Conjugates of 3,5-Bis(arylidene)-4-piperidone and Sesquiterpene Lactones Have an Antitumor Effect via Resetting the Metabolic Phenotype of Cancer Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. Conjugates of Sesquiterpene Lactones and 3,5-Bis(arylidene)piperidin-4-ones Decrease Vitality of Tumor Cells
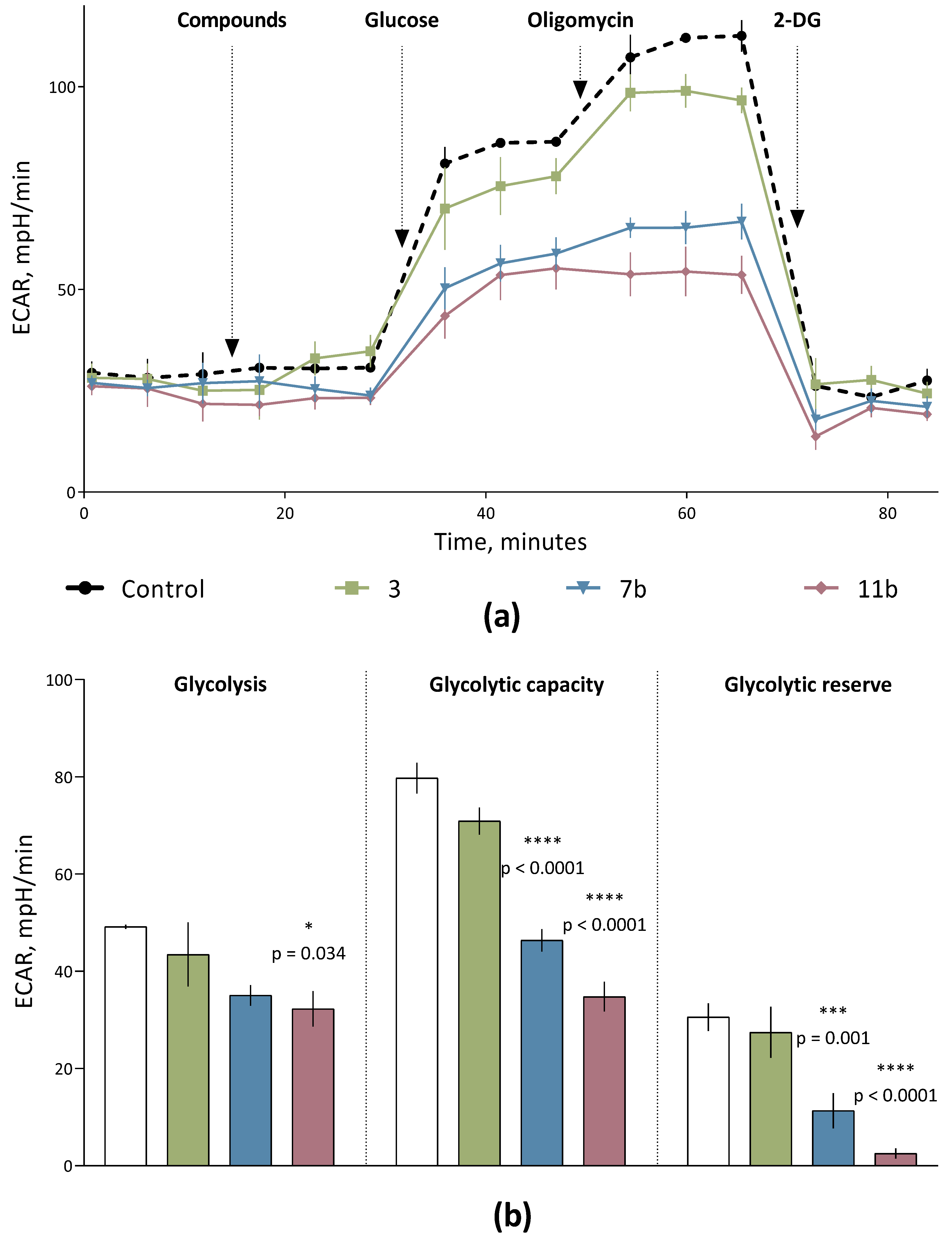
2.2.2. Conjugates of Sesquiterpene Lactones and 3,5-Bis(arylidene)piperidin-4-ones Behave as Negative Regulators of Aerobic Glycolysis in Cells of Human Cervical Carcinoma
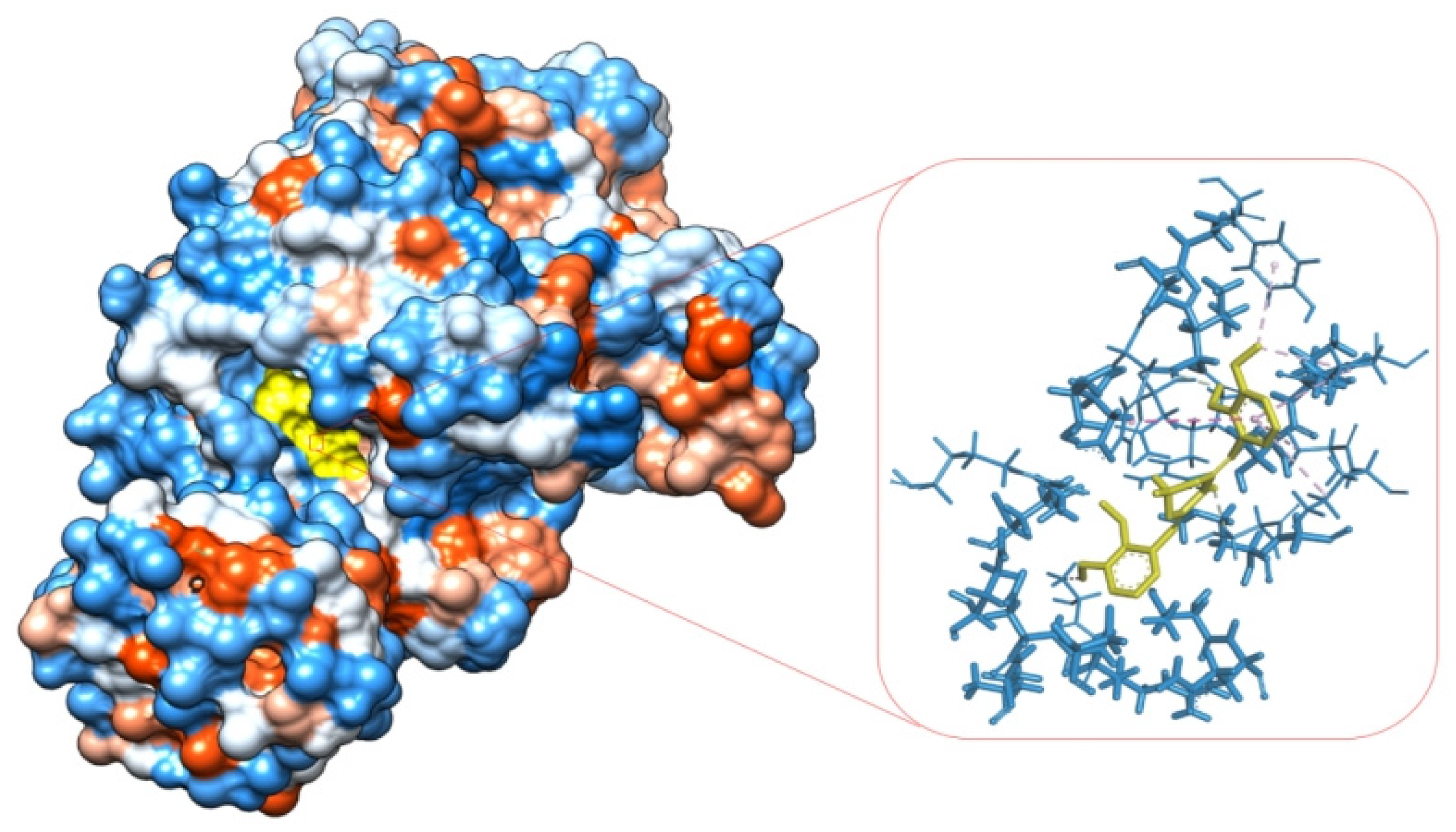
2.2.3. Determination of Allosteric Glycolytic Enzymes Binding Affinities of Compounds by Molecular Docking Analysis
| Hydrogen Bond | ||||||||
|---|---|---|---|---|---|---|---|---|
| Conventional | Carbon | Pi-Donor Hydrogen Bond | Salt Bridge | |||||
| Res. | Dis. | Res. | Dis. | Res. | Dis. | Res. | Dis. | |
| 1 | ARG73 | 2.43 | ||||||
| 2 | LYS270 | 2.92 | ||||||
| 3 | ARG73 | 2.89 | ||||||
| 2.27 | ||||||||
| LYS270 | 2.79 | |||||||
| 2.80 | ||||||||
| 7a | ARG73 | 2.43 | HIS84 | 2.83 | ||||
| SER362 | 2.31 | |||||||
| 7b | ARG73 GLN329 | 2.53 2.36 | SER362 | 2.34 | ||||
| THR328 | 3.75 | |||||||
| ILE51 | 3.50 | |||||||
| 7c | ARG73 | 2.53 | SER362 | 2.57 | ||||
| ASN75 | 3.57 | |||||||
| 9a | ARG73 | 2.48 | SER362 | 2.83 | ||||
| ARG120 | 3.02 | |||||||
| GLN329 | 2.08 | |||||||
| 9b | ARG73 | 2.45 | SER362 | 2.72 | ||||
| ALA293 | 3.76 | |||||||
| 2.04 | ASN75 | 3.26 | ||||||
| HIS78 | 3.44 | |||||||
| 9c | ARG73 | 2.83 | ASN75 | 3.46 | ||||
| LYS270 | 2.41 | GLU118 | 3.76 | |||||
| 2.57 | ASP296 | 3.62 | ||||||
| 10a | ARG120 | 2.98 | GLY79 | 2.38 | ||||
| 10b | ARG294 | 2.82 | LEU180 | 2.89 | ||||
| 2.98 | GLY298 | 3.62 | ||||||
| 2.45 | GLN329 | 3.16 | ||||||
| ALA303 | 2.21 | ASP177 | 3.65 | |||||
| 10c | LYS270 | 2.51 | GLY79 | 2.58 | ||||
| 2.29 | HIS84 | 2.95 | ||||||
| 2.93 | HIS78 | 3.78 | ||||||
| SER362 | 2.76 | GLU118 | 3.66 | |||||
| SER243 | 3.55 | |||||||
| 11a | ARG73 | 2.32 | ASP296 | 3.27 | ASN75 | 2.85 | ||
| ASN75 | 2.44 | 2.95 | ||||||
| 11b | ARG73 | 2.72 | GLU118 | 3.27 | ASP296 | 2.44 | ||
| 2.27 | ||||||||
| 11c | SER77 | 2.47 | GLY79 | 2.32 | ASN75 | 3.11 | ||
| LYS207 | 2.21 | HIS78 | 3.73 | |||||
| LYS270 | 2.20 | GLU118 | 3.62 | |||||
| SER243 | 3.48 | |||||||
| Hydrophobic Interactions | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pi-Pi T-Shaped | Pi-Alkyl/Alkyl | Amide-Pi Stacked | Pi-Pi Stacked | Pi-Sigma | ||||||
| Res. | Dis. | Res. | Dis. | Res. | Dis. | Res. | Dis. | Res. | Dis. | |
| 1 | No electrostatic interactions | |||||||||
| 2 | ||||||||||
| 3 | ||||||||||
| 7a | HIS78 | 5.70 | TYR83 | 4.32 | ||||||
| HIS84 | 4.65 | |||||||||
| ALA366 | 4.73 | |||||||||
| 7b | TYR83 | 4.73 | ||||||||
| HIS84 | 5.31 | |||||||||
| PRO53 | 4.96 | |||||||||
| ALA36 | 4.71 | |||||||||
| LYS367 | 5.37 | |||||||||
| ALA293 | 4.47 | |||||||||
| PRO53 | 4.52 | |||||||||
| LYS367 | 4.06 | |||||||||
| 7c | HIS78 | 5.80 | TYR83 | 4.42 | ||||||
| PRO53 | 4.91 | |||||||||
| ALA366 | 4.70 | |||||||||
| LYS367 | 4.98 | |||||||||
| ALA293 | 3.60 | |||||||||
| PRO53 | 4.29 | |||||||||
| 9a | TYR83 | 4.57 | HIS78 | 5.25 | ||||||
| ALA366 | 4.25 | |||||||||
| 9b | HIS78 | 4.45 | HIS78 | 4.64 | ||||||
| TYR83 | 4.75 | |||||||||
| PRO53 | 5.09 | |||||||||
| 4.31 | ||||||||||
| ALA366 | 4.56 | |||||||||
| LYS367 | 4.64 | |||||||||
| ALA293 | 3.17 | |||||||||
| 9c | HIS78 | 4.96 | ||||||||
| LYS367 | 5.33 | |||||||||
| PRO53 | 4.61 | |||||||||
| ALA293 | 3.67 | |||||||||
| ALA366 | 5.48 | |||||||||
| 4.86 | ||||||||||
| MET291 | 4.98 | |||||||||
| 10a | ALA366 | 4.23 | HIS78 | 3.97 | ||||||
| LYS367 | 5.35 | TYR83 | 3.91 | |||||||
| 10b | ALA303 | 5.47 | TYR175 | 4.89 | GLY298 | 2.79 | ||||
