan, F.; Li, F.; Liu, J.; Ye, S.; Zhang, Y.; Jia, J.; Li, H.; Chen, D.; Mo, X. The formulae and biologically active ingredients of Chinese herbal medicines for the treatment of atopic dermatitis. Biomed. Pharmacother. 2020, 127, 110142. [Google Scholar] [CrossRef]
- Gong, X.; Li, X.; Bo, A.; Shi, R.Y.; Li, Q.Y.; Lei, L.J.; Zhang, L.; Li, M.H. The interactions between gut microbiota and bioactive ingredients of traditional Chinese medicines: A review. Pharmacol. Res. 2020, 157, 104824. [Google Scholar] [CrossRef]
- Li, J.; Bai, Y.; Bai, Y.; Zhu, R.; Liu, W.; Cao, J.; An, M.; Tan, Z.; Chang, Y.X. Pharmacokinetics of Caffeic Acid, Ferulic Acid, Formononetin, Cryptotanshinone, and Tanshinone IIA after Oral Administration of Naoxintong Capsule in Rat by HPLC-MS/MS. Evid. Based Complement. Altern. Med. 2017, 2017, 9057238. [Google Scholar] [CrossRef]
- Zheng, Y.; Cao, C.; Lin, M.; Zhai, Y.; Ge, Z.; ShenTu, J.; Wu, L.; Hu, X. Identification and quantitative analysis of physalin D and its metabolites in rat urine and feces by liquid chromatography with triple quadrupole time-of-flight mass spectrometry. J. Sep. Sci. 2017, 40, 2355–2365. [Google Scholar] [CrossRef]
- Rashid, M.M.; Oh, H.A.; Lee, H.; Jung, B.H. Metabolite identification of AZD8055 in Sprague-Dawley rats after a single oral administration using ultra-performance liquid chromatography and mass spectrometry. J. Pharm. Biomed. Anal. 2017, 145, 473–481. [Google Scholar] [CrossRef]
- Tao, Y.; Du, Y.; Li, W.; Cai, B. Development and validation of an UHPLC-MS/MS approach for simultaneous quantification of five bioactive saponins in rat plasma: Application to a comparative pharmacokinetic study of aqueous extracts of raw and salt-processed Achyranthes bidentata. J. Pharm. Biomed. Anal. 2018, 151, 164–169. [Google Scholar] [CrossRef]
- Kawahara, Y.; Hoshino, T.; Morimoto, H.; Shinizu, T.; Narukawa, Y.; Fuchino, H.; Kawahara, N.; Kiuchi, F. LC-MS-based quantification method for Achyranthes root saponins. J. Nat. Med. 2016, 70, 102–106. [Google Scholar] [CrossRef]
- Qin, J.; Liao, C.N.; Chen, W.W.; Li, H.Y.; Su, J.; Wu, X.D.; He, J.B.; Zhang, G.H. New limonoids and quinolone alkaloids with cytotoxic and anti-platelet aggregation activities from Evodia rutaecarpa (Juss.) Benth. Fitoterapia 2021, 152, 104875. [Google Scholar] [CrossRef]
- Jaiswal, R.; Müller, H.; Müller, A.; Karar, M.G.; Kuhnert, N. Identification and characterization of chlorogenic acids, chlorogenic acid glycosides and flavonoids from Lonicera henryi L. (Caprifoliaceae) leaves by LC-MSn. Phytochemistry 2014, 108, 252–263. [Google Scholar] [CrossRef]
- Thal, S.C.; Neuhaus, W. The blood-brain barrier as a target in traumatic brain injury treatment. Arch. Med. Res. 2014, 45, 698–710. [Google Scholar] [CrossRef]
- Sivandzade, F.; Cucullo, L. Anti-Diabetic Countermeasures Against Tobacco Smoke-Dependent Cerebrovascular Toxicity: Use and Effect of Rosiglitazone. Int. J. Mol. Sci. 2019, 20, 4225. [Google Scholar] [CrossRef]
- Liu, Y.N.; Hu, M.T.; Qian, J.; Wang, Y.; Wang, S.F. Characterization of the chemical constituents of Jie-Geng-Tang and the metabolites in the serums and lungs of mice after oral administration by LC-Q-TOF-MS. Chin. J. Nat. Med. 2021, 19, 284–294. [Google Scholar] [CrossRef]
- Xu, T.; Li, S.; Sun, Y.; Pi, Z.; Liu, S.; Song, F.; Liu, Z. Systematically characterize the absorbed effective substances of Wutou Decoction and their metabolic pathways in rat plasma using UHPLC-Q-TOF-MS combined with a target network pharmacological analysis. J. Pharm. Biomed. Anal. 2017, 141, 95–107. [Google Scholar] [CrossRef]
- Cheng, Q.; Tong, F.; Shen, Y.; He, C.; Wang, C.; Ding, F. Achyranthes bidentata polypeptide k improves long-term neurological outcomes through reducing downstream microvascular thrombosis in experimental ischemic stroke. Brain Res. 2019, 1706, 166–176. [Google Scholar] [CrossRef]
- Cai, E.; Cheng, Q.; Yu, S.; Ding, F. Achyranthes bidentata polypeptide k enhances the survival, growth and axonal regeneration of spinal cord motor neurons in vitro. Neuroreport 2021, 32, 518–524. [Google Scholar] [CrossRef]
- Cheng, Q.; Shen, Y.; Cheng, Z.; Shao, Q.; Wang, C.; Sun, H.; Zhang, Q. Achyranthes bidentata polypeptide k suppresses neuroinflammation in BV2 microglia through Nrf2-dependent mechanism. Ann. Transl. Med. 2019, 7, 575. [Google Scholar] [CrossRef]
- Wang, Y.; Ge, X.; Yu, S.; Cheng, Q. Achyranthes bidentata polypeptide alleviates neurotoxicity of lipopolysaccharide-activated microglia via PI3K/Akt dependent NOX2/ROS pathway. Ann. Transl. Med. 2021, 9, 1522. [Google Scholar] [CrossRef]
- Shen, H.; Wu, X.; Zhu, Y.; Sun, H. Intravenous administration of achyranthes bidentata polypeptides supports recovery from experimental ischemic stroke in vivo. PLoS ONE 2013, 8, e57055. [Google Scholar] [CrossRef]
