Dynamic Interplay between Microbiota Shifts and Differential Metabolites during Dairy Processing and Storage
Abstract
1. Introduction
2. Results
2.1. General DNA Sequencing Observations
2.2. Diversity, Composition, and Difference of the Microbiota in Raw, Pasteurized, and UHT Milk after Storage
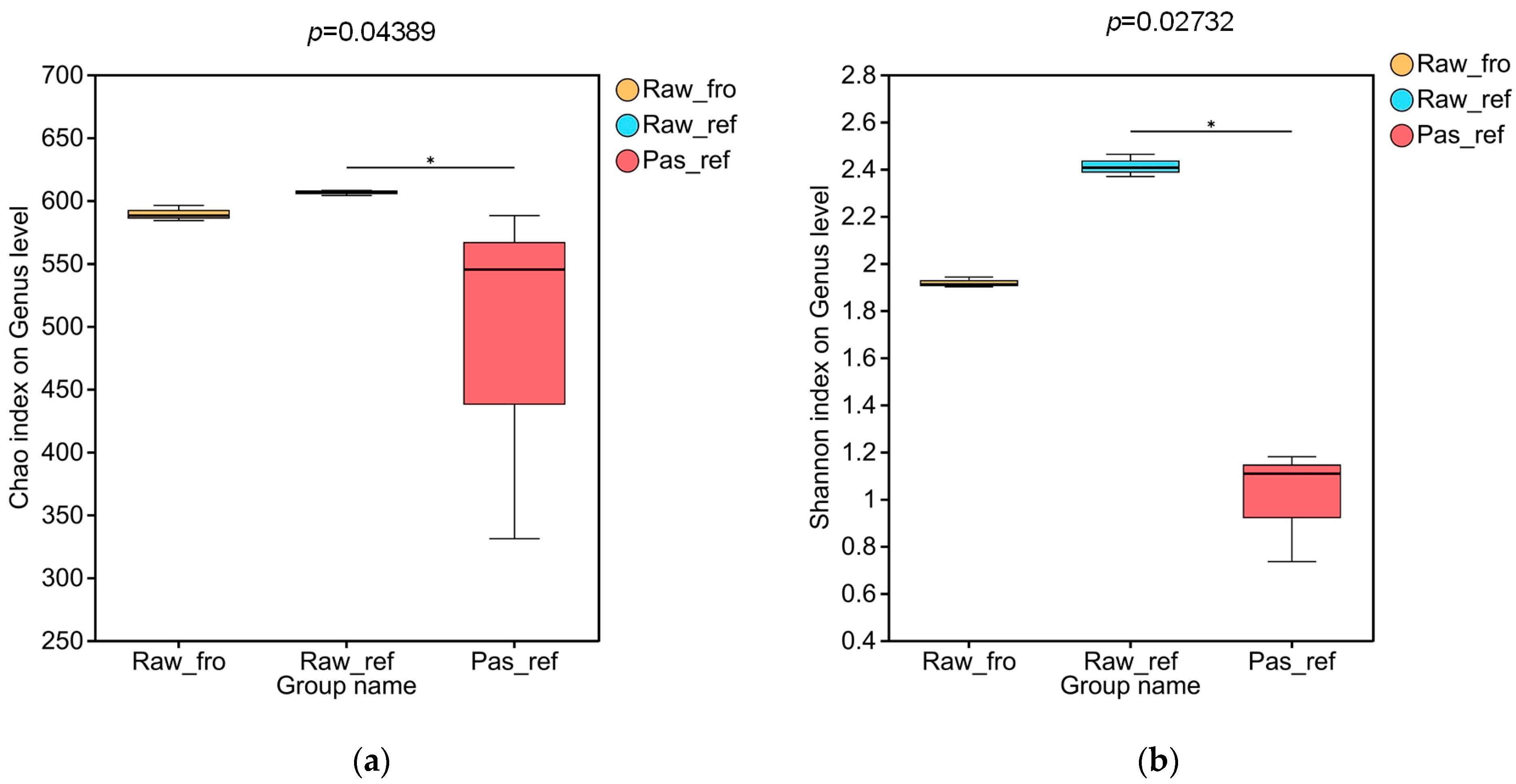