| 4.91 | ||||||||||
| ILE299 | 5.16 | |||||||||
| LEU180 | 4.13 | |||||||||
| ALA343 | 4.26 | |||||||||
| PRO302 | 4.67 | |||||||||
| 10c | ALA366 | 4.62 | ||||||||
| LYS36 | 5.48 | |||||||||
| PRO53 | 5.31 | |||||||||
| 4.54 | ||||||||||
| 3.79 | ||||||||||
| LYS367 | 3.87 | |||||||||
| 11a | HIS78 | 4.80 | ||||||||
| ALA366 | 4.11 | |||||||||
| LEU180 | 4.18 | |||||||||
| ILE299 | 5.35 | |||||||||
| 11b | HIS78 | 4.94 | HIS78 | 5.20 | ASP178 | 3.88 | ||||
| TYR83 | 4.71 | |||||||||
| HIS84 | 4.58 | |||||||||
| ALA366 | 4.64 | |||||||||
| 11c | HIS78 | 5.05 | ||||||||
| 4.40 | ||||||||||
| ALA366 | 4.32 | |||||||||
| PRO53 | 5.34 | |||||||||
| ALA293 | 4.43 | |||||||||
| LYS367 | 4.86 | |||||||||
| Electrostatic Interactions | ||||||
|---|---|---|---|---|---|---|
| Pi-Anion | Pi-Cation | Attractive Charge | ||||
| Res. | Dis. | Res. | Dis. | Res. | Dis. | |
| 1 | No electrostatic interactions | |||||
| 2 | ||||||
| 3 | ||||||
| 7a | ||||||
| 7b | ||||||
| 7c | ||||||
| 9a | GLU332 | 4.04 | ||||
| 9b | ARG120 | 4.27 | ||||
| 9c | ASP296 | 3.64 | ||||
| GLU332 | ||||||
| 10a | No electrostatic interactions | |||||
| 10b | No electrostatic interactions | |||||
| 10c | LYS207 | 3.59 | ||||
| 11a | GLU118 | 4.38 | ARG120 | 3.20 | ||
| 11b | ARG120 | 3.97 | GLU118 | 5.34 | ||
| GLU272 | 4.13 | |||||
| 11c | ASP296 | 4.47 | ||||
3. Materials and Methods
3.1. Reagents and Materials
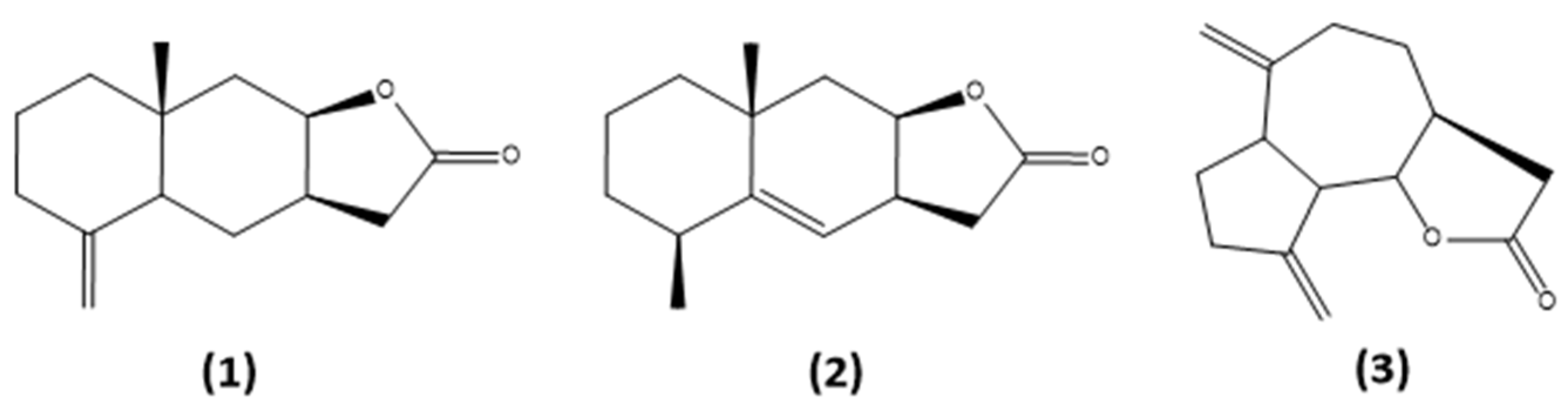
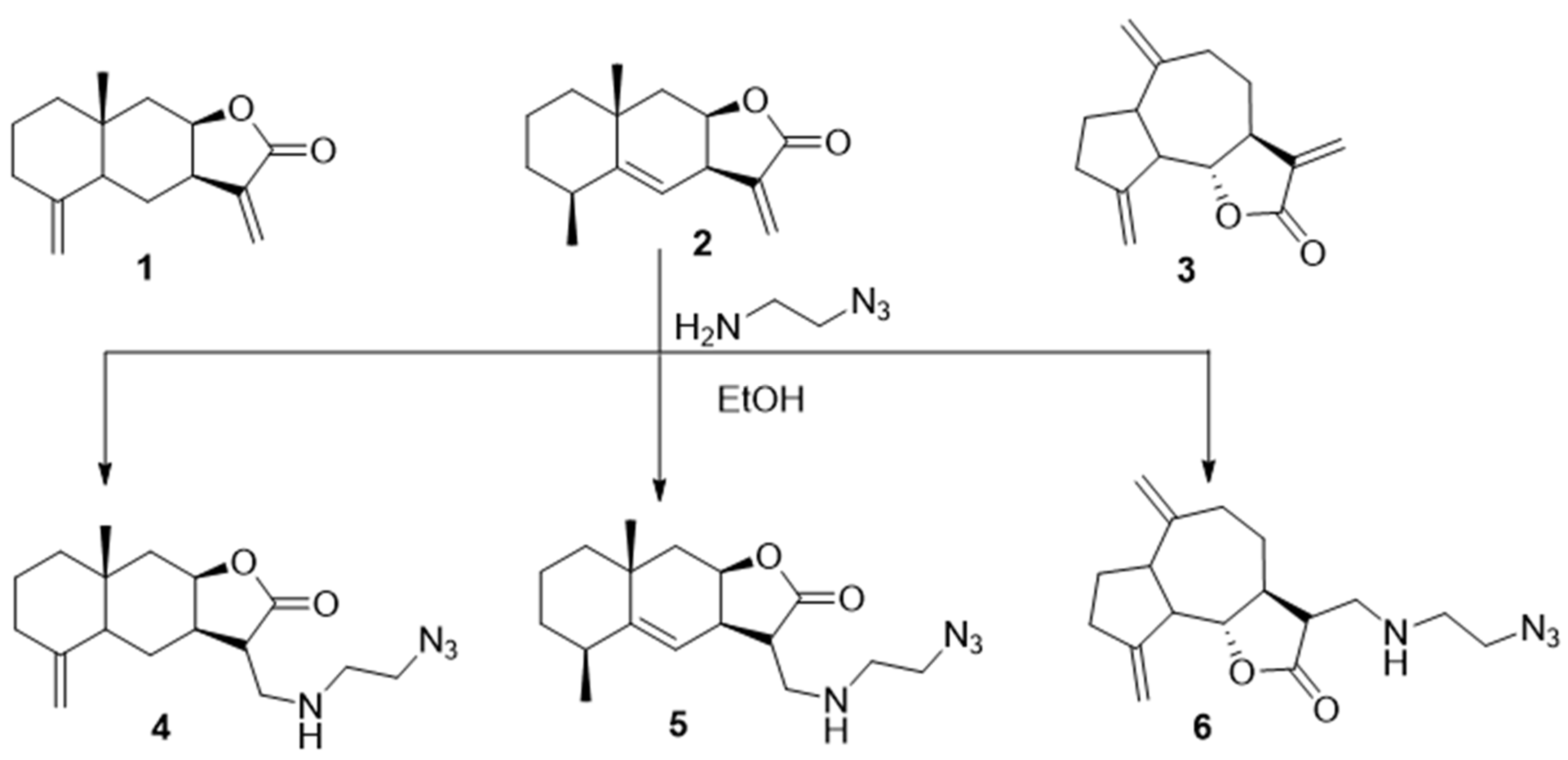
3.2. General Procedure for the Synthesis of Azides 4–6 [52]

- (3S,3aR,8aR,9aR)-3-(((2-azidoethyl)amino)methyl)-8a-methyl-5-methylenedecahydronaphtho[2,3-b]furan-2(3H)-one (4), Pale-yellow powder (83%), m.p. 101–102 °C. 1H NMR (400 MHz, CDCl3) δ 4.79 (1H, br.s, H-14a), 4.51 (1H, br.s, H-9), 4.47 (1H, br.s, H-14b), 3.43 (2H, t, J = 5.6 Hz, H2-17), 3.38 (1H, br.s, NH), 3.06 (1H, dd, J = 11.6 Hz, J = 6.8 Hz, H-15a), 2.95–2.75 (4H, m, H-15b, H2-16, H-8), 2.52 (1H, ddd, J = 12.7 Hz, J = 6.0 Hz, J = 2.0 Hz, H-11), 2.33 (1H, d, J = 12.4 Hz, H-2a), 2.18 (1H, d, J = 15.4 Hz, H-10a), 2.04–1.96 (1H, m, H-2b), 1.80 (1H, d, J = 12.1 Hz, H-6a), 1.70–1.45 (6H, m, H-1, H-4, H-7a, H-10b), 1.28–1.22 (2H, m, H-6b, H-7b), and 0.82 (3H, s, H3-13). 13C NMR (100 MHz, CDCl3) δ 177.85 (C-12), 149.13 (C-3), 106.33 (C-14), 78.20 (C-9), 51.06 (C-17), 48.63 (C-15), 47.42 (C-11), 46.33 (C-4), 44.92 (C-16), 42.07 (C-6), 41.23 (C-10), 38.90 (C-4), 36.59 (C-2), 34.65 (C-5), 22.52 (C-7), 20.91 (C-1), and 17.66 (C-13). IR (KBr) vmax 2932, 2866, 2096 (N3), 1747 (C=O), 1643, 1447, 1297, 1158, 963, and 894 cm−1. HRMS (ESI): m/z calcd. for C17H27N4O2 [M + H]+ 319.2129, found 319.2135. Anal. Calc. for C17H26N4O2 × 0.1 CH2Cl2: C, 62.83; H, 8.08; N, 17.14%. Found: C, 62.85; H, 8.02; N, 17.20%.

- (3S,3aR,5S,8aR,9aR)-3-(((2-Azidoethyl)amino)methyl)-5,8a-dimethyl-3,3a,6,7,8,8a,9,9aoctahydronaphtho[2,3-b]furan-2(5H)-one (5), Yellow oil (85%). 1H NMR (400 MHz, CDCl3) δ 5.15 (1H, d, J = 2.8 Hz, H-7), 4.76 (1H, m, H-9), 3.44 (2H, d, J = 5.6 Hz, H2-17), 3.16–3.13 (1H, m, H-8), 3.02–2.78 (5H, m, H-11, H2-15, H2-16), 2.50–2.47 (1H, m, H-3), 2.11 (1H, dd, J = 14.6 Hz, J = 3.1 Hz, H-10a), 1.87–1.76 (1H, m, H-2a), 1.61–1.42 (6H, m, H-1, H-2b, H-6, H-10b), 1.23 (3H, s, H3-13), and 1.12 (3H, d, J = 7.6 Hz, H3-14). 13C NMR (100 MHz, CDCl3) δ 177.12 (C-12), 150.10 (C-4), 114.58 (C-7), 76.69 (C-9), 50.44 (C-17), 47.93 (C-15), 45.54 (C-16), 45.02 (C-11), 42.04 (C-10), 41.50 (C-6), 37.77 (C-3), 36.92 (C-8), 32.31 (C-5), 32.15 (C-2), 27.99 (C-13), 22.32 (C-14), and 16.18 (C-1). IR (KBr) νmax 2929, 2101, 1762, 1457, 1340, 1180, 1151, 1039, and 733 cm−1. Anal. Calc. for C17H26N4O2·0.15 CH2Cl2: C, 62.20; H, 8.00; N, 16.92%. Found: C, 62.68; H, 8.03; N, 16.80%.