- Liu, P.; Li, R.; Antonov, A.A.; Wang, L.; Li, W.; Hua, Y.; Guo, H.; Wang, L.; Liu, P.; Chen, L.; et al. Discovery of Metabolite Biomarkers for Acute Ischemic Stroke Progression. J. Proteome Res. 2017, 16, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Hu, F.B.; Ruiz-Canela, M.; Clish, C.B.; Dennis, C.; Salas-Salvado, J.; Hruby, A.; Liang, L.; Toledo, E.; Corella, D.; et al. Metabolites of Glutamate Metabolism Are Associated With Incident Cardiovascular Events in the PREDIMED PREvención con DIeta MEDiterránea (PREDIMED) Trial. J. Am. Heart Assoc. 2016, 5, e003755. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Andjelkovic, A.V.; Zhu, L.; Yang, T.; Bennett, M.V.L.; Chen, J.; Keep, R.F.; Shi, Y. Blood-brain barrier dysfunction and recovery after ischemic stroke. Prog. Neurobiol. 2018, 163–164, 144–171. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Gan, L.; Ren, L.; Lin, Y.; Ma, C.; Lin, X. Factors influencing the blood-brain barrier permeability. Brain Res. 2022, 1788, 147937. [Google Scholar] [CrossRef]
- Jin, R.; Yang, G.; Li, G. Inflammatory mechanisms in ischemic stroke: Role of inflammatory cells. J. Leukoc. Biol. 2010, 87, 779–789. [Google Scholar] [CrossRef]
- Jickling, G.C.; Liu, D.; Ander, B.P.; Stamova, B.; Zhan, X.; Sharp, F.R. Targeting neutrophils in ischemic stroke: Translational insights from experimental studies. J. Cereb. Blood Flow. Metab. 2015, 35, 888–901. [Google Scholar] [CrossRef]
- Jin, Y.; Cheng, S.; Liu, R.; Yu, C.; Zhang, L.; Li, X.L.; Yan, G.; Zheng, M.; Zhe Min, J. Comprehensive characterization of the chemical composition of Lurong dabu decoction and its absorbed prototypes and metabolites in rat plasma using UHPLC-Q Exactive Orbitrap-HRMS. Food Res. Int. 2022, 161, 111852. [Google Scholar] [CrossRef]
- AlFaris, N.A.; Wabaidur, S.M.; Alothman, Z.A.; Altamimi, J.Z.; Aldayel, T.S. Fast and efficient immunoaffinity column cleanup and liquid chromatography-tandem mass spectrometry method for the quantitative analysis of aflatoxins in baby food and feeds. J. Sep. Sci. 2020, 43, 2079–2087. [Google Scholar] [CrossRef]

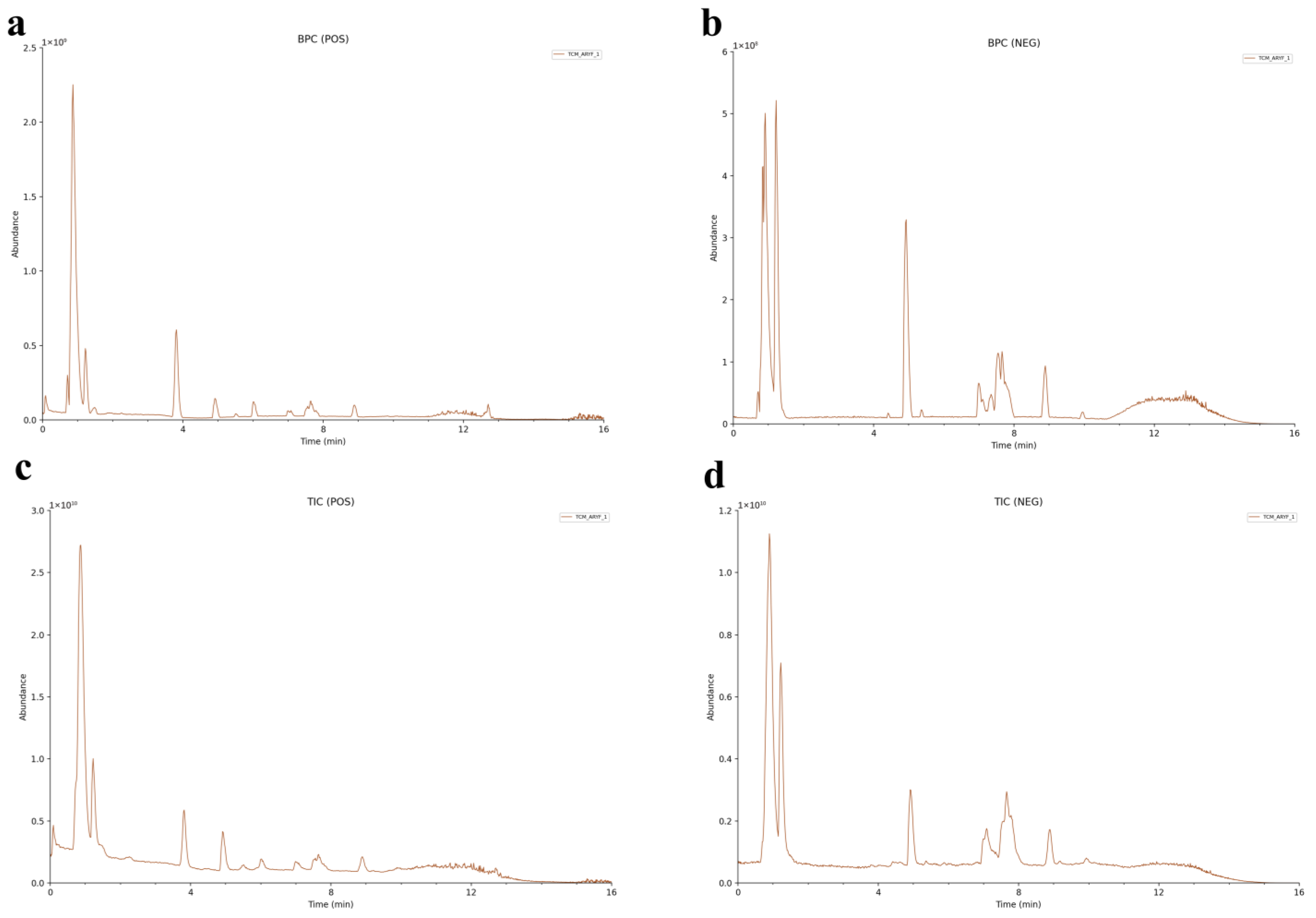



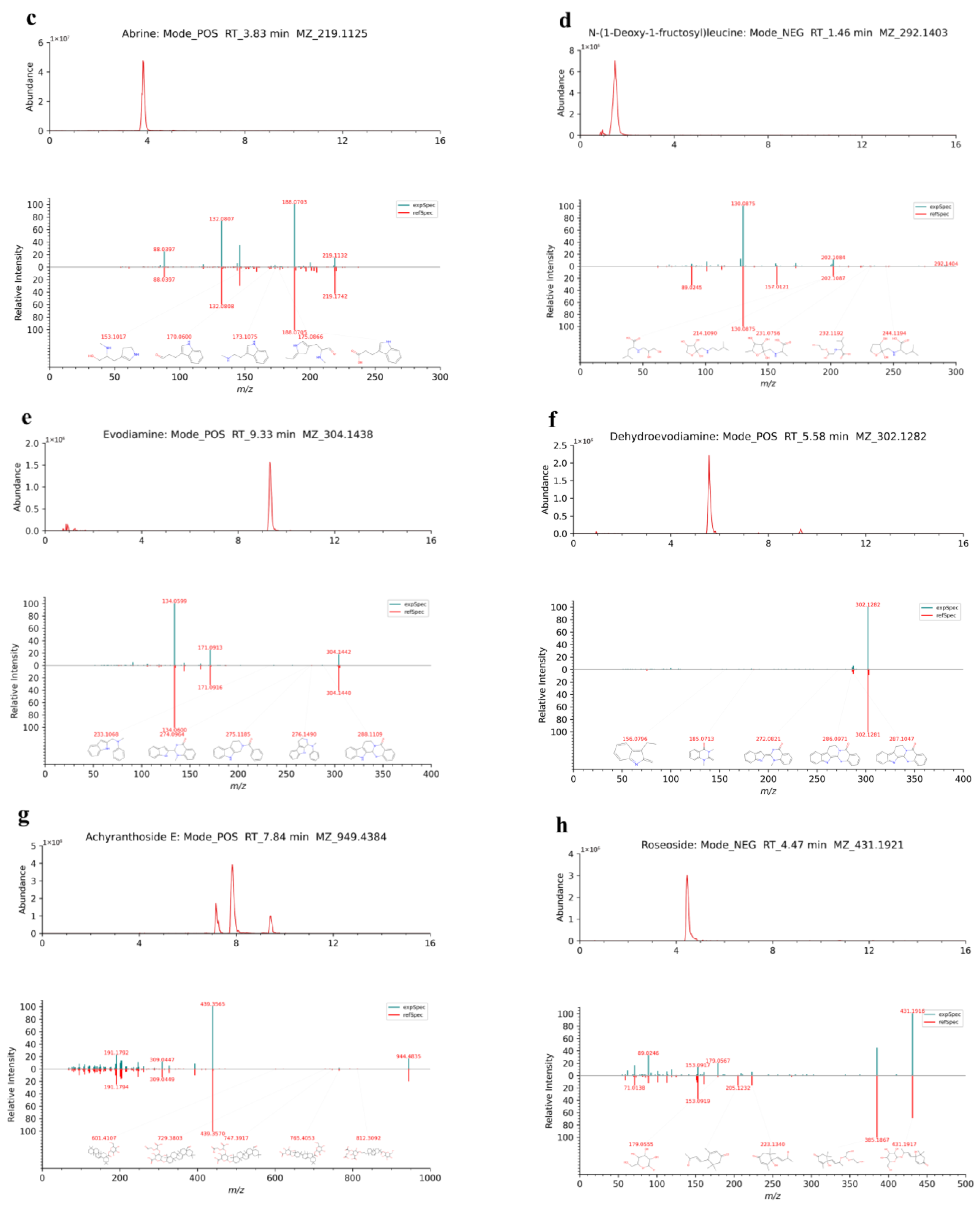

| NO | Adducts | RT (min) | Theoretical m/z | m/z | Mass Error (ppm) | Formula | Metabolites | Ratio of Peak Area % | Fragment Ions | InChIKey |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | [M+H]+ | 3.83 | 219.1128 | 219.1125 | −1.32 | C12H14N2O2 | Abrine | 0.149137 | 88.0397, 132.0807, 144.0807, 146.0598, 188.0703, 200.1278, 219.1132 | CZCIKBSVHDNIDH-NSHDSACASA-N |
| 2 | [M+H]+ | 4.07 | 295.1288 | 295.1286 | −0.86 | C14H18N2O5 | gamma-Glu-Phe | 0.004603 | 84.045, 120.0809, 130.0501, 136.0756, 166.0861, 186.0913, 232.096, 278.1018, 295.1286 | XHHOHZPNYFQJKL-QWRGUYRKSA-N |
| 3 | [M-H]− | 1.34 | 115.0037 | 115.0039 | 1.49 | C4H4O4 | Fumaric acid | 0.008435 | 71.014, 97.9314, 114.9342, 115.0039 | VZCYOOQTPOCHFL-OWOJBTEDSA-N |
| 4 | [M+H]+ | 9.33 | 304.1444 | 304.1438 | −1.97 | C19H17N3O | Evodiamine | 0.009191 | 134.0599, 171.0913, 304.1442 | TXDUTHBFYKGSAH-SFHVURJKSA-N |
| 5 | [M-H, 2M-H]− | 6.1 | 431.0984 | 431.0983 | −0.13 | C21H20O10 | Emodin-8-glucoside | 0.004102 | 269.0454, 431.0979 | HSWIRQIYASIOBE-JNHRPPPUSA-N |
| 6 | [M-H]− | 7.43 | 229.1445 | 229.1446 | 0.24 | C12H22O4 | Dodecanedioic acid | 0.001865 | 99.9259, 112.9857, 116.9293, 130.0875, 167.144, 211.1339, 228.1591, 229.1444 | TVIDDXQYHWJXFK-UHFFFAOYSA-N |
| 7 | [M+H]+ | 5.58 | 302.1288 | 302.1282 | −2.03 | C19H15N3O | Dehydroevodiamine | 0.009602 | 287.1049, 302.1282 | VXHNSVKJHXSKKM-UHFFFAOYSA-N |
| 8 | [M+FA-H]− | 5.37 | 277.0679 | 277.0669 | −3.58 | C8H12N2O6 | Kifunensine | 0.010155 | 209.0795 | OIURYJWYVIAOCW-UHFFFAOYSA-N |
| 9 | [M+H-H2O]+ | 0.71 | 163.0601 | 163.0599 | −0.9 | C6H12O6 | d-Mannose | 0.007622 | 107.0494, 117.0547, 118.0350, 119.0491, 123.0404, 127.0389, 128.0706, 141.0506, 162.0759, 163.0386 | WQZGKKKJIJFFOK-PQMKYFCFSA-N |
| 10 | [M-H]− | 3.87 | 259.1299 | 259.1299 | −0.06 | C11H20N2O5 | gamma-Glutamylleucine | 0.001003 | 128.0355, 130.0873, 146.9383, 173.9944, 179.0359, 197.1286, 223.1078, 241.1191, 259.0633, 259.1295 | MYFMARDICOWMQP-YUMQZZPRSA-N |
| 11 | [M-H2O-H, M-H]− | 1.23 | 191.0197 | 191.0199 | 1.06 | C6H8O7 | Citric acid | 1.885035 | 85.0296, 111.0089, 128.0353, 129.0195 | KRKNYBCHXYNGOX-UHFFFAOYSA-N |
| 12 | [M-H]− | 5.37 | 187.0976 | 187.0978 | 0.95 | C9H16O4 | Azelaic acid | 0.09136 | 97.0658, 125.0973, 187.0978 | BDJRBEYXGGNYIS-UHFFFAOYSA-N |
| 13 | [M+H-H2O]+ | 9.35 | 471.3469 | 471.3464 | −0.96 | C30H48O5 | Arjunolic acid | 0.000739 | 233.1534, 277.1783, 407.3284, 413.3041, 425.3384, 435.3241, 441.3327, 453.3359, 470.3333, 471.3466 | RWNHLTKFBKYDOJ-DDHMHSPCSA-N |
| 14 | [M-H, M+FA-H]− | 0.85 | 195.051 | 195.0511 | 0.59 | C5H10O5 | Arabinose | 0.568192 | 75.0088, 80.9170, 85.0294, 87.0088, 89.0245, 120.9545, 121.0294, 149.0092, 149.0244, 149.0455 | SRBFZHDQGSBBOR-SQOUGZDYSA-N |
| 15 | [M-H]− | 0.91 | 157.0367 | 157.0369 | 0.92 | C4H6N4O3 | Allantoin | 0.079695 | 71.0254, 89.0246, 96.9223, 96.9602, 97.0045, 114.031, 140.0101, 157.0367 | POJWUDADGALRAB-UHFFFAOYSA-N |
| 16 | [M+H]+ | 1.28 | 268.104 | 268.1036 | −1.51 | C10H13N5O4 | Adenosine | 0.091131 | 136.0618, 268.1037 | OIRDTQYFTABQOQ-KQYNXXCUSA-N |
| 17 | [M-H]− | 1.55 | 182.0459 | 182.0461 | 1.15 | C8H9NO4 | 4-Pyridoxic acid | 0.001768 | 112.9858, 115.0401, 119.0353, 121.0447, 124.0072, 138.0561, 138.9071, 163.0612, 181.0720, 182.0457 | HXACOUQIXZGNBF-UHFFFAOYSA-N |
| 18 | [M+H]+ | 1.22 | 140.0342 | 140.0341 | −0.7 | C6H5NO3 | 3-Hydroxypicolinic acid | 0.077143 | 74.0971, 112.0395, 140.034 | BRARRAHGNDUELT-UHFFFAOYSA-N |
| 19 | [M+H]+ | 4.92 | 481.316 | 481.3152 | −1.68 | C27H44O7 | 25R-Inokosterone | 3.047055 | 165.1271, 173.0958, 249.1460, 303.1949, 371.2210, 409.2725, 427.2835, 445.2941, 463.3046, 481.3154 | JQNVCUBPURTQPQ-GYVHUXHASA-N |
| 20 | [M-H]− | 4.12 | 353.0878 | 353.0879 | 0.15 | C16H18O9 | Chlorogenic acid | 0.001511 | 191.0561, 353.0881 | CWVRJTMFETXNAD-JUHZACGLSA-N |
| 21 | [M-H]− | 12.13 | 271.2279 | 271.2278 | −0.31 | C16H32O3 | 2-Hydroxyhexadecanoic acid | 0.002268 | 225.2223, 271.2277 | JGHSBPIZNUXPLA-HNNXBMFYSA-N |
| 22 | [M-H, M+FA-H, 2M-H]− | 0.83 | 179.0561 | 179.0563 | 1.1 | C6H12O6 | Glucose | 1.037797 | 72.9931, 77.0547, 85.0295, 89.0246, 95.0132, 101.0243, 113.0249, 119.0352, 121.0445, 179.0567 | GZCGUPFRVQAUEE-VANKVMQKSA-N |
| 23 | [M+H, M+Na]+ | 4.09 | 247.1441 | 247.1439 | −0.76 | C14H18N2O2 | Hypaphorine | 0.000864 | 59.0611, 60.0816, 85.029, 145.9325, 146.06, 188.0705, 207.1015, 246.1242, 246.1698, 247.144 | AOHCBEAZXHZMOR-ZDUSSCGKSA-N |
| 24 | [M-H]− | 6.8 | 215.1289 | 215.1289 | 0.22 | C11H20O4 | Undecanedioic acid | 0.004839 | 153.1286, 197.1187, 215.0101, 215.129 | LWBHHRRTOZQPDM-UHFFFAOYSA-N |
| 25 | [M+H]+ | 4.52 | 497.3109 | 497.3103 | −1.24 | C27H44O8 | Turkesterone | 0.124028 | 345.2051, 371.2209, 385.2366, 387.2155, 407.2562, 425.2677, 443.2786, 461.2887, 479.3007, 497.3104 | WSBAGDDNVWTLOM-XHZKDPLLSA-N |
| 26 | [M+NH4]+ | 0.93 | 360.1501 | 360.1492 | −2.31 | C12H22O11 | Turanose | 0.478689 | 69.0342, 85.0288, 97.0287, 127.0389, 145.0493, 163.0597, 180.0863, 289.0909, 325.1123 | RULSWEULPANCDV-PIXUTMIVSA-N |
| 27 | [M+FA-H]− | 4.68 | 186.1135 | 186.1138 | 1.36 | C8H15NO | Tropine | 0.001222 | 79.9574, 80.9652, 97.0658, 107.0503, 125.0973, 142.1239, 186.1136 | CYHOMWAPJJPNMW-DHBOJHSNSA-N |
| 28 | [M-H]− | 8.48 | 243.1602 | 243.1602 | 0.11 | C13H24O4 | Tridecanedioic acid | 0.001745 | 146.9611, 174.9559, 181.1595, 225.1494, 242.1766, 243.1597 | DXNCZXXFRKPEPY-UHFFFAOYSA-N |
| 29 | [M-H]− | 9.28 | 257.1758 | 257.176 | 0.82 | C14H26O4 | Tetradecanedioic acid | 0.000628 | 195.1759, 239.1652, 257.1758 | HQHCYKULIHKCEB-UHFFFAOYSA-N |
| 30 | [M-H]− | 4.85 | 173.0819 | 173.0821 | 0.99 | C8H14O4 | Suberic acid | 0.008054 | 93.0346, 99.9259, 104.9538, 111.0817, 115.9207, 116.9283, 129.0923, 130.0875, 172.0978, 173.0820 | TYFQFVWCELRYAO-UHFFFAOYSA-N |
| 31 | [M+NH4]+ | 9.64 | 302.3054 | 302.3049 | −1.49 | C18H36O2 | Stearic acid | 0.007109 | 88.0763, 91.0547, 302.305 | QIQXTHQIDYTFRH-UHFFFAOYSA-N |
| 32 | [M-H]− | 10.29 | 285.2071 | 285.2071 | −0.16 | C16H30O4 | Hexadecanedioic acid | 0.002157 | 223.2067, 267.197, 285.2072 | QQHJDPROMQRDLA-UHFFFAOYSA-N |
| 33 | [M-H, M+FA-H]− | 0.8 | 711.2201 | 711.2198 | −0.33 | C24H42O21 | Stachyose | 3.048184 | 89.0245, 101.0244, 113.0246, 125.0241, 143.0347, 161.0451, 179.0560, 221.0663, 341.1080, 665.2141 | UQZIYBXSHAGNOE-XNSRJBNMSA-N |
| 34 | [M+H, M+Na]+ | 8.97 | 343.1176 | 343.1169 | −2.01 | C19H18O6 | Scutellarein tetramethyl ether | 0.154561 | 157.0128, 313.0701, 343.1171 | URSUMOWUGDXZHU-UHFFFAOYSA-N |
| 35 | [M-H]− | 5.59 | 137.0244 | 137.0246 | 1.42 | C7H6O3 | Salicylic acid | 0.021021 | 93.0348, 137.0247 | YGSDEFSMJLZEOE-UHFFFAOYSA-N |
| 36 | [M+FA-H]− | 4.47 | 431.1923 | 431.1921 | −0.38 | C19H30O8 | Roseoside | 0.027857 | 71.0140, 89.0246, 101.0249, 113.0244, 119.0355, 153.0917, 179.0567, 223.1350, 385.1871, 431.1916 | SWYRVCGNMNAFEK-MHXFFUGFSA-N |
| 37 | [M+H]+ | 1.22 | 130.0499 | 130.0499 | 0.07 | C5H7NO3 | Pyroglutamic acid | 1.325712 | 84.045, 84.0814, 87.0046, 96.0098, 98.5125, 113.9639, 130.05 | ODHCTXKNWHHXJC-VKHMYHEASA-N |
| 38 | [M-H, M+FA-H]− | 4.82 | 435.1297 | 435.1296 | −0.28 | C20H22O8 | Polydatin | 0.001234 | 185.0606, 227.0715 | HSTZMXCBWJGKHG-CUYWLFDKSA-N |
| 39 | [M+H]+ | 4.96 | 153.0546 | 153.0545 | −1.03 | C8H8O3 | Isovanillin | 0.022396 | 111.0205, 111.0442, 111.9685, 125.0597, 126.0548, 129.9789, 134.0598, 136.0755, 152.0704, 153.0545 | JVTZFYYHCGSXJV-UHFFFAOYSA-N |
| 40 | [M+H-H2O, M+NH4]+ | 0.92 | 684.2557 | 684.2544 | −1.88 | C24H42O21 | Isomaltotetraose | 0.761064 | 162.0757, 163.0597, 180.0863, 259.0810, 271.0804, 289.0912, 325.1121, 343.1226, 487.1653, 684.2551 | DFKPJBWUFOESDV-NGZVDTABSA-N |
| 41 | [2M-H]− | 10.53 | 291.1615 | 291.1601 | −4.77 | C9H10N2 | Indole-3-methanamine | 0.013972 | 219.1752, 235.171, 263.1653, 291.1603 | JXYGLMATGAAIBU-UHFFFAOYSA-N |
| 42 | [M-H]− | 6.01 | 201.1132 | 201.1134 | 0.89 | C10H18O4 | Sebacic acid | 0.008393 | 89.0245, 116.9289, 121.066, 139.113, 183.103, 201.0226, 201.1131 | CXMXRPHRNRROMY-UHFFFAOYSA-N |