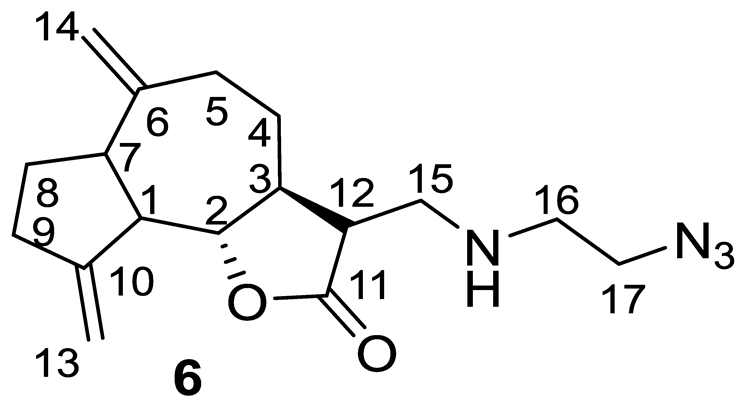
- (3R,3aS,9bS)-3-(((2-Azidoethyl)amino)methyl)-6,9-dimethylenedecahydroazuleno[4,5-b]furan-2(9bH)-one (6), Yellow oil (82%). 1H NMR (400 MHz, CDCl3) δ 5.30 (1H, s, H-13a), 5.17 (1H, s, H-13b), 4.87 (1H, s, H-14a), 4.76 (1H, s, H-14b), 3.96 (1H, t, J = 9.1 Hz, H-2), 3.37 (2H, t, J = 5.6 Hz, H2-17), 2.97 (1H, dd, J = 12.1 Hz, J = 3.8 Hz, H-15a), 2.89–2.81 (4H, m, H-1, H-7, H-12, H-15b), 2.51–2.45 (3H, m, H-5a, H-9), 2.39–2.34 (2H, m, H2-16), 2.15–1.85 (5H, m, H-3, H-4a, H-5b, H-8), and 1.39–1.24 (1H, m, H-4b). 13C NMR (100 MHz, CDCl3) δ 177.51 (C-11), 151.56 (C-10), 149.65 (C-6), 111.64 (C-13), 108.85 (C-14), 85.62 (C-2), 51.64 (C-12), 50.96 (C-17), 48.82 (C-15), 47.30 (C-1), 47.11 (C-16), 46.74 (C-7), 44.57 (C-3), 37.46 (C-9), 32.40 (C-5), 32.33 (C-4), and 29.95 (C-8). IR (KBr) νmax 2929, 2100, 1767, 1456, 1340, 1176, 1004, and 894 cm−1. Anal. Calc. for C17H24N4O2·0.1 CH2Cl2: C, 63.22; H, 7.51; N, 17.24%. Found: C, 63.15; H, 7.42; N, 17.09%.
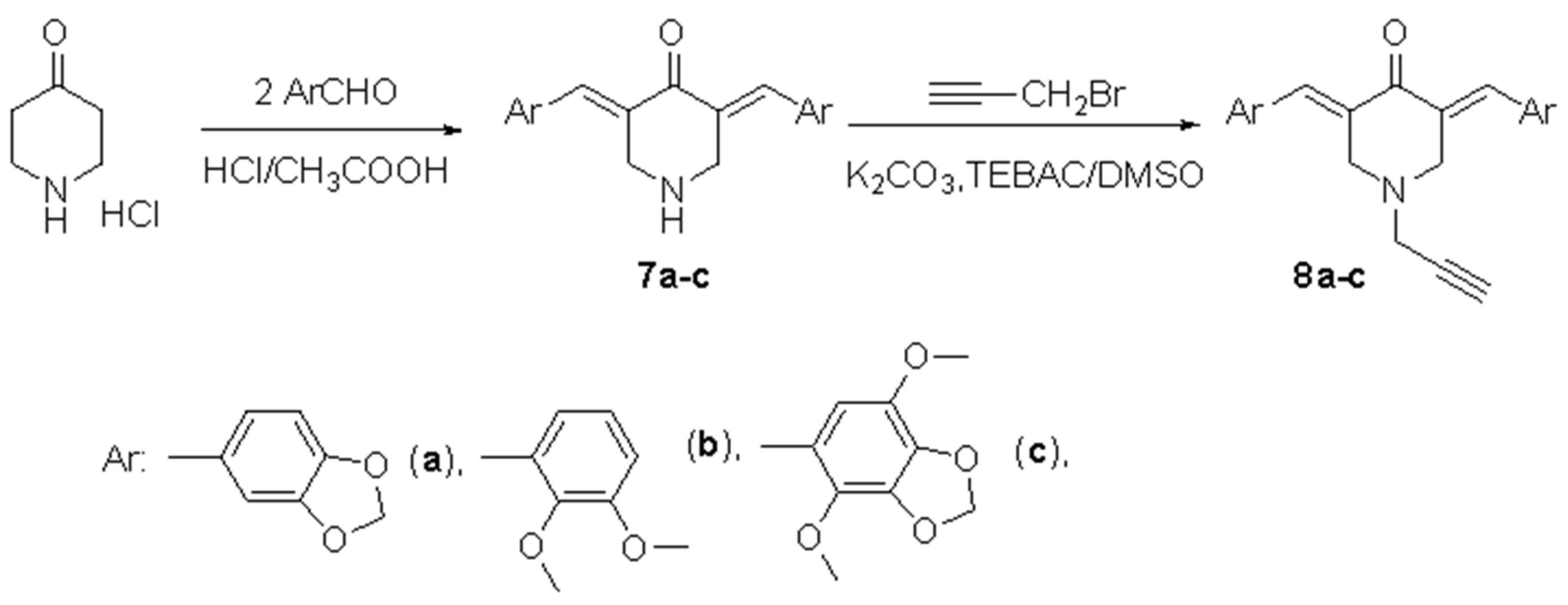
3.3. General Procedure for the Synthesis of 3,5-Bis(arylidene)piperidin-4-ones (7a–c)
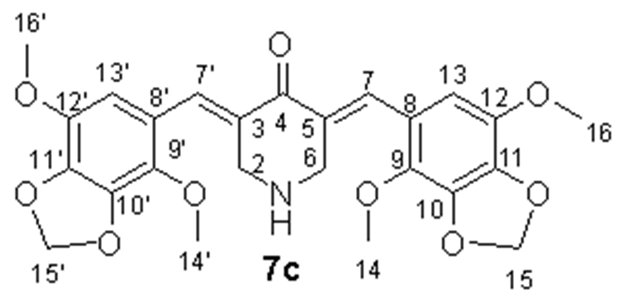
- (3E,5E)-3,5-Bis((4,7-dimethoxybenzo[d][1,3]dioxol-5-yl)methylene)piperidin-4-one (7c), Yellow-green crystals (95%), m.p. 163–164 °C. 1H NMR (400 MHz, DMSO-d6) δ 7.68 (2H, s, H-7, H-7′), 6.52 (2H, s, H-13, H-13′), 6.09 (4H, s, H-15, H-15′), 3.92 (4H, s, H-2, H-6), and 3.88 and 3.83 (12H, both s, H3-14, H3-14′, H3-16, H3-16′). 13C NMR (100 MHz, DMSO-d6) δ 187.77 (C-4), 138.89 (C-11, C-11′), 138.81 (C-10, C-10′), 138.32 (C-12, C-12′), 137.71 (C-9, C-9′), 135.48 (C-7, C-7′), 129.18 (C-3, C-5), 121.25 (C-8, C-8′), 109.87 (C-13, C-13′), 102.58 (C-15, C-15′), 60.64 (C-14, C-14′), 57.07 (C-16, C-16′), and 48.08 (C-2, C-6). IR (KBr) vmax 2945, 2840, 1631, 1589, 1492, 1456, 1358, 1237, 1141, 1068, 958, and 521 cm−1. HRMS (ESI): m/z calcd. for C25H26NO9 [M + H]+ 484.1602, found 484.1614. Anal. Calc. for C25H25NO9 × 0.35 H2O: C, 61.31; H, 5.29; N, 2.86%. Found: C, 61.23; H, 5.48; N, 2.88%.
3.4. General Procedure for the Synthesis of N-Propargyl-3,5-bis(arylidene)piperidin-4-ones (8a–c)
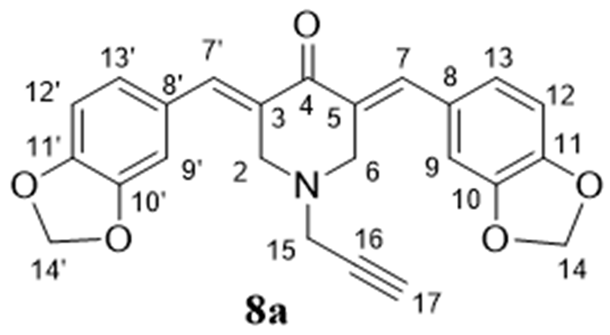
- (3E,5E)-3,5-Bis(benzo[d][1,3]dioxol-5-ylmethylene)-1-(prop-2-yn-1-yl)piperidin-4-one (8a), Yellow crystals (82%), m.p. > 192 °C (decomp.). 1H NMR (400 MHz, CDCl3) δ 7.76 (2H, s, H-7, H-7′), 6.97 (2H, d, J = 8.0 Hz, H-12, H-12′), 6.91 (2H, s, H-9, H-9′), 6.90 (2H, d, J = 8.0 Hz, H-13, H-13′), 6.05 (4H, s H-14, H-14′), 3.93 (4H, s, H-2, H-6), 3.58 (2H, d, J = 1.8 Hz, H2-15), and 2.41 (1H, t, J = 1.8 Hz, H-17). 13C NMR (100 MHz, CDCl3) δ 186.16 (C-4), 148.42 (C-11, C-11′), 147.83 (C-10, C-10′), 136.70 (C-7, C-7′), 130.93 (C-8, C-8′), 129.19 (C-3, C-5), 125.95 (C-13, C-13′), 109.92 (C-12, C-12′), 108.55 (C-9, C-9′), 101.41 (C-14, C-14′), 77.12 (C-16), 74.89 (C-17), 53.22 (C-2, C-6), and 46.33 (C-15). HRMS (ESI): m/z calcd. for C24H20NO5 [M + H]+ 402.1336, found 402.1346. Anal. Calc. for C24H19NO5× 0.5 H2O: C, 70.23; H, 4.91; N, 3.41%. Found: C, 70.06; H, 4.81; N, 3.55%.
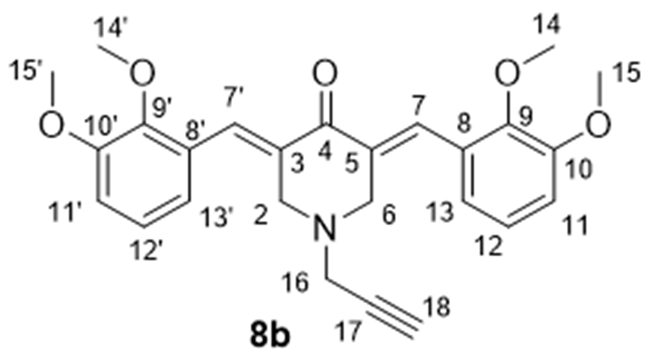
- 3,5-Bis((E)-2,3-dimethoxybenzylidene)-1-(prop-2-yn-1-yl)piperidin-4-one (8b), Yellow crystals (87%), m.p. 60–61 °C. 1H NMR (400 MHz, CDCl3) δ 8.07 (2H, s, H-7, H-7′), 7.11 (2H, t, J = 8.0 Hz, H-12, H-12′), 6.97 (2H, d, J = 8.0 Hz, H-11, H-11′), 6.87 (2H, d, J = 8.0 Hz, H-13, H-13′), 3.90 (6H, s, H3-15, H3-15′), 3.83 (10H, br.s, H3-14, H3-14′, H-2, H-6), 3.47 (2H, d, J = 1.6 Hz, H2-16), and 2.31 (1H, t, J = 1.6 Hz, H-18). 13C NMR (100 MHz, CDCl3) δ 186.26 (C-4), 152.79 (C-9, C-9′), 148.39 (C-10, C-10′), 133.43 (C-8, C-8′), 132.58 (C-7, C-7′), 129.41 (C-3, C-5), 123.61 (C-13, C-13′), 121.85 (C-12, C-12′), 113.12 (C-11, C-11′), 77.41 (C-17), 74.43 (C-18), 61.27 (C-16, C-16′), 55.72 (C-15, C-15′), 53.41 (C-2, C-6), and 46.13 (C-16). HRMS (ESI): m/z calcd. for C26H28NO5 [M + H]+ 434.1962, found 434.1971. Anal. Calc. for C26H27NO5 × 1.5 H2O: C, 68.54; H, 6.68; N, 3.04%. Found: C, 68.59; H, 6.31; N, 3.10%.