| 43 | [M+H]+ | 14.88 | 338.3417 | 338.3411 | −1.84 | C22H43NO | 13-Docosenamide | 0.062269 | 69.0705, 71.0861, 81.0704, 83.0861, 95.0858, 97.1015, 111.1174, 303.3048, 321.3154, 338.3410 | UAUDZVJPLUQNMU-UHFFFAOYSA-N |
| 44 | [M-H]− | 1.39 | 282.0844 | 282.0844 | −0.03 | C10H13N5O5 | Guanosine | 0.009596 | 133.0159, 150.0421, 282.0841 | NYHBQMYGNKIUIF-UUOKFMHZSA-N |
| 45 | [M-H, M+FA-H]− | 1.21 | 243.0623 | 243.0622 | −0.3 | C9H12N2O6 | 1-β-d-Arabinofuranosyluracil | 0.012177 | 128.0360, 152.0354, 153.0305, 158.6527, 174.8874, 185.9930, 200.0569, 213.3648, 216.4419, 243.0624 | DRTQHJPVMGBUCF-CCXZUQQUSA-N |
| 46 | [M+H-H2O, M+H]+ | 0.89 | 147.0764 | 147.0763 | −1 | C5H10N2O3 | l-Glutamine | 0.50798 | 83.0609, 84.045, 84.0813, 129.0659 | ZDXPYRJPNDTMRX-VKHMYHEASA-N |
| 47 | [M-H]− | 0.78 | 154.0622 | 154.0624 | 1.45 | C6H9N3O2 | l-Histidine | 0.00218 | 93.0459, 94.9252, 96.9222, 96.9602, 96.969, 110.0725, 137.0358, 154.0623 | HNDVDQJCIGZPNO-YFKPBYRVSA-N |
| 48 | [M+H]+ | 1.5 | 132.1019 | 132.1019 | −0.07 | C6H13NO2 | l-Leucine | 0.482585 | 69.0706, 72.9378, 86.0969, 88.0047, 97.0099, 113.9641, 132.1021 | ROHFNLRQFUQHCH-YFKPBYRVSA-N |
| 49 | [M+H-H2O, M+H]+ | 0.73 | 147.1128 | 147.1127 | −0.55 | C6H14N2O2 | l-Lysine | 0.008875 | 84.0449, 84.0813 | KDXKERNSBIXSRK-YFKPBYRVSA-N |
| 50 | [M+H]+ | 1.14 | 150.0583 | 150.0582 | −0.52 | C5H11NO2S | l-Methionine | 0.001225 | 56.0503, 61.0115, 74.0244, 74.0608, 76.0764, 87.0269, 102.0554, 104.0532, 133.0318, 150.0582 | FFEARJCKVFRZRR-BYPYZUCNSA-N |
| 51 | [M-H]− | 2.26 | 164.0717 | 164.0719 | 1.37 | C9H11NO2 | l-Phenylalanine | 0.002097 | 96.9602, 96.9693, 119.0502, 120.0456, 121.0293, 136.9325, 147.0453, 163.0616, 164.0355, 164.0723 | COLNVLDHVKWLRT-QMMMGPOBSA-N |
| 52 | [M+H, M+NH4]+ | 10.78 | 279.2319 | 279.2313 | −1.99 | C18H30O2 | Punicic acid | 0.044704 | 57.0706, 67.0548, 81.0703, 95.0859, 109.1014, 123.1167, 137.1319, 149.0231, 173.1325, 279.2314 | CUXYLFPMQMFGPL-MRZTUZPCSA-N |
| 53 | [M-H, M+FA-H]− | 0.78 | 873.2729 | 873.2725 | −0.46 | C30H52O26 | Maltopentaose | 2.750419 | 71.0140, 89.0245, 101.0246, 113.0244, 125.0212, 143.0350, 161.0451, 179.0564, 221.0666, 827.2667 | FJCUPROCOFFUSR-GMMZZHHDSA-N |
| 54 | [M-H, M+FA-H]− | 0.8 | 549.1672 | 549.1671 | −0.25 | C18H32O16 | Manninotriose | 3.098184 | 113.0244, 119.0348, 143.0353, 161.0453, 179.0562, 221.0661, 323.0975, 341.1084, 383.1195, 503.1615 | FZWBNHMXJMCXLU-YRBKNLIBSA-N |
| 55 | [M+H-H2O, M+H, M+K, M+Na]+ | 0.76 | 162.0761 | 162.0758 | −1.35 | C6H13NO5 | Mannosamine | 0.970436 | 101.0237, 102.0552, 103.0393, 114.0550, 115.0391, 116.0708, 126.0549, 127.0389, 144.0652, 162.0758 | MSWZFWKMSRAUBD-CBPJZXOFSA-N |
| 56 | [M+H]+ | 4.23 | 154.0499 | 154.0498 | −0.32 | C7H7NO3 | Methyl 5-hydroxypyridine-2-carboxylate | 0.021269 | 72.9378, 90.9481, 112.0395, 113.9638, 131.9742, 140.034, 154.0505 | YYAYXDDHGPXWTA-UHFFFAOYSA-N |
| 57 | [M-H]− | 1.36 | 117.0193 | 117.0195 | 1.81 | C4H6O4 | Methylmalonic acid | 0.050209 | 73.0296, 99.0089, 99.926, 116.9287, 117.0195 | ZIYVHBGGAOATLY-UHFFFAOYSA-N |
| 58 | [M-H]− | 1.46 | 292.1402 | 292.1403 | 0.37 | C12H23NO7 | N-(1-Deoxy-1-fructosyl)leucine | 0.051817 | 101.0246, 128.0356, 130.0875, 202.1084 | KGTRBDVOUPALMB-PPNLDZOPSA-N |
| 59 | [M-H2O-H, M-H]− | 0.96 | 133.0142 | 133.0145 | 1.63 | C4H6O5 | Malic acid | 1.602263 | 71.014, 114.9343, 115.0038 | BJEPYKJPYRNKOW-REOHCLBHSA-N |
| 60 | [M+NH4, M+Na]+ | 7.84 | 949.4404 | 949.4384 | −2.11 | C46H70O19 | Achyranthoside E | 0.392011 | 189.1633, 191.1792, 201.1634, 203.1791, 205.1948, 247.1689, 309.0447, 393.3502, 439.3565, 944.4835 | DVEJWYUSLPQXTD-UHFFFAOYSA-N |
| 61 | [M-H2O-H, M-H]− | 0.93 | 189.0041 | 189.0043 | 0.96 | C6H8O8 | Hydroxycitric acid | 0.480247 | 73.0296, 83.0139, 85.0296, 87.0088, 99.0088, 111.0088, 127.0038, 129.0195, 189.0041 | ZMJBYMUCKBYSCP-CVYQJGLWSA-N |
| 62 | [M-H]− | 9.9 | 313.2384 | 313.2384 | 0.04 | C18H34O4 | 12,13-DHOME | 0.06907 | 129.0922, 183.1393, 201.1133, 313.238 | CQSLTKIXAJTQGA-BTDPBSJTSA-N |
| 63 | [M+H-H2O, M+H]+ | 2.28 | 328.1391 | 328.1383 | −2.45 | C15H21NO7 | N-(1-Deoxy-1-fructosyl)phenylalanine | 0.231268 | 97.0288, 120.081, 127.0392, 132.0808, 166.0864, 178.0862, 264.1228, 292.118, 310.0899, 310.1281 | FAVRCIXPIVJIPN-VJDSNFAGSA-N |
| 64 | [M-H]− | 1.43 | 130.051 | 130.0512 | 1.72 | C5H9NO3 | N-Acetyl-l-alanine | 0.011574 | 71.0141, 74.0248, 85.0296, 86.0612, 86.9402, 87.0452, 88.0405, 101.0246, 129.0197, 130.0512 | KTHDTJVBEPMMGL-VKHMYHEASA-N |
| 65 | [M+H, 2M+H, 2M+Na, M+Na]+ | 3.76 | 205.0972 | 205.0969 | −1.27 | C11H12N2O2 | l-Tryptophan | 0.060655 | 118.0651, 144.0807, 146.06, 159.0917, 188.0705 | QIVBCDIJIAJPQS-VIFPVBQESA-N |
| 66 | [M-H]− | 1.23 | 147.0299 | 147.0301 | 1.64 | C5H8O5 | l-2-Hydroxyglutaric acid | 0.020338 | 87.0088, 87.0453, 89.0246, 101.0245, 101.0610, 102.9490, 103.0401, 129.0195, 129.0558, 147.0301 | HWXBTNAVRSUOJR-VKHMYHEASA-N |
| 67 | [M+FA-H]− | 0.86 | 135.0298 | 135.0301 | 2.72 | C3H6O3 | d-Lactic acid | 0.019063 | 61.9884, 72.9932, 75.0088, 89.0246, 117.0195, 134.0473, 135.0301 | JVTAAEKCZFNVCJ-UWTATZPHSA-N |
| 68 | [M-H]− | 4.98 | 206.0823 | 206.0824 | 0.41 | C11H13NO3 | N-Acetyl-l-phenylalanine | 0.007067 | 73.0296, 79.9576, 85.0296, 89.0248, 131.0352, 147.0453, 161.0457, 164.0718, 166.0000, 206.0823 | CBQJSKKFNMDLON-JTQLQIEISA-N |
| 69 | [M+H, M+K]+ | 0.76 | 175.119 | 175.1188 | −0.68 | C6H14N4O2 | l-Arginine | 0.314091 | 112.0869, 113.0711, 116.0709, 129.1024, 130.0977, 135.0028, 151.9380, 158.0925, 159.0765, 175.1190 | ODKSFYDXXFIFQN-BYPYZUCNSA-N |
| 70 | [M+H-H2O]+ | 0.92 | 130.0499 | 130.0499 | 0.07 | C5H9NO4 | l-Glutamic acid | 0.771336 | 83.061, 84.0451, 84.0814, 130.0501 | WHUUTDBJXJRKMK-VKHMYHEASA-N |
| 71 | [M+FA-H]− | 5.51 | 507.2964 | 507.2961 | −0.45 | C27H42O6 | Podecdysone B | 0.096931 | 75.009, 159.103, 301.1814, 461.2914, 507.2961 | AEFMTBQZWMUASH-IILZZRPCSA-N |
| 72 | [M-H]− | 2.76 | 218.1034 | 218.1035 | 0.26 | C9H17NO5 | Pantothenic acid | 0.025937 | 71.0139, 88.0405, 92.9281, 116.9069, 146.0823, 159.8601, 218.1028 | GHOKWGTUZJEAQD-ZETCQYMHSA-N |
| 73 | [M-H2O-H, M+FA-H, M-H]− | 0.82 | 387.1144 | 387.1143 | −0.38 | C12H22O11 | Palatinose | 3.615434 | 59.014, 71.014, 89.0246, 101.0245, 113.0246, 119.0351, 179.0563, 341.1089 | PVXPPJIGRGXGCY-UHFFFAOYSA-N |
| 74 | [M-H2O-H, M-H]− | 0.91 | 145.0142 | 145.0144 | 1.35 | C5H6O5 | Oxoglutaric acid | 0.016136 | 101.0608, 101.0721, 102.0562, 107.0252, 109.0409, 125.0360, 126.0196, 127.0515, 128.0356, 145.0143 | KPGXRSRHYNQIFN-UHFFFAOYSA-N |
| 75 | [M-H]− | 11.03 | 313.2384 | 313.2384 | −0.02 | C18H34O4 | Octadecanedioic acid | 0.001225 | 251.2379, 295.2279, 313.2384 | BNJOQKFENDDGSC-UHFFFAOYSA-N |
| 76 | [2M-H]− | 10.4 | 315.2541 | 315.2539 | −0.55 | C9H18O2 | Pelargonic acid | 0.010138 | 112.9855, 246.9448, 315.2537 | FBUKVWPVBMHYJY-UHFFFAOYSA-N |
| 77 | [M+Na]+ | 0.95 | 689.2111 | 689.2098 | −1.92 | C24H42O21 | Nystose | 4.056686 | 185.042, 203.0528, 347.094, 365.1043, 527.1553, 689.209 | FLDFNEBHEXLZRX-DLQNOBSRSA-N |
| 78 | [M+H, M+Na]+ | 0.76 | 189.1598 | 189.1596 | −1.04 | C9H20N2O2 | N6,N6,N6-Trimethyl-l-lysine | 0.010983 | 60.0816, 80.9485, 84.0814, 130.0865, 143.118, 144.1126, 188.1395, 189.1334, 189.1597 | MXNRLFUSFKVQSK-QMMMGPOBSA-N |
| 79 | [M-H, M+FA-H]− | 6.03 | 342.1347 | 342.1345 | −0.61 | C19H21NO5 | N-trans-sinapoyltyramine | 0.102446 | 135.045, 148.0532, 178.0507, 190.0516, 327.1111, 342.1354 | IEDBNTAKVGBZEP-VMPITWQZSA-N |
| 80 | [M+H, 2M+Na, M+K]+ | 5.9 | 314.1387 | 314.138 | −2.05 | C18H19NO4 | N-Feruloyltyramine | 0.241944 | 121.0648, 145.0281, 177.0543, 314.138 | NPNNKDMSXVRADT-WEVVVXLNSA-N |
| 81 | [M-H]− | 5.07 | 328.119 | 328.119 | −0.04 | C18H19NO5 | N-Feruloyloctopamine | 0.01335 | 133.0535, 161.0245, 295.0848, 297.0404, 310.1085, 328.1202 | VJSCHQMOTSXAKB-YCRREMRBSA-N |
| 82 | [M+H]+ | 5.1 | 247.1077 | 247.1075 | −1.03 | C13H14N2O3 | N-Acetyltryptophan | 0.00161 | 176.9718, 187.0865, 188.0704, 201.1019, 205.0978, 206.0813, 207.1012, 229.0990, 246.1325, 247.0998 | DZTHIGRZJZPRDV-LBPRGKRZSA-N |
| 83 | [M+H, 2M+H]+ | 0.89 | 148.0604 | 148.0602 | −1.36 | C5H9NO4 | N-Acetylserine | 0.200878 | 84.045, 84.0814, 102.0554, 130.05 | JJIHLJJYMXLCOY-BYPYZUCNSA-N |
| 84 | [M-H]− | 4.74 | 172.0979 | 172.0982 | 1.55 | C8H15NO3 | N-Acetylleucine | 0.006483 | 111.0818, 130.0875, 172.098 | WXNXCEHXYPACJF-ZETCQYMHSA-N |
| 85 | [M+Na]+ | 6.75 | 387.2142 | 387.2135 | −1.97 | C21H32O5 | Pergularin | 0.011047 | 57.0707, 73.0655, 89.0602, 101.0965, 145.1222, 243.0989, 347.2213, 387.2136 | VJMNSJUASLIQEP-UPFSRWTJSA-N |
| 86 | [M+NH4]+ | 10.46 | 241.2275 | 241.2269 | −2.54 | C14H25NO | Pellitorine | 0.005497 | 200.2005 | MAGQQZHFHJDIRE-BNFZFUHLSA-N |
| 87 | [M+H-H2O]+ | 7.12 | 633.3997 | 633.3984 | −2.07 | C36H58O10 | Pedunculoside | 0.013272 | LARPFJIXBULVPK-FBAXZNBGSA-N | |
| 88 | [M+H]+ | 10.39 | 277.2162 | 277.2157 | −1.9 | C18H28O2 | Parinaric acid | 0.020866 | 119.0856, 121.1013, 133.1010, 135.1166, 137.0596, 147.1167, 149.1324, 235.1693, 277.1795, 277.2158 | IJTNSXPMYKJZPR-UHFFFAOYSA-N |
| 89 | [M-H]− | 5.73 | 282.1136 | 282.1134 | −0.56 | C17H17NO3 | p-Coumaroyltyramine | 0.000862 | RXGUTQNKCXHALN-BJMVGYQFSA-N | |
| 90 | [M-H2O-H]− | 6.45 | 535.1821 | 535.1823 | 0.35 | C26H34O13 | osthenol-7-o-β-gentiobioside | 0.01308 | 78.9529, 95.0134, 96.9601, 104.6861, 110.2844, 137.3326, 152.9869, 241.0017, 513.1867, 535.1809 | LCNBLLDTRINYAW-NXEOTYAVSA-N |
| 91 | [M+Na]+ | 9.11 | 374.0999 | 374.1001 | 0.61 | C20H17NO5 | Oxyberberine | 0.000787 | ZHYQCBCBTQWPLC-UHFFFAOYSA-N | |
| 92 | [M+FA-H]− | 4.71 | 475.1821 | 475.182 | −0.23 | C20H30O10 | Phenethyl rutinoside | 0.009461 | 163.0612, 167.7898, 189.8643, 205.0708, 258.2900, 269.4057, 272.7622, 429.1785, 460.7686, 475.1847 | OKUGUNDXBGUFPA-UHFFFAOYSA-N |
| 93 | [M+FA-H]− | 4.64 | 525.1614 | 525.1612 | −0.3 | C23H28O11 | Paeoniflorin | 0.001499 | YKRGDOXKVOZESV-UHFFFAOYSA-N | |
| 94 | [M+H-H2O]+ | 6.75 | 455.3519 | 455.351 | −2.16 | C30H48O4 | Phlegmaric acid | 0.060044 | 203.1792, 205.1588, 205.1951, 207.1739, 249.1849, 397.3095, 409.3452, 425.3403, 437.3408, 455.3515 | UCBRMUIDZFUDIJ-KBUITVGKSA-N |