- (3E,5E)-3,5-Bis((4,7-dimethoxybenzo[d][1,3]dioxol-5-yl)methylene)-1-(prop-2-yn-1-yl)piperidin-4-one (8c), Yellow crystals (95%), m.p. 167–168 °C. 1H NMR (400 MHz, CDCl3) δ 7.96 (2H, s, H-7, H-7′), 6.43 (2H, s, H-13, H-13′), 6.04 (4H, s, H-15, H-15′), 3.93 and 3.87 (12H, both s, H3-14, H3-14′, H3-16, H3-16′), 3.82 (4H, s, H-2, H-6), 3.49 (2H, d, J = 2.0 Hz, H2-17), and 2.31 (1H, t, J = 2.0 Hz, H-19). 13C NMR (100 MHz, CDCl3) δ 186.20 (C-4), 138.60 (C-11, C-11′), 138.57 (C-10, C-10′), 138.04 (C-12, C-12′), 137.89 (C-9, C-9′), 132.40 (C-3, C-5), 132.24 (C-7, C-7′), 121.44 (C-8, C-8′), 109.41 (C-13, C-13′), 101.92 (C-15, C-15′), 77.52 (C-18), 74.14 (C-19), 60.40 (C-14, C-14′), 56.92 (C-16, C-16′), 53.65 (C-2, C-6), and 46.25 (C-17). HRMS (ESI): m/z calcd. for C28H28NO9 [M + H]+ 522.1759, found 522.1765. Anal. Calc. for C28H27NO9 × 0.25 H2O: C, 63.93; H, 5.27; N, 2.66%. Found: C, 63.90; H, 5.29; N, 2.57%.
3.5. General Procedure for the “Click”-Reactions

- (3E,5E)-3,5-bis(benzo[d][1,3]dioxol-5-ylmethylene)-1-((1-(2-((((3S,3aR,8aR,9aR)-8a-methyl-5-methylene-2-oxododecahydronaphtho[2,3-b]furan-3-yl)methyl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)piperidin-4-one (9a), Yellow oil (68%). 1H NMR (400 MHz, CDCl3) δ 7.63 (2H, s, H-26, H-26′), 7.52 (1H, s, H-18), 6.88 (2H, d, J = 8.0 Hz, H-32, H-32′), 6.82–6.77 (4H, m, H-28, H-28′, H-31, H-31′), 5.97 (4H, s, H2-33, H2-33′), 4.72 (1H, s, H-14a), 4.38–4.35 (4H, m, H-9, H-14b, H2-17), 3.86–3.82 (6H, m, H2-20, H2-21, H2-25), 3.10–3.06 (1H, m, H-8), 2.99–2.95 (2H, m, H2-16), 2.78–2.73 and 2.70–2.63 (2H, both m, H2-15), 2.41–2.35 (1H, m, H-11), 2.28–2.25 (1H, m, H-2a), 2.10 (1H, d, J = 15.4 Hz, H-10a), 1.96–1.92 (1H, m, H-2b), 1.71 (1H, d, J = 12.0 Hz, H-6a), 1.55–1.42 (5H, m, H-1, H-4, H-7a, H-10b), 1.22–1.05 (2H, m, H-6b, H-7b), and 0.74 (3H, s, H3-13). 13C NMR (100 MHz, CDCl3) δ 186.72 (C-23), 177.69 (C-12), 148.93 (C-3), 148.21 (C-30, C-30′), 147.66 (C-29, C-29′), 143.86 (C-19), 135.84 (C-26, C-26′), 131.35 (C-27, C-27′), 129.02 (C-22, C-24), 125.67 (C-32, C-32′), 123.14 (C-18), 109.77 (C-31, C-31′), 108.34 (C-28, C-28′), 106.22 (C-14), 101.27 (C-33, C-33′), 78.05 (C-9), 54.33 (C-21, C-25), 52.25 (C-20), 49.76 (C-17), 48.92 (C-15), 47.02 (C-11), 46.10 (C-4), 44.63 (C-16), 41.86 (C-6), 41.08 (C-10), 38.70 (C-8), 36.41 (C-2), 34.47 (C-5), 22.37 (C-7), 20.75 (C-1), and 17.51 (C-13). IR (film) vmax 2930, 1761 (C=O), 1596, 1504, 1489, 1446, 1263, 1235, 1039, 931, and 756 cm−1. HRMS (ESI): m/z calcd. for C41H46N5O7 [M + H]+: 720.3397, found 720.3395.

- (3E,5E)-3,5-bis(2,3-dimethoxybenzylidene)-1-((1-(2-((((3S,3aR,8aR,9aR)-8a-methyl-5-methylene-2-oxododecahydronaphtho[2,3-b]furan-3-yl)methyl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)piperidin-4-one (9b), Yellow oil (62%). 1H NMR (400 MHz, CDCl3) δ 8.05 (2H, s, H-26, H-26′), 7.47 (1H, s, H-18), 7.08 (2H, t, J = 7.6 Hz, H-31, H-31′), 6.96 (2H, d, J = 8.0 Hz, H-30, H-30′), 6.81 (2H, d, J = 7.6 Hz, H-32, H-32′), 4.76 (1H, s, H-14a), 4.48–4.37 (4H, m, H-9, H-14b, H2-17), 3.90–3.82 (18H, m, H3-33, H3-33′, H3-34, H3-34′, H2-20, H2-21, H2-25), 3.10–3.05 (1H, m, H-8), 3.05–2.99 (2H, m, H2-16), 2.88–2.83 and 2.79–2.69 (2H, both m, H2-15), 2.50–2.40 (1H, m, H-2a), 2.33–2.30 (1H, m, H-11), 2.16 (1H, d, J = 15.6 Hz, H-10a), 2.00–1.95 (1H, m, H-2b), 1.75 (1H, d, J = 12.0 Hz, H-6a), 1.65–1.46 (5H, m, H-1, H-4, H-7a, H-10b), 1.30–1.15 (2H, m, H-6b, H-7b), and 0.80 (3H, s, H3-13). 13C NMR (100 MHz, CDCl3) δ 187.17 (C-23), 177.87 (C-12), 152.74 (C-3), 149.08 (C-28, C-28′), 148.24 (C-29, C-29′), 144.02 (C-19), 133.61 (C-22, C-24), 132.88 (C-26, C-26′), 129.44 (C-27, C-27′), 123.82 (C-30, C-30′), 123.30 (C-18), 122.01 (C-30, C-30′), 113.37 (C-32, C-32′), 106.49 (C-14), 78.47 (C-9), 61.28 (C-33, C-33′), 55.89 (C-34, C-34′), 54.37 (C-21, C-25), 51.61 (C-20), 49.60 (C-17), 49.09 (C-15), 46.38 (C-11), 46.24 (C-4), 44.91 (C-16), 42.17 (C-6), 41.35 (C-10), 38.98 (C-8), 36.70 (C-2), 34.47 (C-5), 22.67 (C-7), 21.06 (C-1), and 17.81 (C-13). IR (film) vmax 2934, 1762 (C=O), 1576, 1477, 1278, 1223, 1076, 1004, and 753 cm−1. HRMS (ESI): m/z calcd. for C43H54N5O7 [M + H]+: 752.4023, found 752.4015.
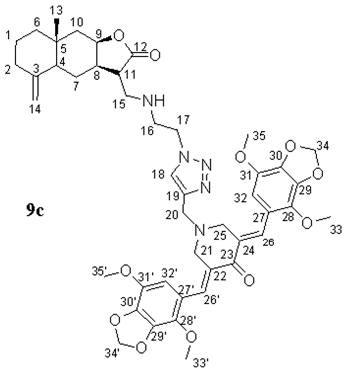
- (3E,5E)-3,5-bis((4,7-dimethoxybenzo[d][1,3]dioxol-5-yl)methylene)-1-((1-(2-((((3S,3aR,8aR,9aR)-8a-methyl-5-methylene-2-oxododecahydronaphtho[2,3-b]furan-3-yl)methyl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)piperidin-4-one (9c), Yellow oil (64%). 1H NMR (400 MHz, CDCl3) δ 7.96 (2H, s, H-26, H-26′), 7.50 (1H, s, H-18), 6.38 (2H, s, H-32, H-32′), 6.03 (4H, s, H-34, H-34′), 4.76 (1H, s, H-14a), 4.48–4.38 (4H, m, H-9, H-14b, H2-17), 3.94 (6H, s, H3-33, H3-33′), 3.90 (6H, s, H3-35, H3-35′), 3.83 (2H, s H2-20), 3.80 (4H, s, H2-21, H2-25), 3.15-3.12 (1H, m, H-8), 3.02-3.00 (2H, m, H2-16), 2.84–2.82 and 2.75–2.70 (2H, both m, H2-15), 2.45–2.43 (1H, m, H-2a), 2.33–2.30 (1H, m, H-11), 2.15 (1H, d, J = 15.2 Hz, H-10a), 2.01–1.93 (1H, m, H-2b), 1.75 (1H, d, J = 12.0 Hz, H-6a), 1.60–1.45 (5H, m, H-1, H-4, H-7a, H-10b), 1.30–1.15 (2H, m, H-6b, H-7b), and 0.79 (3H, s, H3-13). 13C NMR (100 MHz, CDCl3) δ 186.65 (C-23), 177.86 (C-12), 149.08 (C-3), 143.79 (C-19), 138.62 (C-30, C-30′), 138.47 (C-29, C-29′), 138.05 (C-31, C-31′), 137.79 (C-28, C-28′), 132.15 (C-26, C-26′), 131.97 (C-22, C-24), 123.45 (C-18), 121.28 (C-27, C-27′), 109.29 (C-32, C-32′), 106.32 (C-14), 101.90 (C-34, C-34′), 78.26 (C-9), 60.33 (C-33, C-33′), 56.94 (C-35, C-35′), 54.41 (C-21, C-25), 51.65 (C-20), 49.75 (C-17), 49.01 (C-15), 47.06 (C-11), 46.24 (C-4), 44.80 (C-16), 42.02 (C-6), 41.21 (C-10), 38.86 (C-8), 36.55 (C-2), 34.61 (C-5), 22.51 (C-7), 20.91 (C-1), and 17.64 (C-13). IR (film) vmax 2931, 1762 (C=O), 1599, 1493, 1456, 1242, 1143, 1067, 963, and 755 cm−1. HRMS (ESI): m/z calcd. for C45H54N5O11 [M + H]+: 840.3820, found 840.3807.
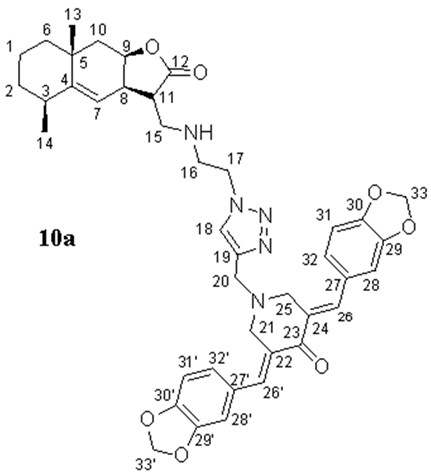
- (3E,5E)-3,5-bis(benzo[d][1,3]dioxol-5-ylmethylene)-1-((1-(2-((((3S,3aR,5S,8aR,9aR)-5,8a-dimethyl-2-oxo-2,3,3a,5,6,7,8,8a,9,9a-decahydronaphtho[2,3-b]furan-3-yl)methyl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)piperidin-4-one (10a), Yellow oil (62%). 1H NMR (400 MHz, CDCl3) δ 7.69 (2H, s, H-26, H-26′), 7.55 (1H, s, H-18), 6.93–6.85 (6H, m, H-28, H-28′, H-31, H-31′, H-32, H-32′), 6.01 (4H, s, H-33, H-33′), 5.05 (1H, br.s, H-7), 4.72 (1H, br.s, H-9), 4.43–4.39 (2H, m, H2-17), 3.91 (2H, s, H2-20), 3.87 (4H, s, H2-21, H2-25), 3.13–2.88 (5H, m, H2-16, H2-15, H-8), 2.76–2.70 (1H, m, H-11), 2.45–2.44 (1H, m, H-3), 2.09 (1H, dd, J = 14.8 Hz, J = 2.8 Hz, H-10a), 1.81–1.75 (1H, m, H-2a), 1.60–1.43 (6H, m, H-1, H-2b, H-6, H-10b), 1.21 (3H, s, H3-13), and 1.10 (3H, d, J = 7.4 Hz, H3-14). 13C NMR (100 MHz, CDCl3) δ 186.91 (C-23), 177.66 (C-12), 151.16 (C-4), 148.33 (C-30, C-30′), 147.78 (C-29, C-29′), 144.04 (C-19), 136.05 (C-26, C-26′), 131.43 (C-27, C-27′), 129.17 (C-22, C-24), 125.72 (C-32, C-32′), 123.25 (C-18), 114.58 (C-7), 109.91 (C-31, C-31′), 108.47 (C-28, C-28′), 101.36 (C-33, C-33′), 77.29 (C-9), 54.48 (C-21, C-25), 52.40 (C-20), 49.85 (C-17), 48.98 (C-15), 46.04 (C-16), 45.43 (C-11), 42.48 (C-10), 42.00 (C-6), 38.30 (C-3), 37.46 (C-8), 32.86 (C-5), 32.64 (C-2), 28.49 (C-13), 22.80 (C-14), and 16.65 (C-1). IR (film) vmax 2927, 1758 (C=O), 1596, 1503, 1489, 1446, 1234, 1038, 929, and 734 cm−1. HRMS (ESI): m/z calcd. for C41H46N5O7 [M + H]+: 720.3397, found 720.3380.