| 95 | [M-H, M+FA-H]− | 4.61 | 541.3018 | 541.3018 | −0.07 | C27H44O8 | 5-β-hydroxyecdysterone | 0.398516 | 83.0504, 85.0298, 87.0452, 99.0452, 145.0872, 157.087, 175.0977, 319.1918, 495.2964, 541.3013 | GMFLGNRCCFYOKL-ACCCYTKYSA-N |
| 96 | [M+FA-H]− | 4.38 | 727.2455 | 727.2456 | 0.2 | C32H42O16 | Pinoresinol Diglucoside | 0.00149 | ZJSJQWDXAYNLNS-FUPWJLLWSA-N | |
| 97 | [2M+H]+ | 8.93 | 377.102 | 377.1034 | 3.93 | C11H8O3 | Plumbagin | 0.002221 | VCMMXZQDRFWYSE-UHFFFAOYSA-N | |
| 98 | [M+H]+ | 10.43 | 293.2111 | 293.2105 | −2.22 | C18H28O3 | Polyacetylene PQ-1 | 0.029545 | 81.0702, 95.0495, 99.0806, 151.1114, 163.1114, 223.1324, 257.1910, 275.2001, 293.1755, 293.2101 | QSLYECSTHSYXDL-KSZLIROESA-N |
| 99 | [M-H2O-H]− | 6.08 | 969.4701 | 969.4712 | 1.11 | C48H76O21 | Bayogenin-3-O-[β-d-Galactose-(1→3)-β-d-glucuronic acid-28-O-β-d-glucopyranoside | 0.013741 | SQVBXHAEJALFEQ-RYEUOLHJSA-N | |
| 100 | [M+H]+ | 5.63 | 195.1016 | 195.1013 | −1.47 | C11H14O3 | 3,4-Dimethoxycinnamyl alcohol | 0.002244 | OYICGYUCCHVYRR-ONEGZZNKSA-N |
| No | Adducts | RT (min) | Theoretical m/z | m/z | Mass Error (ppm) | Formula | Metabolites | Ratio of Peak Area % | Fragment Ions | InChIKey |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | [M+H]+ | 3.83 | 219.1128 | 219.1125 | −1.32 | C12H14N2O2 | Abrine | 0.149136 | 88.0397, 132.0807, 144.0807, 146.0598, 188.0703, 200.1278, 219.1132 | CZCIKBSVHDNIDH-NSHDSACASA-N |
| 2 | [M-H2O-H, M-H]− | 0.93 | 189.0041 | 189.0043 | 0.96 | C6H8O8 | Hydroxycitric acid | 0.480244 | 73.0296, 83.0139, 85.0296, 87.0088, 99.0088, 111.0088, 127.0038, 129.0195, 189.0041 | ZMJBYMUCKBYSCP-CVYQJGLWSA-N |
| 3 | [M+FA-H]− | 1.27 | 221.0303 | 221.0303 | 0.02 | C6H8O6 | Ascorbyl | 0.007983 | 59.0139, 72.9933, 73.0297, 83.014, 99.009, 103.0039, 127.0039, 159.0298, 189.0042, 221.0295 | CIWBSHSKHKDKBQ-UHFFFAOYSA-N |
| 4 | [M-H]− | 9.19 | 955.4544 | 955.4541 | −0.37 | C47H72O20 | Betavulgaroside III | 1.005851 | 71.0139, 89.0245, 101.0244, 113.0245, 455.3524, 569.3864, 793.437, 835.4518, 955.4528 | GNCYMXULNXKROG-UHFFFAOYSA-N |
| 5 | [M-H2O-H, M-H]− | 0.91 | 189.0041 | 189.0043 | 0.89 | C6H8O8 | Hydroxycitric acid | 0.542071 | 73.0296, 83.0139, 85.0296, 87.0088, 99.0089, 111.0088, 127.0037, 129.0194, 189.0041 | ZMJBYMUCKBYSCP-CVYQJGLWSA-N |
| 6 | [M+H-H2O, M+H]+ | 9.97 | 399.3257 | 399.325 | −1.74 | C27H44O3 | Sarsasapogenin | 0.018722 | 115.0756, 121.1013, 147.1165, 159.1167, 161.1324, 255.2105, 285.2569, 359.2144, 381.3141, 399.3253 | GMBQZIIUCVWOCD-WWASVFFGSA-N |
| 7 | [M+FA-H]− | 1.27 | 221.0303 | 221.0302 | −0.48 | C6H8O6 | Ascorbyl | 0.009148 | 72.9931, 73.0296, 83.0140, 94.9252, 99.0086, 103.0037, 127.0037, 159.0297, 189.0039, 221.0292 | CIWBSHSKHKDKBQ-UHFFFAOYSA-N |
| 8 | [M-H]− | 0.92 | 157.0367 | 157.0367 | 0.19 | C4H6N4O3 | Allantoin | 0.072752 | 71.0251, 89.0244, 96.0457, 97.0044, 114.0309, 140.0101, 157.0368 | POJWUDADGALRAB-UHFFFAOYSA-N |
| 9 | [M+H, M+Na, M+NH4]+ | 0.91 | 365.1055 | 365.1048 | −1.94 | C12H22O11 | Sucrose | 2.398857 | 185.0419, 203.0522, 365.1049 | CZMRCDWAGMRECN-UGDNZRGBSA-N |
| 10 | [M-H, M+FA-H, M-H2O-H]− | 0.81 | 387.1144 | 387.1142 | −0.6 | C12H22O11 | Palatinose | 3.321968 | 59.014, 71.0139, 89.0245, 101.0244, 113.0243, 119.0349, 161.0456, 179.0562, 341.1085 | PVXPPJIGRGXGCY-UHFFFAOYSA-N |
| 11 | [M+NH4, M+Na]+ | 0.91 | 527.1583 | 527.1574 | −1.72 | C18H32O16 | Melezitose | 2.832384 | 144.0652, 145.0493, 162.0757, 163.0597, 180.0864, 259.0801, 289.0912, 325.1122, 343.1228, 522.2019 | QWIZNVHXZXRPDR-WSCXOGSTSA-N |
| 12 | [M+FA-H]− | 4.46 | 431.1923 | 431.1921 | −0.42 | C19H30O8 | Roseoside | 0.0257 | 61.9884, 71.0139, 89.0249, 101.0244, 119.0354, 153.0924, 161.044, 179.0566, 385.1876, 431.1905 | SWYRVCGNMNAFEK-MHXFFUGFSA-N |
| 13 | [M-H]− | 6.12 | 301.0354 | 301.0354 | 0.11 | C15H10O7 | Morin | 0.000739 | 107.0139, 121.0295, 151.0037, 178.9985, 301.0351 | YXOLAZRVSSWPPT-UHFFFAOYSA-N |
| No | Adducts | RT (min) | m/z | Mass Error (ppm) | Parent Compound | Formula | Metabolites | Transformations | Metabolism Type |
|---|---|---|---|---|---|---|---|---|---|
| 1 | [M-H2O-H]− | 5.20 | 529.1331 | −3.69 | (+)-Balanophonin | C26H28O13 | (+)-Balanophonin_M1 | Hydroxylation, glucuronidation | I, II |
| 2 | [M+H-H2O]+ | 8.54 | 427.2838 | −1.17 | (25R)-Spirosta-1,4-diene-3,12-dione | C27H40O5 | (25R)-Spirosta-1,4-diene-3,12-dione_M1 | Deglycosidation, reduction | I |
| 3 | [M-H]− | 6.51 | 469.2081 | 0.41 | 1-Dehydro-6-gingerdione | C23H34O10 | 1-Dehydro-6-gingerdione_M1 | Reduction, reduction, glucuronidation | I, II |
| 4 | [M-H2O-H]− | 6.56 | 567.2991 | −1.09 | 16-Oxoalisol A | C30H50O9S | 16-Oxoalisol A_M1 | Reduction, sulfation | I, II |
| 5 | [M+H]+ | 9.42 | 469.3306 | −1.39 | 16α-Hydroxydehydrotrametenolic acid | C30H44O4 | 16α-Hydroxydehydrotrametenolic acid_M1 | Oxidation | I |
| 6 | [M+FA-H]− | 9.41 | 531.3330 | 0.62 | 16α-Hydroxydehydrotrametenolic acid | C30H46O5 | 16α-Hydroxydehydrotrametenolic acid_M2-1 | Hydroxylation | I |
| 7 | [M-H]− | 9.95 | 485.3271 | −0.39 | 16α-Hydroxydehydrotrametenolic acid | C30H46O5 | 16α-Hydroxydehydrotrametenolic acid_M2-2 | Hydroxylation | I |
| 8 | [M+FA-H]− | 9.95 | 531.3330 | 0.52 | 16α-Hydroxydehydrotrametenolic acid | C30H46O5 | 16α-Hydroxydehydrotrametenolic acid_M2-3 | Hydroxylation | I |
| 9 | [M-H]− | 7.15 | 565.2842 | 0.18 | 16α-Hydroxydehydrotrametenolic acid | C30H46O8S | 16α-Hydroxydehydrotrametenolic acid_M3 | Hydroxylation, sulfation | I, II |
| 10 | [M+H]+ | 9.26 | 503.3358 | −1.73 | 16α-Hydroxydehydrotrametenolic acid | C30H46O6 | 16α-Hydroxydehydrotrametenolic acid_M4 | Hydroxylation, hydroxylation | I |
| 11 | [M-H]− | 10.01 | 483.3118 | 0.43 | 18α-Glycyrrhetinic acid | C30H44O5 | 18α-Glycyrrhetinic acid_M1 | Oxidation, hydroxylation | I |
| 12 | [M-H2O-H]− | 9.03 | 423.1462 | 2.86 | 3-Isomangostin | C24H26O8 | 3-Isomangostin_M1 | Hydroxylation, hydroxylation | I |
| 13 | [M-H]− | 5.75 | 473.1091 | 0.28 | 5,6,7-Trimethoxyflavone | C23H22O11 | 5,6,7-Trimethoxyflavone_M1 | Demethylation, glucuronidation | I, II |
| 14 | [M-H]− | 4.45 | 337.0934 | 1.43 | 5-Hydroxy-1-tetralone | C16H18O8 | 5-Hydroxy-1-tetralone_M1 | Glucuronidation | II |
| 15 | [M-H2O-H]− | 6.30 | 491.2629 | −4.24 | 5α-Pregnane-3β,6α-diol-20-one | C27H42O9 | 5α-Pregnane-3β,6α-diol-20-one_M1 | Glucuronidation | II |
| 16 | [M+FA-H]− | 9.36 | 533.3490 | 1.28 | Ainidiol | C30H48O5 | Ainidiol_M1-1 | Carboxylation, hydroxylation | I |
| 17 | [M-H]− | 9.38 | 487.3436 | 1.37 | Ainidiol | C30H48O5 | Ainidiol_M1-2 | Carboxylation, hydroxylation | I |
| 18 | [2M-H]− | 8.59 | 567.3178 | 0.65 | Artemether | C15H24O5 | Artemether_M1 | Demethylation | I |
| 19 | [2M-H]− | 9.49 | 499.3069 | 0.71 | Costunolide | C15H22O3 | Costunolide_M1 | Hydroxylation, reduction | I |
| 20 | [M-H2O-H]− | 5.24 | 427.0650 | −4.70 | Damnacanthol | C21H18O11 | Damnacanthol_M1 | Demethylation, glucuronidation | I, II |
| 21 | [M+H-H2O]+ | 9.48 | 297.1845 | −1.16 | Dehydroabietic acid | C20H26O3 | Dehydroabietic acid_M1 | Hydroxylation, oxidation | I |
| 22 | [M-H]− | 7.32 | 491.2288 | 0.31 | Dehydroabietic acid | C26H36O9 | Dehydroabietic acid_M2 | Hydroxylation, glucuronidation | I, II |
| 23 | [M-H]− | 5.77 | 531.1512 | 0.67 | Dehydrodiisoeugenol | C26H28O12 | Dehydrodiisoeugenol_M1 | Carboxylation, glucuronidation | I, II |
| 24 | [M+H-H2O]+ | 4.80 | 485.0763 | 2.92 | Demethylsuberosin | C20H22O13S | Demethylsuberosin_M1 | Hydroxylation, glucuronidation, sulfation | I, II |
| 25 | [M+NH4]+ | 6.84 | 566.2227 | −0.83 | Dihydrocurcumin | C27H32O12 | Dihydrocurcumin_M1 | Reduction, glucuronidation | I, II |
| 26 | [M+Na]+ | 5.12 | 471.0896 | −0.51 | Flavokawain C | C21H20O11 | Flavokawain C_M1 | Demethylation, demethylation, glucuronidation | I, II |
| 27 | [M+FA-H]− | 9.56 | 393.1925 | 1.93 | Jolkinolide B | C20H28O5 | Jolkinolide B_M1 | Epoxide hydrolysis | I |
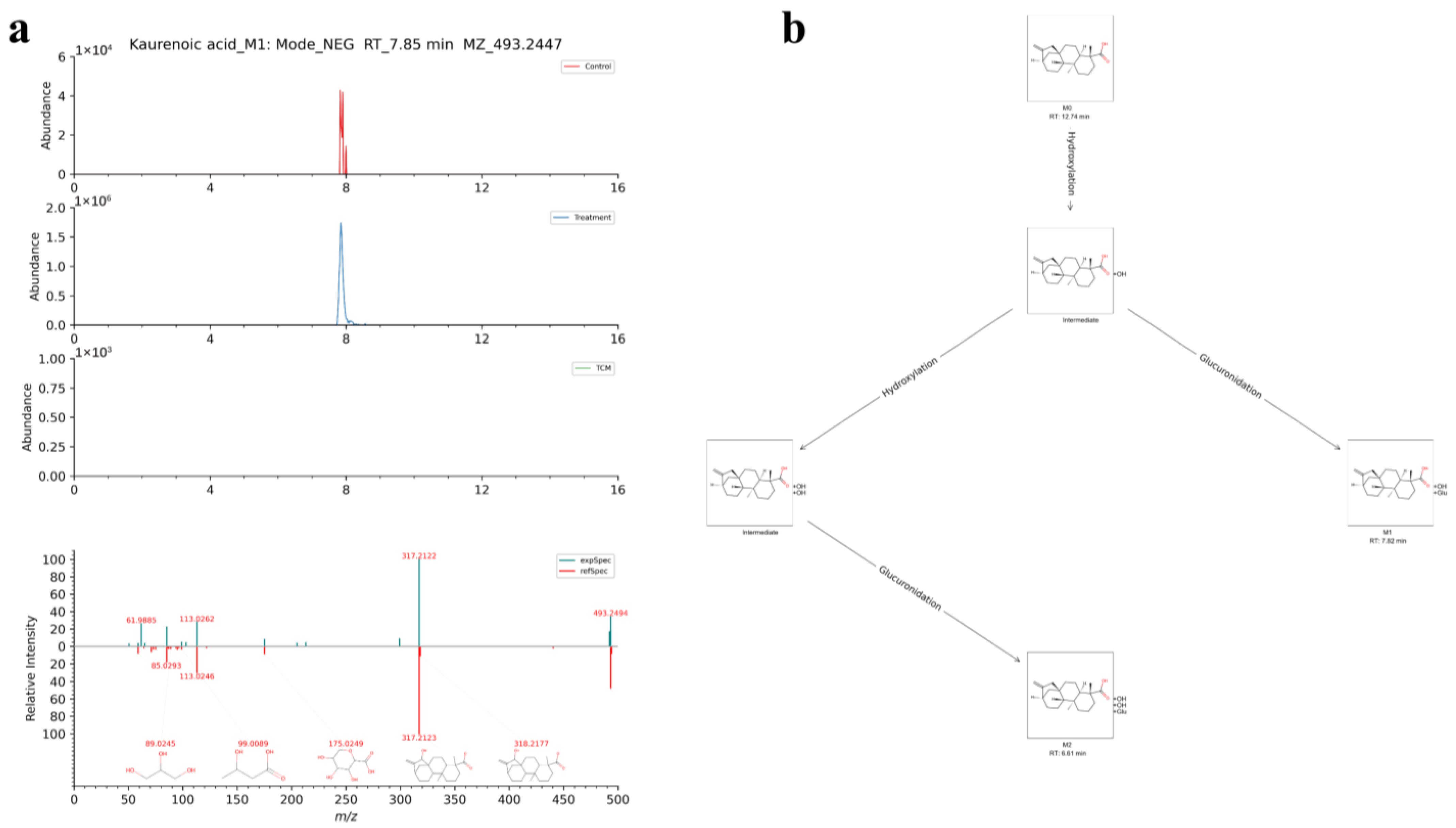
| 28 | [M-H]− | 7.85 | 493.2447 | 0.75 | Kaurenoic acid | C26H38O9 | Kaurenoic acid_M1 | Hydroxylation, glucuronidation | I, II |
| 29 | [M-H]− | 6.58 | 509.2388 | −0.83 | Kaurenoic acid | C26H38O10 | Kaurenoic acid_M2 | Hydroxylation, hydroxylation, glucuronidation | I, II |
| 30 | [2M+H]+ | 9.52 | 501.3203 | −1.61 | Kissoone A | C15H22O3 | Kissoone A_M1 | Hydroxylation, hydroxylation | I |
| 31 | [M+H-H2O]+ | 8.26 | 405.1679 | −4.15 | Licoflavone B | C25H26O6 | Licoflavone B_M1 | Hydroxylation, hydroxylation | I |
| 32 | [M-H2O-H]− | 5.84 | 403.1400 | 0.36 | Lindenenol | C21H26O9 | Lindenenol_M1 | Hydroxylation, glucuronidation | I, II |
| 33 | [M+Na]+ | 5.52 | 519.1495 | 4.36 | Methyl mycophenolate | C23H28O12 | Methyl mycophenolate_M1 | Demethylation, glucuronidation | I, II |
| 34 | [M-H]− | 7.23 | 567.3003 | 0.95 | Momordicine I | C30H48O8S | Momordicine I_M1 | Hydroxylation, sulfation | I, II |
| 35 | [M+H-H2O]+ | 9.79 | 430.2947 | −1.06 | N-Benzyloleamide | C26H41NO5 | N-Benzyloleamide_M1 | Carboxylation, hydroxylation | I |
| 36 | [M+H]+ | 4.89 | 490.1702 | −1.18 | N-Feruloyltyramine | C24H27NO10 | N-Feruloyltyramine_M1 | Glucuronidation | II |
| 37 | [M-H]− | 4.96 | 518.1671 | 0.52 | N-trans-sinapoyltyramine | C25H29NO11 | N-trans-sinapoyltyramine_M1-1 | Glucuronidation | II |
| 38 | [M+H]+ | 4.98 | 520.1808 | −0.95 | N-trans-sinapoyltyramine | C25H29NO11 | N-trans-sinapoyltyramine_M1-2 | Glucuronidation | II |
| 39 | [M-H]− | 11.09 | 485.3274 | 0.23 | Nigranoic acid | C30H46O5 | Nigranoic acid_M1 | Hydroxylation | I |
| 40 | [2M+H]+ | 5.59 | 623.3127 | 1.79 | Nuciferine | C19H21NO3 | Nuciferine_M1 | Oxidation | I |
| 41 | [M-H]− | 4.91 | 349.0569 | 1.07 | Padmatin | C16H14O9 | Padmatin_M1 | Hydroxylation, hydroxylation | I |
| 42 | [M-H2O-H]− | 5.49 | 517.1354 | 0.43 | Peucedanocoumarin II | C25H28O13 | Peucedanocoumarin II_M1 | Deacetylation, hydroxylation, glucuronidation | I, II |
| 43 | [M-H]− | 4.68 | 537.1982 | 0.82 | Secoisolariciresinol | C26H34O12 | Secoisolariciresinol_M1 | Glucuronidation | II |