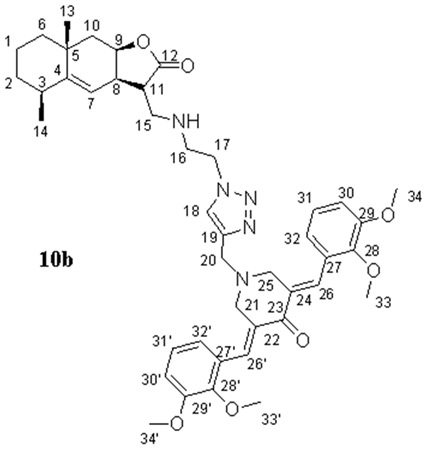
- (3E,5E)-3,5-bis(2,3-dimethoxybenzylidene)-1-((1-(2-((((3S,3aR,5S,8aR,9aR)-5,8a-dimethyl-2-oxo-2,3,3a,5,6,7,8,8a,9,9a-decahydronaphtho[2,3-b]furan-3-yl)methyl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)piperidin-4-one (10b), Yellow oil (59%). 1H NMR (400 MHz, CDCl3) δ 8.03 (2H, s, H-26, H-26′), 7.44 (1H, s, H-18), 7.06 (2H, t, J = 7.9 Hz, H-31, H-31′), 6.95 (2H, d, J = 7.9 Hz, H-30, H-30′), 6.80 (2H, d, J = 7.6 Hz, H-32, H-32′), 5.06 (1H, s, H-7), 4.73 (1H, s, H-9), 4.35–4.33 (2H, m, H2-17), 3.89–3.80 (18H, m, H3-33, H3-33′, H3-34, H3-34′, H2-20, H2-21, H2-25), 3.07 (2H, br.s, H2-16), 3.02–2.91 (3H, m, H2-15, H-8), 2.74 (1H, br.s, H-11), 2.48–2.43 (1H, m, H-3), 2.09 (1H, dd, J = 14.7 Hz, J = 2.9 Hz, H-10a), 1.86–1.76 (1H, m, H-2a), 1.60–1.48 (6H, m, H-1, H-2b, H-6, H-10b), 1.21 (3H, s, H3-13), and 1.10 (3H, d, J = 7.4 Hz, H3-14). 13C NMR (100 MHz, CDCl3) δ 187.06 (C-23), 177.66 (C-12), 152.70 (C-28, C-28′), 151.09 (C-4), 148.22 (C-29, C-29′), 144.09 (C-19), 133.67 (C-27, C-27′), 132.43 (C-26, C-26′), 129.33 (C-22, C-24), 123.70 (C-18), 123.62 (C-32, C-32′), 121.83 (C-31, C-31′), 114.63 (C-7), 113.13 (C-30, C-30′), 77.31 (C-9), 61.10 (C-33, C-33′), 55.69 (C-34, C-34′), 54.31 (C-21, C-25), 51.58 (C-20), 49.70 (C-17), 49.01 (C-15), 45.98 (C-16), 45.49 (C-11), 42.50 (C-10), 42.01 (C-6), 38.29 (C-3), 37.44 (C-8), 32.86 (C-5), 32.64 (C-2), 28.50 (C-13), 22.81 (C-14), and 16.65 (C-1). IR (film) vmax 2930, 1759 (C=O), 1576, 1477, 1263, 1224, 1076, 1006, and 753 cm−1. HRMS (ESI): m/z calcd. for C43H54N5O7 [M + H]+: 752.4023, found 752.4018.
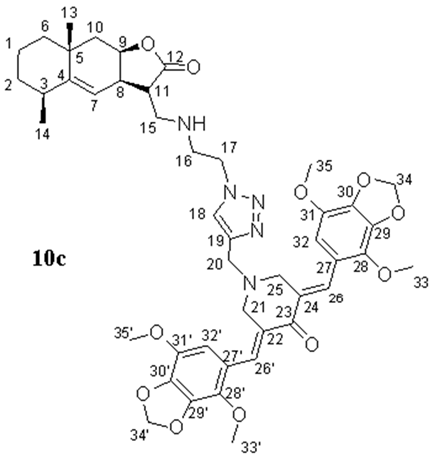
- (3E,5E)-3,5-bis((4,7-dimethoxybenzo[d][1,3]dioxol-5-yl)methylene)-1-((1-(2-((((3S,3aR,5S,8aR,9aR)-5,8a-dimethyl-2-oxo-2,3,3a,5,6,7,8,8a,9,9a-decahydronaphtho[2,3-b]furan-3-yl)methyl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)piperidin-4-one (10c), Yellow oil (65%). 1H NMR (400 MHz, CDCl3) δ 7.93 (2H, s, H-26, H-26′), 7.50 (1H, s, H-18), 6.36 (2H, s, H-32, H-32′), 6.00 (4H, s, H-34, H-34′), 5.04 (1H, br.s, H-7), 4.72 (1H, br.s, H-9), 4.40–4.31 (2H, m, H2-17), 3.91–3.77 (18H, m, H3-33, H3-33′, H3-35, H3-35′, H2-20, H2-21, H2-25), 3.10–3.00 (3H, m, H2-16, H-8), 2.98–2.90 (2H, m, H2-15), 2.73–2.71 (1H, m, H-11), 2.43–2.41 (1H, m, H-3), 2.09–1.99 (1H, m, H-10a), 1.84–1.75 (1H, m, H-2a), 1.59–1.42 (6H, m, H-1, H-2b, H-6, H-10b), 1.19 (3H, s, H3-13), and 1.08 (3H, d, J = 7.4 Hz, H3-14). 13C NMR (100 MHz, CDCl3) δ 186.66 (C-23), 177.72 (C-12), 151.09 (C-4), 143.71 (C-19), 138.56 (C-30, C-30′), 138.42 (C-29, C-29′), 138.00 (C-28, C-28′), 132.05 (C-26, C-26′), 131.96 (C-22, C-24), 123.39 (C-18), 121.22 (C-27, C-27′), 114.58 (C-7), 109.24 (C-32, C-32′), 101.85 (C-34, C-34′), 101.36 (C-33, C-33′), 77.29 (C-9), 60.27 (C-33, C-33′), 56.87 (C-35, C-35′), 54.39 (C-21, C-25), 51.66 (C-20), 49.79 (C-17), 48.93 (C-15), 45.98 (C-16), 45.39 (C-11), 42.44 (C-10), 41.96 (C-6), 38.26 (C-3), 37.42 (C-8), 32.81 (C-5), 32.60 (C-2), 28.44 (C-13), 18.19 (C-14), and 16.61 (C-1). IR (film) vmax 2931, 1760 (C=O), 1600, 1495, 1456, 1245, 1143, 1067, and 736 cm−1. HRMS (ESI): m/z calcd. for C45H54N5O11 [M + H]+: 840.3820, found 840.3822.
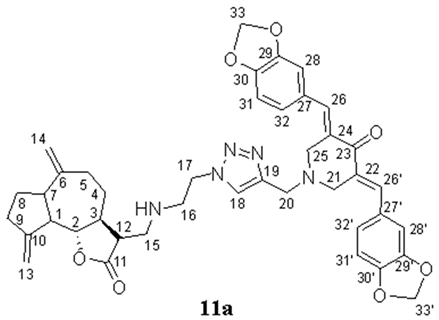
- (3E,5E)-3,5-bis(benzo[d][1,3]dioxol-5-ylmethylene)-1-((1-(2-((((3R,3aS)-6,9-dimethylene-2-oxododecahydroazuleno[4,5-b]furan-3-yl)methyl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)piperidin-4-one (11a), Yellow oil (62%). 1H NMR (400 MHz, CDCl3) δ 7.72 (2H, s, H-26, H-26′), 7.57 (1H, s, H-18), 6.95–6.87 (6H, m, H-28, H-28′, H-31, H-31′, H-32, H-32′), 6.03 (4H, s, H-33, H-33′), 5.15 (1H, s, H-13a), 5.04 (1H, s, H-13b), 4.87 (1H, s, H-14a), 4.77 (1H, s, H-14b), 4.44–4.42 (2H, m, H2-17), 3.96–3.90 (7H, m, H-2, H2-20, H2-21, H2-25), 3.12–3.11 (2H, m, H2-16), 2.90–2.78 (4H, m, H2-15, H-12, H-7), 2.51–2.43 (3H, m, H-1, H-9), 2.37–2.35 (1H, m, H-5a,), 2.23–2.15 (1H, m, H-4a), 2.04–1.83 (4H, m, H-3, H-5b, H-8), and 1.34–1.24 (1H, m, H-4b). 13C NMR (100 MHz, CDCl3) δ 186.96 (C-23), 177.44 (C-11), 151.51 (C-10), 149.54 (C-6), 148.41 (C-30, C-30′), 147.83 (C-29, C-29′), 143.87 (C-19), 136.43 (C-26, C-26′), 131.10 (C-27, C-27′), 129.15 (C-22, C-24), 125.87 (C-32, C-32′), 123.47 (C-18), 111.88 (C-13), 109.97 (C-31, C-31′), 109.06 (C-14), 108.54 (C-28, C-28′), 101.49 (C-33, C-33′), 85.73 (C-2), 54.37 (C-21, C-25), 52.24 (C-20), 51.67 (C-12), 49.60 (C-17), 48.96 (C-15), 46.97 (C-1), 46.86 (C-7), 46.80 (C-3), 44.82 (C-16), 37.39 (C-9), 32.39 (C-5), 32.34 (C-4), and 30.03 (C-8). IR (film) vmax 2934, 1762 (C=O), 1576, 1477, 1263, 1223, 1076, 1004, and 753 cm−1. HRMS (ESI): m/z calcd. for C41H44N5O7 [M + H]+: 718.3240, found 718.3227.
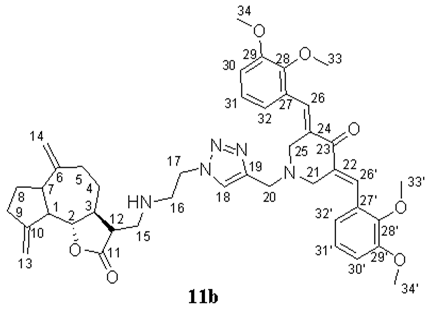
- (3E,5E)-3,5-bis(2,3-dimethoxybenzylidene)-1-((1-(2-((((3R,3aS)-6,9-dimethylene-2-oxododecahydroazuleno[4,5-b]furan-3-yl)methyl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)piperidin-4-one (11b), Yellow oil (59%). 1H NMR (400 MHz, CDCl3) δ 8.06 (2H, s, H-26, H-26′), 7.46 (1H, s, H-18), 7.09–7.07 (2H, m, H-31, H-31′), 6.96 (2H, d, J = 7.6 Hz, H-30, H-30′), 6.82–6.81 (2H, m, H-32, H-32′), 5.17 (1H, s, H-13a), 5.05 (1H, s, H-13b), 4.87 (1H, s, H-14a), 4.77 (1H, s, H-14b), 4.34 (2H, br.s, H2-17), 3.91–3.85 (19H, m, H3-33, H3-33′, H3-34, H3-34′, H-2, H2-20, H2-21, H2-25), 3.03 (2H, br.s, H2-16), 2.87–2.77 (4H, m, H2-15, H-12, H-7), 2.52–2.44 (3H, m, H-1, H-9), 2.35 (1H, br.s, H-5a), 2.19 (1H, br.s, H-4a), 2.02–1.86 (4H, m, H-3, H-5b, H-8), and 1.29–1.27 (1H, m, H-4b). 13C NMR (100 MHz, CDCl3) δ 187.17 (C-23), 177.66 (C-11), 152.83 (C-28, C-28′), 151.76 (C-10), 149.76 (C-6), 148.35 (C-29, C-29′), 144.15 (C-19), 133.78 (C-27, C-27′), 132.59 (C-26, C-26′), 129.45 (C-22, C-24), 123.81 (C-32, C-32′), 123.33 (C-18), 121.99 (C-31, C-31′), 113.28 (C-30, C-30′), 111.87 (C-13), 109.05 (C-14), 85.77 (C-2), 61.27 (C-33, C-33′), 55.84 (C-34, C-34′), 54.45 (C-21, C-25), 51.81 (C-12), 51.67 (C-20), 49.93 (C-17), 49.17 (C-15), 47.39 (C-1), 46.94 (C-7), 46.81 (C-16), 44.85 (C-3), 37.55 (C-9), 32.54 (C-5), 32.50 (C-4), and 30.14 (C-8). IR (film) vmax 2933, 1764 (C=O), 1575, 1476, 1455, 1263, 1224, 1075, 1004, and 753 cm−1. HRMS (ESI): m/z calcd. for C43H52N5O7 [M + H]+: 750.3866, found 750.3855.

- (3E,5E)-3,5-bis((4,7-dimethoxybenzo[d][1,3]dioxol-5-yl)methylene)-1-((1-(2-((((3R,3aS)-6,9-dimethylene-2-oxododecahydroazuleno[4,5-b]furan-3-yl)methyl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)piperidin-4-one (11c), Yellow oil (57%). 1H NMR (400 MHz, CDCl3) δ 7.92 (2H, s, H-26, H-26′), 7.46 (1H, s, H-18), 6.35 (2H, s, H-32, H-32′), 5.99 (4H, s, H-34, H-34′), 5.12 (1H, s, H-13a), 4.99 (1H, s, H-13b), 4.83 (1H, s, H-14a), 4.73 (1H, s, H-14b), 4.32 (2H, br.s, H2-17), 3.90–3.77 (19H, m, H-33, H-33′, H-35, H-35′, H-2, H2-20, H2-21, H2-25), 2.99–2.82 (6H, m, H2-16, H2-15, H-12, H-7), 2.48–2.40 (3H, m, H-1, H-9), 2.27–1.80 (6H, m, H-5a, H-4a, H-3, H-5b, H-8), and 1.27–1.22 (1H, m, H-4b). 13C NMR (100 MHz, CDCl3) δ 186.56 (C-23), 177.42 (C-11), 151.51 (C-10), 149.54 (C-6), 143.67 (C-19), 138.49 (C-30, C-30′), 138.35 (C-29, C-29′), 137.91 (C-31, C-31′), 137.69 (C-28, C-28′), 131.96 (C-22, C-24), 131.90 (C-26, C-26′), 123.26 (C-18), 121.18 (C-27, C-27′), 111.63 (C-13), 109.14 (C-32, C-32′), 108.88 (C-14), 101.82 (C-34, C-34′), 85.52 (C-2), 60.25 (C-33, C-33′), 56.80 (C-35, C-35′), 54.34 (C-21, C-25), 51.60 (C-20), 51.56 (C-12), 49.78 (C-17), 49.00 (C-15), 47.15 (C-1), 47.01(C-16), 46.71 (C-7), 44.63 (C-3), 37.33 (C-9), 32.29 (C-5), 32.24 (C-4), and 29.90 (C-8). IR (film) vmax 2937, 1763 (C=O), 1599, 1495, 1456, 1193, 1066, and 755 cm−1. HRMS (ESI): m/z calcd. for C45H52N5O11 [M + H]+: 838.3663, found 838.3661.
3.6. Cell Culture
3.7. Assessment of Cells Vitality
3.8. Measuring Glycolytic Function in Cells
3.9. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Saadet, E.D.; Tek, I. Evaluation of chemotherapy-induced cutaneous side effects in cancer patients. Int. J. Dermatol. 2022, 61, 1519–1526. [Google Scholar] [CrossRef] [PubMed]
- Schirrmacher, V. From chemotherapy to biological therapy: A review of novel concepts to reduce the side effects of systemic cancer treatment (Review). Int. J. Oncol. 2019, 54, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Di Nardo, P.; Lisanti, C.; Garutti, M.; Buriolla, S.; Alberti, M.; Mazzeo, R.; Puglisi, F. Chemotherapy in patients with early breast cancer: Clinical overview and management of long-term side effects. Expert. Opin. Drug Saf. 2022, 21, 1341–1355. [Google Scholar] [CrossRef] [PubMed]
- Yavarpour-Bali, H.; Ghasemi-Kasman, M.; Pirzadeh, M. Curcumin-loaded nanoparticles: A novel therapeutic strategy in treatment of central nervous system disorders. Int. J. Nanomed. 2019, 14, 4449–4460. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.C.; Kamarudin, M.N.A.; Naidu, R. Anticancer Mechanism of Curcumin on Human Glioblastoma. Nutrients 2021, 13, 950. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.; Joshi, S.; Semwal, D.; Bisht, A.; Paliwal, S.; Dwivedi, J.; Sharma, S. Curcumin: An Insight into Molecular Pathways Involved in Anticancer Activity. Mini Rev. Med. Chem. 2021, 21, 2420–2457. [Google Scholar] [CrossRef]
- Mohanty, C.; Sahoo, S.K. The in vitro stability and in vivo pharmacokinetics of curcumin prepared as an aqueous nanoparticulate formulation. Biomaterials 2010, 31, 6597–6611. [Google Scholar] [CrossRef]
- Jurenka, J.S. Anti-inflammatory properties of curcumin, a major constituent of Curcuma longa: A review of preclinical and clinical research. Altern. Med. Rev. 2009, 14, 141–153. [Google Scholar] [PubMed]
- Gordon, O.N.; Luis, P.B.; Sintim, H.O.; Schneider, C. Unraveling curcumin degradation: Autoxidation proceeds through spiroepoxide and vinylether intermediates en route to the main bicyclopentadione. J. Biol. Chem. 2015, 290, 4817–4828. [Google Scholar] [CrossRef] [PubMed]
- Shehzad, A.; Wahid, F.; Lee, Y.S. Curcumin in cancer chemoprevention: Molecular targets, pharmacokinetics, bioavailability, and clinical trials. Arch. Pharm. 2010, 343, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Tomeh, M.A.; Hadianamrei, R.; Zhao, X. A Review of Curcumin and Its Derivatives as Anticancer Agents. Int. J. Mol. Sci. 2019, 20, 1033. [Google Scholar] [CrossRef] [PubMed]
- Razali, N.S.C.; Lam, K.W.; Rajab, N.F.; AR, A.J.; Kamaluddin, N.F.; Chan, K.M. Curcumin piperidone derivatives induce anti-proliferative and anti-migratory effects in LN-18 human glioblastoma cells. Sci. Rep. 2022, 12, 13131. [Google Scholar] [CrossRef] [PubMed]
- Razali, N.S.C.; Lam, K.W.; Rajab, N.F.; Jamal, A.R.A.; Kamaludin, N.F.; Chan, K.M. Curcumin piperidone derivatives induce caspase-dependent apoptosis and suppress miRNA-21 expression in LN-18 human glioblastoma cells. Genes Environ. 2024, 46, 4. [Google Scholar] [CrossRef] [PubMed]
- Olivera, A.; Moore, T.W.; Hu, F.; Brown, A.P.; Sun, A.; Liotta, D.C.; Snyder, J.P.; Yoon, Y.; Shim, H.; Marcus, A.I.; et al. Inhibition of the NF-kappaB signaling pathway by the curcumin analog, 3,5-Bis(2-pyridinylmethylidene)-4-piperidone (EF31): Anti-inflammatory and anti-cancer properties. Int. Immunopharmacol. 2012, 12, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, F.; Subramaniam, D.; Anant, S.; Padhye, S.; Begemann, G.; Schobert, R.; Biersack, B. Halogenated Bis(methoxybenzylidene)-4-piperidone Curcuminoids with Improved Anticancer Activity. ChemMedChem 2018, 13, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- Hazafa, A.; Rehman, K.U.; Jahan, N.; Jabeen, Z. The Role of Polyphenol (Flavonoids) Compounds in the Treatment of Cancer Cells. Nutr. Cancer 2020, 72, 386–397. [Google Scholar] [CrossRef]
- Rahman, H.S. Preclinical Drug Discovery in Colorectal Cancer: A Focus on Natural Compounds. Curr. Drug Targets 2021, 22, 977–997. [Google Scholar] [CrossRef]
- Shafabakhsh, R.; Asemi, Z. Quercetin: A natural compound for ovarian cancer treatment. J. Ovarian. Res. 2019, 12, 55. [Google Scholar] [CrossRef]
- Zhu, Z.; Turak, A.; Aisa, H.A. Sesquiterpene lactones from Artemisia mongolica. Phytochemistry 2022, 199, 113158. [Google Scholar] [CrossRef] [PubMed]
- Laurella, L.C.; Mirakian, N.T.; Garcia, M.N.; Grasso, D.H.; Sulsen, V.P.; Papademetrio, D.L. Sesquiterpene Lactones as Promising Candidates for Cancer Therapy: Focus on Pancreatic Cancer. Molecules 2022, 27, 3492. [Google Scholar] [CrossRef] [PubMed]
- Kreuger, M.R.; Grootjans, S.; Biavatti, M.W.; Vandenabeele, P.; D’Herde, K. Sesquiterpene lactones as drugs with multiple targets in cancer treatment: Focus on parthenolide. Anticancer Drugs 2012, 23, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Cheikh, I.A.; El-Baba, C.; Youssef, A.; Saliba, N.A.; Ghantous, A.; Darwiche, N. Lessons learned from the discovery and development of the sesquiterpene lactones in cancer therapy and prevention. Expert. Opin. Drug Discov. 2022, 17, 1377–1405. [Google Scholar] [CrossRef]
- Kim, D.Y.; Choi, B.Y. Costunolide-A Bioactive Sesquiterpene Lactone with Diverse Therapeutic Potential. Int. J. Mol. Sci. 2019, 20, 2926. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Park, S.; Zhang, H.; Park, S.; Kwon, W.; Kim, E.; Zhang, X.; Jang, S.; Yoon, D.; Choi, S.K.; et al. Targeting AKT with costunolide suppresses the growth of colorectal cancer cells and induces apoptosis in vitro and in vivo. J. Exp. Clin. Cancer Res. 2021, 40, 114. [Google Scholar] [CrossRef]
- Wei, M.; Li, J.; Qiu, J.; Yan, Y.; Wang, H.; Wu, Z.; Liu, Y.; Shen, X.; Su, C.; Guo, Q.; et al. Costunolide induces apoptosis and inhibits migration and invasion in H1299 lung cancer cells. Oncol. Rep. 2020, 43, 1986–1994. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Sheng, B.; Li, R.; Xu, Q.; Zhang, L.; Lu, Z. Dehydrocostus Lactone Induces Apoptosis and Cell Cycle Arrest through Regulation of JAK2/STAT3/PLK1 Signaling Pathway in Human Esophageal Squamous Cell Carcinoma Cells. Anticancer Agents Med. Chem. 2022, 22, 1742–1752. [Google Scholar] [CrossRef]
- Peng, Y.; Zhou, T.; Wang, S.; Bahetjan, Y.; Li, X.; Yang, X. Dehydrocostus lactone inhibits the proliferation of esophageal cancer cells in vivo and in vitro through ROS-mediated apoptosis and autophagy. Food Chem. Toxicol. 2022, 170, 113453. [Google Scholar] [CrossRef]
- Babaei, G.; Gholizadeh-Ghaleh Aziz, S.; Rajabi Bazl, M.; Khadem Ansari, M.H. A comprehensive review of anticancer mechanisms of action of Alantolactone. Biomed. Pharmacother. 2021, 136, 111231. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Gao, K.; Peng, B.; Xu, Z.; Peng, J.; Li, J.; Chen, X.; Zeng, S.; Hu, K.; Yan, Y. Alantolactone: A Natural Plant Extract as a Potential Therapeutic Agent for Cancer. Front. Pharmacol. 2021, 12, 781033. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wen, Q.; Cai, Y.; Liu, Y.; Ma, W.; Li, Q.; Song, F.; Guo, Y.; Zhu, L.; Ge, J.; et al. Alantolactone induces concurrent apoptosis and GSDME-dependent pyroptosis of anaplastic thyroid cancer through ROS mitochondria-dependent caspase pathway. Phytomedicine 2023, 108, 154528. [Google Scholar] [CrossRef] [PubMed]