| 44 | [M-H]− | 5.09 | 593.1880 | 0.78 | Syringaresinol | C28H34O14 | Syringaresinol_M1 | Glucuronidation | II |
| 45 | [M-H]− | 9.58 | 485.3275 | 0.57 | Ursolic acid acetic acid | C30H46O5 | Ursolic acid acetic acid_M1 | Deacetylation, carboxylation | I |
| 46 | [M-H]− | 10.34 | 485.3271 | −0.21 | Ursolic aldehyde | C30H46O5 | Ursolic aldehyde_M1 | Carboxylation, hydroxylation | I |
| 47 | [M-H]− | 4.43 | 399.1661 | 0.06 | Vomifoliol | C19H28O9 | Vomifoliol_M1 | Glucuronidation | II |
| 48 | [M+FA-H]− | 4.69 | 481.0989 | 0.29 | p-hydroxy-5,6-dehydrokawain | C20H20O11 | p-hydroxy-5,6-dehydrokawain_M1 | Hydroxylation, glucuronidation | I, II |
| 49 | [2M+NH4]+ | 11.86 | 1042.7327 | −1.41 | (3α,4β)-3-(Acetyloxy)ursa-5,12-dien-23-oic acid | C32H48O5 | (3α,4β)-3-(Acetyloxy)ursa-5,12-dien-23-oic acid_M1 | Hydroxylation | I |
| 50 | [2M+NH4]+ | 8.67 | 354.2632 | −2.03 | 1,8-Cineole | C10H16O2 | 1,8-Cineole_M1 | Hydroxylation, oxidation | I |
| 51 | [M+FA-H]− | 4.03 | 305.0335 | −0.45 | 3,4-Dimethoxycinnamyl alcohol | C10H12O6S | 3,4-Dimethoxycinnamyl alcohol_M1 | Demethylation, sulfation | I, II |
| 52 | [2M+H]+ | 5.53 | 697.0680 | −0.03 | 7-Hydroxyisoflavone | C16H12O7S | 7-Hydroxyisoflavone_M1 | Hydroxylation, methylation, sulfation | I, II |
| 53 | [M+NH4]+ | 5.75 | 570.2174 | −1.30 | Angelol A | C26H32O13 | Angelol A_M1 | Glucuronidation | II |
| 54 | [M-H]− | 7.21 | 351.1812 | −0.31 | Arnicolide C | C19H28O6 | Arnicolide C_M1-1 | Reduction, hydroxylation | I |
| 55 | [M+H-H2O]+ | 7.19 | 335.1846 | −2.08 | Arnicolide C | C19H28O6 | Arnicolide C_M1-2 | Reduction, hydroxylation | I |
| 56 | [M+NH4]+ | 7.77 | 554.2225 | −1.34 | Guaiacin | C26H32O12 | Guaiacin_M1 | Hydroxylation, hydroxylation, glucuronidation | I, II |
| 57 | [M+NH4]+ | 10.92 | 334.2734 | −1.98 | Guggulsterone | C21H32O2 | Guggulsterone_M1 | Reduction, reduction | I |
| 58 | [2M-H]− | 5.79 | 599.2627 | −3.92 | Honokiol | C18H20O4 | Honokiol_M1 | Vinyl oxidation | I |
| 59 | [M-H2O-H]− | 11.07 | 317.2119 | −0.80 | Incensole Acetic acid | C20H32O4 | Incensole Acetic acid_M1 | Deacetylation, carboxylation | I |
| 60 | [M-H]− | 4.98 | 518.1664 | −0.74 | N-trans-sinapoyltyramine | C25H29NO11 | N-trans-sinapoyltyramine_M1 | Glucuronidation | II |
| 61 | [2M+H]+ | 5.60 | 623.3119 | 0.58 | Nuciferine | C19H21NO3 | Nuciferine_M1 | Oxidation | I |
| 62 | [M-H2O-H]− | 4.71 | 447.0571 | 0.50 | 3,4,8,9,10-Pentahydroxy Urolithin | C20H18O13 | 3,4,8,9,10-Pentahydroxy Urolithin_M1 | Glucuronidation, methylation | II |
| 63 | [M-H2O-H]− | 4.15 | 335.0776 | 1.01 | 3-O-Caffeoylquinic acid methyl ester | C16H18O9 | 3-O-Caffeoylquinic acid methyl ester_M1 | Hydrolysis | I |
| 64 | [M-H]− | 10.23 | 437.2912 | 0.69 | Methyl cholate | C25H42O6 | Methyl cholate_M1 | Hydroxylation | I |
| 65 | [M+H-H2O]+ | 6.88 | 489.3569 | −1.03 | Olean-12-ene-3β,16β,21β,23,28-pentol | C30H50O6 | Olean-12-ene-3β,16β,21β,23,28-pentol_M1 | Hydroxylation | I |
| 66 | [M-H2O-H]− | 4.30 | 401.0880 | 0.51 | Oxyresveratrol | C20H20O10 | Oxyresveratrol_M1 | Glucuronidation | II |
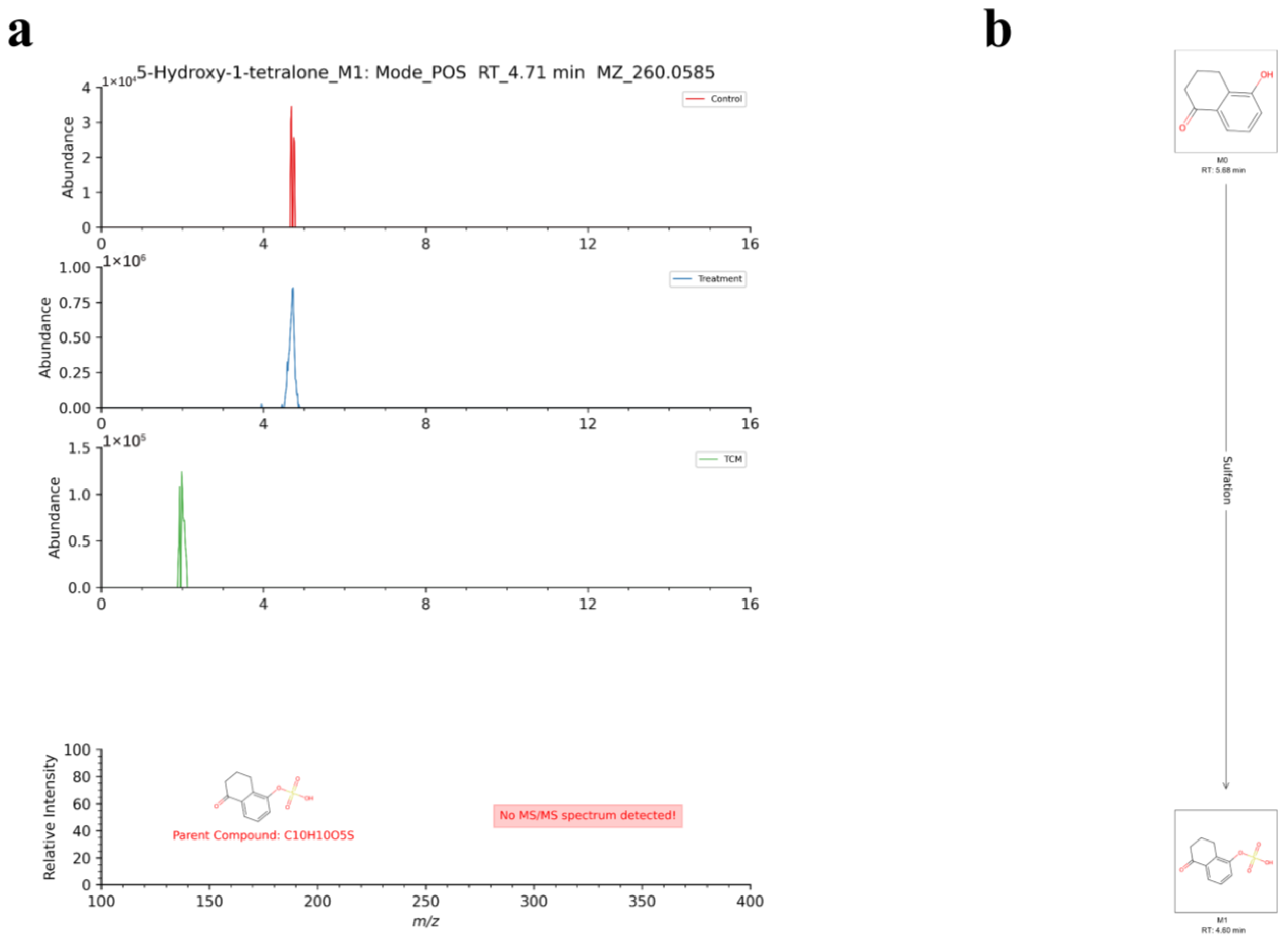
| 67 | [M+NH4]+ | 4.71 | 260.0585 | −0.99 | 5-Hydroxy-1-tetralone | C10H10O5S | 5-Hydroxy-1-tetralone_M1 | Sulfation | II |
| 68 | [M-H2O-H]− | 9.73 | 333.2071 | −0.09 | Ginkgolic Acid (C13:0) | C20H32O5 | Ginkgolic Acid (C13:0)_M1 | Hydroxylation, hydroxylation | I |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, M.; Yang, P.; Wang, J.; Yang, R.; Chen, Y.; Liu, K.; Yuan, Y.; Zhang, L. Characterization of the Components and Metabolites of Achyranthes Bidentata in the Plasma and Brain Tissue of Rats Based on Ultrahigh Performance Liquid Chromatography–High-Resolution Mass Spectrometry (UHPLC–HR-MS). Molecules 2024, 29, 2840. https://doi.org/10.3390/molecules29122840
Wu M, Yang P, Wang J, Yang R, Chen Y, Liu K, Yuan Y, Zhang L. Characterization of the Components and Metabolites of Achyranthes Bidentata in the Plasma and Brain Tissue of Rats Based on Ultrahigh Performance Liquid Chromatography–High-Resolution Mass Spectrometry (UHPLC–HR-MS). Molecules. 2024; 29(12):2840. https://doi.org/10.3390/molecules29122840
Chicago/Turabian StyleWu, Mengting, Peilin Yang, Jianying Wang, Ruoyan Yang, Yingyuan Chen, Kun Liu, Ying Yuan, and Lei Zhang. 2024. "Characterization of the Components and Metabolites of Achyranthes Bidentata in the Plasma and Brain Tissue of Rats Based on Ultrahigh Performance Liquid Chromatography–High-Resolution Mass Spectrometry (UHPLC–HR-MS)" Molecules 29, no. 12: 2840. https://doi.org/10.3390/molecules29122840
APA StyleWu, M., Yang, P., Wang, J., Yang, R., Chen, Y., Liu, K., Yuan, Y., & Zhang, L. (2024). Characterization of the Components and Metabolites of Achyranthes Bidentata in the Plasma and Brain Tissue of Rats Based on Ultrahigh Performance Liquid Chromatography–High-Resolution Mass Spectrometry (UHPLC–HR-MS). Molecules, 29(12), 2840. https://doi.org/10.3390/molecules29122840