- Sztiller-Sikorska, M.; Czyz, M. Parthenolide as Cooperating Agent for Anti-Cancer Treatment of Various Malignancies. Pharmaceuticals 2020, 13, 194. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Nguyen, M.T.; Little, P.J.; Do, A.T.; Tran, P.T.; Vo, X.N.; Do, B.H. Vernolide-A and Vernodaline: Sesquiterpene Lactones with Cytotoxicity against Cancer. J. Environ. Pathol. Toxicol. Oncol. 2020, 39, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Lin, Y.; Zhu, X.; Cai, X. Isoalantolactone suppresses gallbladder cancer progression via inhibiting the ERK signalling pathway. Pharm. Biol. 2023, 61, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Yang, P. Isoalantolactone exerts anticancer effects on human HEC-1-B endometrial cancer cells via induction of ROS mediated apoptosis and inhibition of MEK/ERK signalling pathway. Acta Biochim. Pol. 2022, 69, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, P.; Chen, Y.; Zhang, S.; Zhang, Z.; Zhang, Z.; Wang, Y.; Jiang, X.; Lin, K.; Wu, W.; et al. Isoalantolactone Induces Cell Cycle Arrest, Apoptosis and Autophagy in Colorectal Cancer Cells. Front. Pharmacol. 2022, 13, 903599. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.C.; Hui, X.G.; Huo, L.; Sun, D.X.; Peng, W.; Zhang, Y.; Li, X.B.; Ma, T.; Li, W.H.; Liang, J.; et al. Antiproliferative effects of isoalantolactone in human liver cancer cells are mediated through caspase-dependent apoptosis, ROS generation, suppression of cell migration and invasion and targeting Ras/Raf/MEK signalling pathway. Acta Biochim. Pol. 2022, 69, 299–304. [Google Scholar] [CrossRef]
- Li, Z.; Qin, B.; Qi, X.; Mao, J.; Wu, D. Isoalantolactone induces apoptosis in human breast cancer cells via ROS-mediated mitochondrial pathway and downregulation of SIRT1. Arch. Pharm. Res. 2016, 39, 1441–1453. [Google Scholar] [CrossRef]
- Huang, H.; Li, P.; Ye, X.; Zhang, F.; Lin, Q.; Wu, K.; Chen, W. Isoalantolactone Increases the Sensitivity of Prostate Cancer Cells to Cisplatin Treatment by Inducing Oxidative Stress. Front. Cell Dev. Biol. 2021, 9, 632779. [Google Scholar] [CrossRef]
- Chun, J. Isoalantolactone Suppresses Glycolysis and Resensitizes Cisplatin-Based Chemotherapy in Cisplatin-Resistant Ovarian Cancer Cells. Int. J. Mol. Sci. 2023, 24, 12397. [Google Scholar] [CrossRef]
- Wang, Z.; Hu, Q.; Chen, H.; Shi, L.; He, M.; Liu, H.; Li, T.; Lu, M.; Deng, M.; Luo, G. Inhibition of Growth of Esophageal Cancer by Alantolactone via Wnt/beta- Catenin Signaling. Anticancer Agents Med. Chem. 2021, 21, 2525–2535. [Google Scholar] [CrossRef] [PubMed]
- Nasirzadeh, M.; Atari Hajipirloo, S.; Gholizadeh-Ghaleh Aziz, S.; Rasmi, Y.; Babaei, G.; Alipour, S. Alantolactone triggers oxeiptosis in human ovarian cancer cells via Nrf2 signaling pathway. Biochem. Biophys. Rep. 2023, 35, 101537. [Google Scholar] [CrossRef]
- He, Y.; Cao, X.; Kong, Y.; Wang, S.; Xia, Y.; Bi, R.; Liu, J. Apoptosis-promoting and migration-suppressing effect of alantolactone on gastric cancer cell lines BGC-823 and SGC-7901 via regulating p38MAPK and NF-kappaB pathways. Hum. Exp. Toxicol. 2019, 38, 1132–1144. [Google Scholar] [CrossRef]
- Ren, Y.; Lv, C.; Zhang, J.; Zhang, B.; Yue, B.; Luo, X.; Yu, Z.; Wang, H.; Ren, J.; Wang, Z.; et al. Alantolactone exhibits antiproliferative and apoptosis-promoting properties in colon cancer model via activation of the MAPK-JNK/c-Jun signaling pathway. Mol. Cell Biochem. 2021, 476, 4387–4403. [Google Scholar] [CrossRef]
- Kim, E.J.; Hong, J.E.; Lim, S.S.; Kwon, G.T.; Kim, J.; Kim, J.S.; Lee, K.W.; Park, J.H. The hexane extract of Saussurea lappa and its active principle, dehydrocostus lactone, inhibit prostate cancer cell migration. J. Med. Food 2012, 15, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; Dai, J.; Gan, A.; Wang, J.; Lin, F.; Zhang, X.; Lv, X.; Wu, B.; Yan, T.; Jia, Y. A network pharmacology approach to investigate dehydrocostus lactone inhibits the proliferation and epithelial-mesenchymal transition of human gastric cancer cells via regulating the PI3K/Akt and extracellular signal-regulated kinases/mitogen-activated protein kinase signalling pathways. J. Pharm. Pharmacol. 2023, 75, 1344–1356. [Google Scholar] [PubMed]
- Long, H.Y.; Huang, Q.X.; Yu, Y.Y.; Zhang, Z.B.; Yao, Z.W.; Chen, H.B.; Feng, J.W. Dehydrocostus lactone inhibits in vitro gastrinoma cancer cell growth through apoptosis induction, sub-G1 cell cycle arrest, DNA damage and loss of mitochondrial membrane potential. Arch. Med. Sci. 2019, 15, 765–773. [Google Scholar] [CrossRef]
- Guo, H.Y.; Chen, Z.A.; Shen, Q.K.; Quan, Z.S. Application of triazoles in the structural modification of natural products. J. Enzyme. Inhib. Med. Chem. 2021, 36, 1115–1144. [Google Scholar] [CrossRef]
- Kolb, H.C.; Sharpless, K.B. The growing impact of click chemistry on drug discovery. Drug Discov. Today 2003, 8, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Neganova, M.E.; Smirnova, E.V.; Sharova, E.V.; Artyushin, O.I.; Aleksandrova, Y.R.; Yandulova, E.Y.; Nikolaeva, N.S.; Brel, V.K. Design of Conjugates Based on Sesquiterpene Lactones with Polyalkoxybenzenes by “Click” Chemistry to Create Potential Anticancer Agents. Molecules 2022, 27, 8411. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, A.S.; Yarovaya, O.I.; Artyushin, O.I.; Sharova, E.V.; Baev, D.S.; Mordvinova, E.D.; Shcherbakov, D.N.; Shnaider, T.A.; Nikitina, T.V.; Esaulkova, I.L.; et al. Design, synthesis and antiviral evaluation of novel conjugates of the 1,7,7-trimethylbicyclo[2.2.1]heptane scaffold and saturated N-heterocycles via 1,2,3-triazole linker. Arch. Pharm. 2024, 357, e2300549. [Google Scholar] [CrossRef] [PubMed]
- Artyushin, O.I.; Moiseeva, A.A.; Zarubaev, V.V.; Slita, A.V.; Galochkina, A.V.; Muryleva, A.A.; Borisevich, S.S.; Yarovaya, O.I.; Salakhutdinov, N.F.; Brel, V.K. Synthesis of Camphecene and Cytisine Conjugates Using Click Chemistry Methodology and Study of Their Antiviral Activity. Chem. Biodivers 2019, 16, e1900340. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Liu, X.; Shen, J.; Zhao, G.; Wang, P.G. The impact of click chemistry in medicinal chemistry. Expert Opin. Drug Discov. 2012, 7, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.; Duffield, K.M.; Ness, M.R.; Ravoori, S.; Andrews, G.; Bhullar, K.S.; Rupasinghe, H.P.; Balzarini, J. Curcumin-inspired cytotoxic 3,5-bis(arylmethylene)-1-(N-(ortho-substituted aryl)maleamoyl)-4-piperidones: A novel group of topoisomerase II alpha inhibitors. Bioorg. Med. Chem. 2015, 23, 6404–6417. [Google Scholar] [CrossRef]
- Adekenov, S.; Spiwok, V.; Beutler, J.; Maslova, O.; Rakhimov, K. Cytotoxicity and Antitumor Activity of Arglabin and its Derivatives. Open Access Maced. J. Med. Sci. 2023, 11, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Giordano, A.; Tommonaro, G. Curcumin and Cancer. Nutrients 2019, 11, 2376. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, M.; Wang, S.; He, Y.; Huo, Y.; Yang, Z.; Cao, X. Alantolactone induces apoptosis and suppresses migration in MCF-7 human breast cancer cells via the p38 MAPK, NF-kappaB and Nrf2 signaling pathways. Int. J. Mol. Med. 2018, 42, 1847–1856. [Google Scholar]
- Sun, X.; Xu, H.; Dai, T.; Xie, L.; Zhao, Q.; Hao, X.; Sun, Y.; Wang, X.; Jiang, N.; Sang, M. Alantolactone inhibits cervical cancer progression by downregulating BMI1. Sci. Rep. 2021, 11, 9251. [Google Scholar] [CrossRef]
- Kemboi, D.; Langat, M.K.; Siwe-Noundou, X.; Tshiwawa, T.; Krause, R.W.M.; Davison, C.; Smit, C.J.; de la Mare, J.A.; Tembu, V.J. 13-amino derivatives of dehydrocostus lactone display greatly enhanced selective toxicity against breast cancer cells and improved binding energies to protein kinases in silico. PLoS ONE 2022, 17, e0271389. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Multhoff, G. Revisiting the Warburg effect: Historical dogma versus current understanding. J. Physiol. 2021, 599, 1745–1757. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhai, Z.; Duan, J.; Wang, X.; Zhong, J.; Wu, L.; Li, A.; Cao, M.; Wu, Y.; Shi, H.; et al. Lactate: The Mediator of Metabolism and Immunosuppression. Front. Endocrinol. 2022, 13, 901495. [Google Scholar] [CrossRef]
- Icard, P.; Shulman, S.; Farhat, D.; Steyaert, J.M.; Alifano, M.; Lincet, H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist. Updat. 2018, 38, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, Y.; Yuan, M.; Song, Q.; Liu, M. Correlation between the Warburg effect and progression of triple-negative breast cancer. Front. Oncol. 2022, 12, 1060495. [Google Scholar] [CrossRef] [PubMed]
- Nantasupha, C.; Thonusin, C.; Charoenkwan, K.; Chattipakorn, S.; Chattipakorn, N. Metabolic reprogramming in epithelial ovarian cancer. Am. J. Transl. Res. 2021, 13, 9950–9973. [Google Scholar] [PubMed]
- Duan, W.; Liu, W.; Xia, S.; Zhou, Y.; Tang, M.; Xu, M.; Lin, M.; Li, X.; Wang, Q. Warburg effect enhanced by AKR1B10 promotes acquired resistance to pemetrexed in lung cancer-derived brain metastasis. J. Transl. Med. 2023, 21, 547. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Teng, Y.; Li, X.; Zhong, Y.; Li, C.; Yan, F.; He, X. Hypoxia promotes non-small cell lung cancer cell stemness, migration, and invasion via promoting glycolysis by lactylation of SOX9. Cancer Biol. Ther. 2024, 25, 2304161. [Google Scholar] [CrossRef]
- Li, Y.; Ma, H. circRNA PLOD2 promotes tumorigenesis and Warburg effect in colon cancer by the miR-513a-5p/SIX1/LDHA axis. Cell Cycle 2022, 21, 2484–2498. [Google Scholar] [CrossRef]
- Zhong, X.; He, X.; Wang, Y.; Hu, Z.; Huang, H.; Zhao, S.; Wei, P.; Li, D. Warburg effect in colorectal cancer: The emerging roles in tumor microenvironment and therapeutic implications. J. Hematol. Oncol. 2022, 15, 160. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Lee, H.; Jo, W.Y.; Byun, Y.H.; Shin, K.W.; Choi, S.; Oh, H.; Park, C.K.; Park, H.P. The Warburg effect in patients with brain tumors: A comprehensive analysis of clinical significance. J. Neurooncol. 2023, 165, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Shi, L.; Wang, L.; Zhang, D.; Li, Y. Dihydroartemisinin enhances the anti-tumour effect of photodynamic therapy by targeting PKM2-mediated glycolysis in oesophageal cancer cell. J. Enzyme. Inhib. Med. Chem. 2024, 39, 2296695. [Google Scholar] [CrossRef]
- Zhu, S.; Guo, Y.; Zhang, X.; Liu, H.; Yin, M.; Chen, X.; Peng, C. Pyruvate kinase M2 (PKM2) in cancer and cancer therapeutics. Cancer Lett. 2021, 503, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Escara-Wilke, J.; Keller, J.M.; Jung, Y.; Taichman, R.S.; Pienta, K.J.; Keller, E.T. Primary prostate cancer educates bone stroma through exosomal pyruvate kinase M2 to promote bone metastasis. J. Exp. Med. 2019, 216, 2883–2899. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhang, Z.; Li, G.; Lai, X.; Gu, R.; Xu, W.; Chen, H.; Xing, Z.; Chen, L.; Qian, J.; et al. Pyruvate Kinase M2 Promotes Prostate Cancer Metastasis Through Regulating ERK1/2-COX-2 Signaling. Front. Oncol. 2020, 10, 544288. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, H.; Xing, R.; Pan, Y.; Li, W.; Cui, J.; Lu, Y. Pyruvate kinase M2 contributes to cell growth in gastric cancer via aerobic glycolysis. Pathol. Res. Pract. 2019, 215, 152409. [Google Scholar] [CrossRef] [PubMed]
- Yavuz, B.B.; Kilinc, F.; Kanyilmaz, G.; Aktan, M. Pyruvate kinase M2 (PKM-2) expression and prognostic significance in glioblastoma patients. J. Neurooncol. 2023, 165, 527–533. [Google Scholar] [CrossRef]
- Sun, X.; Wang, M.; Wang, M.; Yu, X.; Guo, J.; Sun, T.; Li, X.; Yao, L.; Dong, H.; Xu, Y. Metabolic Reprogramming in Triple-Negative Breast Cancer. Front. Oncol. 2020, 10, 428. [Google Scholar] [CrossRef]
- Semakov, A.V.; Klochkov, S.G. Methods of preparative isolation of isoalantholactone and alantholactone from elecampane root. Chem. Plant Raw Mater. 2020, 3, 145–154. [Google Scholar] [CrossRef]
- Semakov, A.V.; Anikina, L.V.; Klochkov, S.G. Synthesis and cytotoxic activity of the products of addition of thiophenol to sesquiterpene lactones. Russ. J. Bioorg. Chem. 2021, 47, 906–917. [Google Scholar] [CrossRef]
- Gregory, M.; Dandavati, A.; Lee, M.; Tzou, S.; Savagian, M.; Brien, K.A.; Satam, V.; Patil, P.; Lee, M. Synthesis, cytotoxicity, and structure–activity insight of NH- and N-methyl-3,5-bis-(arylidenyl)-4-piperidones. Med. Chem. Res. 2013, 22, 5588–5597. [Google Scholar]
- Aditama, R.; Eryanti, Y.; Mujahidin, D.; Syah, Y.M.; Hertadi, R. Determination of activities of human carbonic anhydrase II inhibitors from curcumin analogs. Trop. J. Pharm. Res. 2017, 16, 849–854. [Google Scholar] [CrossRef]
- Tanbin, S.; Ahmad Fuad, F.A.; Abdul Hamid, A.A. Virtual Screening for Potential Inhibitors of Human Hexokinase II for the Development of Anti-Dengue Therapeutics. BioTech 2020, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.H.; Ferreira, J.C.; Nedyalkova, L.; Zhu, H.; Carrasco-Lopez, C.; Kirmizialtin, S.; Rabeh, W.M. The catalytic inactivation of the N-half of human hexokinase 2 and structural and biochemical characterization of its mitochondrial conformation. Biosci. Rep. 2018, 38, BSR20171666. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Li, Y.; Uyeda, K.; Hasemann, C.A. Tissue-specific structure/function differentiation of the liver isoform of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. J. Biol. Chem. 2003, 278, 523–530. [Google Scholar] [CrossRef]
- Nandi, S.; Dey, M. Biochemical and structural insights into how amino acids regulate pyruvate kinase muscle isoform 2. J. Biol. Chem. 2020, 295, 5390–5403. [Google Scholar] [CrossRef]


| IC50 of Cytotoxic Effect, µM | |||||
|---|---|---|---|---|---|
| MCF-7 | SH-SY5Y | HeLa | IMR-32 | WI-38 | |
| 1 | 27.51 ± 0.20 | 19.22 ± 0.1 | 13.41 ± 0.12 | 26.83 ± 0.27 | 74.03 ± 0.11 |
| 2 | 13.15 ± 0.13 | 15.31 ± 0.09 | 10.05 ± 0.03 | 30.26 ± 1.10 | 48.45 ± 0.14 |
| 3 | 10.56 ± 0.07 | 9.15 ± 0.11 | 7.24 ± 0.06 | 22.00 ± 0.98 | 38.55 ± 0.86 |
| 7a | 23.93 ± 0.08 | 4.75 ± 0.03 | 9.21 ± 0.03 | 25.66 ± 0.63 | 5.26 ± 0.07 |
| 7b | 6.73 ± 0.05 | 4.83 ± 0.02 | 43.99 ± 0.37 | 9.05 ± 0.09 | 2.63 ± 0.08 |
| 7c | 6.95 ± 0.02 | 5.55 ± 0.05 | 28.09 ± 0.35 | 24.14 ± 0.08 | 0.66 ± 0.05 |
| 9a | 8.90 ± 0.18 | 8.84 ± 0.01 | 57.29 ± 1.69 | 8.73 ± 0.02 | 39.47 ± 0.07 |
| SI = 4 | SI = 4 | SI = 5 | |||
| 9b | 9.44 ± 0.13 | 8.30 ± 0.01 | 8.77 ± 0.02 | 6.72 ± 0.07 | 28.08 ± 0.03 |
| SI = 3 | SI = 3 | SI = 3 | SI = 4 | ||
| 9c | 24.88 ± 0.21 | 21.95 ± 0.23 | 19.81 ± 0.17 | 21.95 ± 0.21 | 35.65 ± 0.08 |
| 10a | 8.60 ± 0.16 | 10.01 ± 0.09 | 9.97 ± 0.13 | 16.41 ± 0.07 | 18.48 ± 0.12 |
| 10b | 8.17 ± 0.05 | 7.93 ± 0.05 | 22.68 ± 0.08 | 5.76 ± 0.03 | 29.53 ± 0.03 |
| SI = 4 | SI = 4 | SI = 5 | |||
| 10c | 18.60 ± 0.11 | 19.01 ± 0.08 | 21.31 ± 0.17 | 17.38 ± 0.07 | 28.44 ± 0.13 |
| 11a | 8.91 ± 0.08 | 22.75 ± 0.34 | 21.06 ± 0.04 | 8.64 ± 0.07 | 32.75 ± 0.05 |
| SI = 4 | SI = 4 | ||||
| 11b | 8.07 ± 0.02 | 7.41 ± 0.09 | 6.58 ± 0.04 | 6.07 ± 0.06 | 40.36 ± 0.09 |
| SI = 5 | SI = 5 | SI = 6 | SI = 7 | ||
| 11c | 11.46 ± 0.21 | 14.38 ± 0.11 | 22.73 ± 0.12 | 17.12 ± 0.17 | 22.38 ± 0.08 |
| Arglabin | 21.82 ± 0.34 | 15.06 ± 0.22 | 25.09 ± 0.45 | 30.23 ± 1.35 | 9.32 ± 0.01 |
| Curcumin | 15.24 ± 0.11 | 11.78 ± 0.34 | 12.76 ± 0.52 | 21.82 ± 0.91 | 26.12 ± 0.24 |
| Glycolysis | Glycolytic Capacity | Glycolytic Reserve | |
|---|---|---|---|
| Control | 49.18 ± 0.43 | 79.74 ± 3.14 | 30.56 ± 2.85 |
| 1 | 54.30 ± 2.46 | 77.18 ± 1.56 | 22.88 ± 0.90 |
| 2 | 53.51 ± 2.81 | 79.38 ± 2.50 | 25.87 ± 2.60 |
| 3 | 43.46 ± 6.58 | 77.18 ± 1.56 | 27.44 ± 5.27 |
| 7a | 46.29 ± 2.53 | 62.68 ± 3.95 ** | 16.39 ± 1.75 * |
| 7b | 37.44 ± 4.10 | 54.29 ± 0.72 **** | 16.85 ± 4.01 * |
| 7c | 35.03 ± 2.11 | 46.35 ± 2.99 **** | 11.32 ± 3.63 *** |
| 9a | 41.37 ± 1.10 | 51.82 ± 3.82 **** | 10.45 ± 0.52 *** |
| 9b | 35.79 ± 1.71 | 40.02 ± 3.88 **** | 4.23 ± 2.32 **** |
| 9c | 36.07 ± 4.59 | 48.54 ± 2.55 **** | 12.47 ± 4.31 ** |
| 10a | 38.39 ± 4.74 | 56.49 ± 2.98 **** | 14.08 ± 2.84 ** |
| 10b | 37.11 ± 0.49 | 42.52 ± 1.35 **** | 5.41 ± 0.87 **** |
| 10c | 35.46 ± 1.10 | 46.53 ± 2.37 **** | 11.07 ± 1.57 *** |
| 11a | 34.32 ± 1.90 | 50.34 ± 4.93 **** | 16.02 ± 4.49 * |
| 11b | 32.26 ± 3.66 * | 34.77 ± 3.05 **** | 2.51 ± 1.07 **** |
| 11c | 35.17 ± 1.39 | 44.02 ± 4.19 **** | 8.86 ± 2.93 *** |
| Binding Energy, kcal/mol | |||
|---|---|---|---|
| Hexokinase 2 | 6-Phosphofructo-2-Kinase | Pyruvate Kinase M2 | |
| 1 | −5.2 | −4.1 | −5.8 |
| 2 | −4.8 | −3.9 | −6.0 |
| 3 | −5.5 | −4.2 | −6.1 |
| 7a | −7.0 | −7.3 | −7.4 |
| 7b | −6.9 | −7.0 | −7.6 |
| 7c | −7.3 | −7.8 | −7.9 |
| 9a | −6.4 | −6.2 | −8.7 |
| 9b | −6.5 | −6.5 | −9.2 |
| 9c | −5.9 | −6.1 | −8.5 |
| 10a | −6.0 | −6.4 | −8.4 |
| 10b | −6.4 | −6.2 | −9.4 |
| 10c | −6.3 | −5.9 | −9.0 |
| 11a | −6.5 | −6.5 | −8.7 |
| 11b | −6.0 | −6.3 | −9.9 |
| 11c | −5.8 | −6.0 | −8.9 |
| Reference compound * | −7.6 | - | −8.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neganova, M.E.; Aleksandrova, Y.R.; Sharova, E.V.; Smirnova, E.V.; Artyushin, O.I.; Nikolaeva, N.S.; Semakov, A.V.; Schagina, I.A.; Akylbekov, N.; Kurmanbayev, R.; et al. Conjugates of 3,5-Bis(arylidene)-4-piperidone and Sesquiterpene Lactones Have an Antitumor Effect via Resetting the Metabolic Phenotype of Cancer Cells. Molecules 2024, 29, 2765. https://doi.org/10.3390/molecules29122765
Neganova ME, Aleksandrova YR, Sharova EV, Smirnova EV, Artyushin OI, Nikolaeva NS, Semakov AV, Schagina IA, Akylbekov N, Kurmanbayev R, et al. Conjugates of 3,5-Bis(arylidene)-4-piperidone and Sesquiterpene Lactones Have an Antitumor Effect via Resetting the Metabolic Phenotype of Cancer Cells. Molecules. 2024; 29(12):2765. https://doi.org/10.3390/molecules29122765
Chicago/Turabian StyleNeganova, M. E., Yu. R. Aleksandrova, E. V. Sharova, E. V. Smirnova, O. I. Artyushin, N. S. Nikolaeva, A. V. Semakov, I. A. Schagina, N. Akylbekov, R. Kurmanbayev, and et al. 2024. "Conjugates of 3,5-Bis(arylidene)-4-piperidone and Sesquiterpene Lactones Have an Antitumor Effect via Resetting the Metabolic Phenotype of Cancer Cells" Molecules 29, no. 12: 2765. https://doi.org/10.3390/molecules29122765
APA StyleNeganova, M. E., Aleksandrova, Y. R., Sharova, E. V., Smirnova, E. V., Artyushin, O. I., Nikolaeva, N. S., Semakov, A. V., Schagina, I. A., Akylbekov, N., Kurmanbayev, R., Orynbekov, D., & Brel, V. K. (2024). Conjugates of 3,5-Bis(arylidene)-4-piperidone and Sesquiterpene Lactones Have an Antitumor Effect via Resetting the Metabolic Phenotype of Cancer Cells. Molecules, 29(12), 2765. https://doi.org/10.3390/molecules29122765