2.2.1. The Diversity of the Processed Milk Samples Microbiota
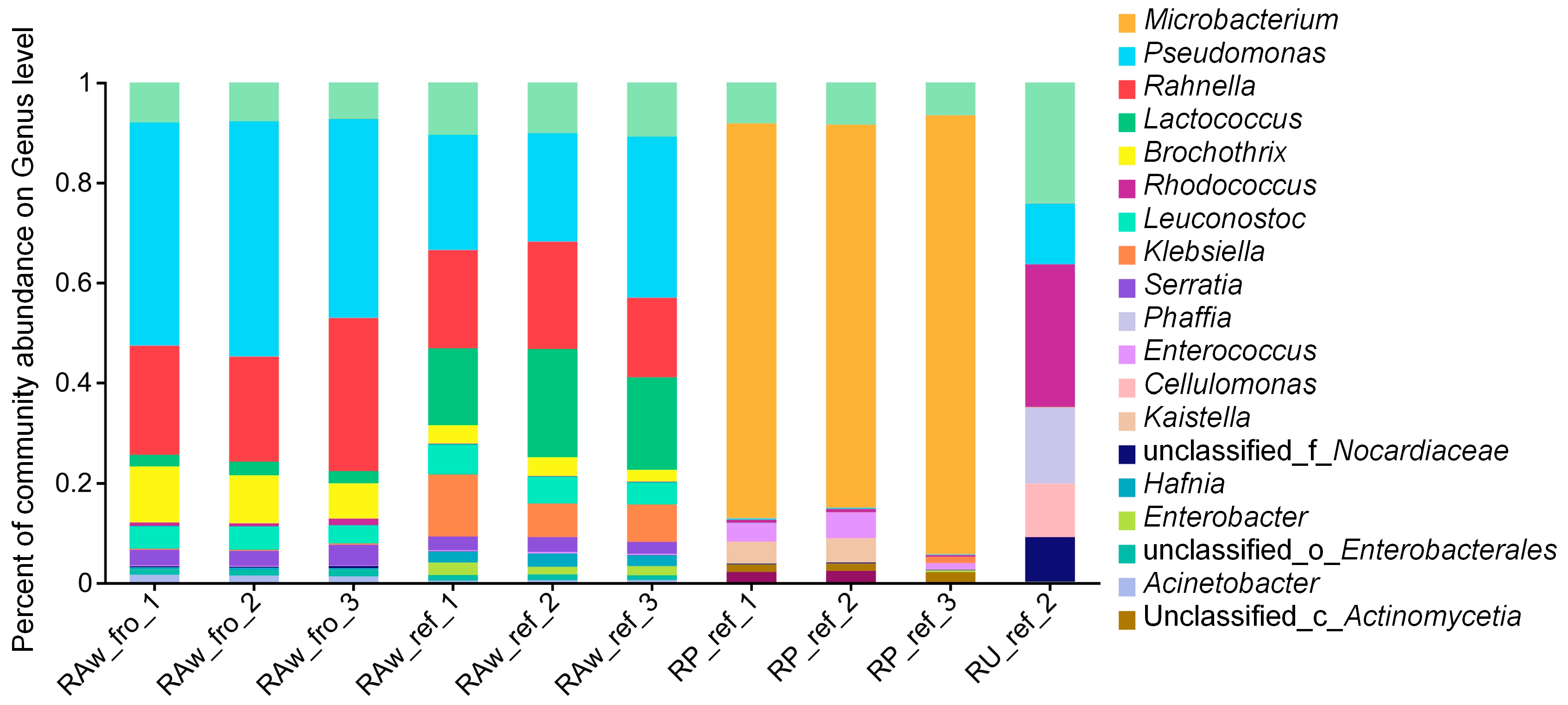
2.2.2. The Composition of the Milk Microbiota
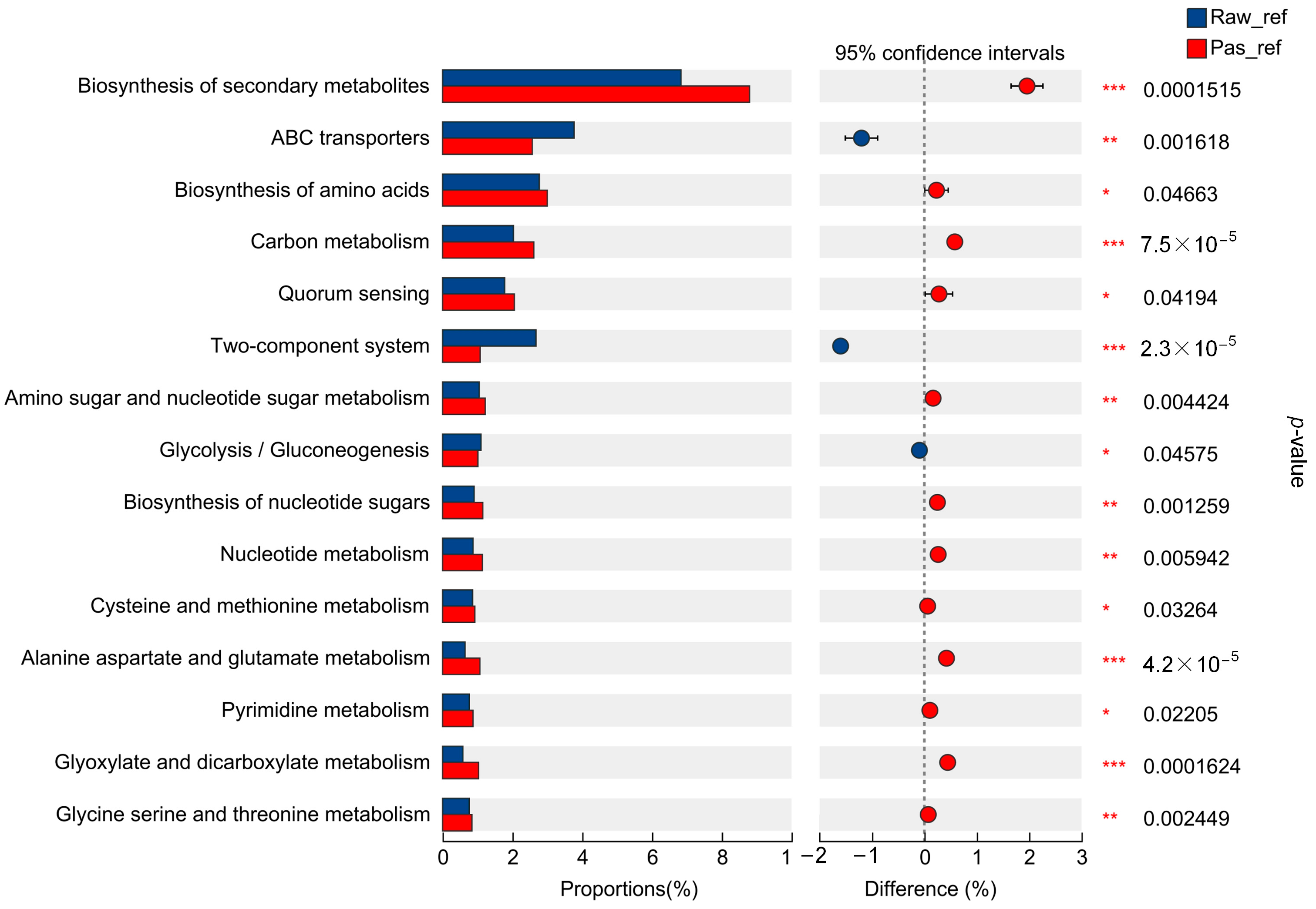
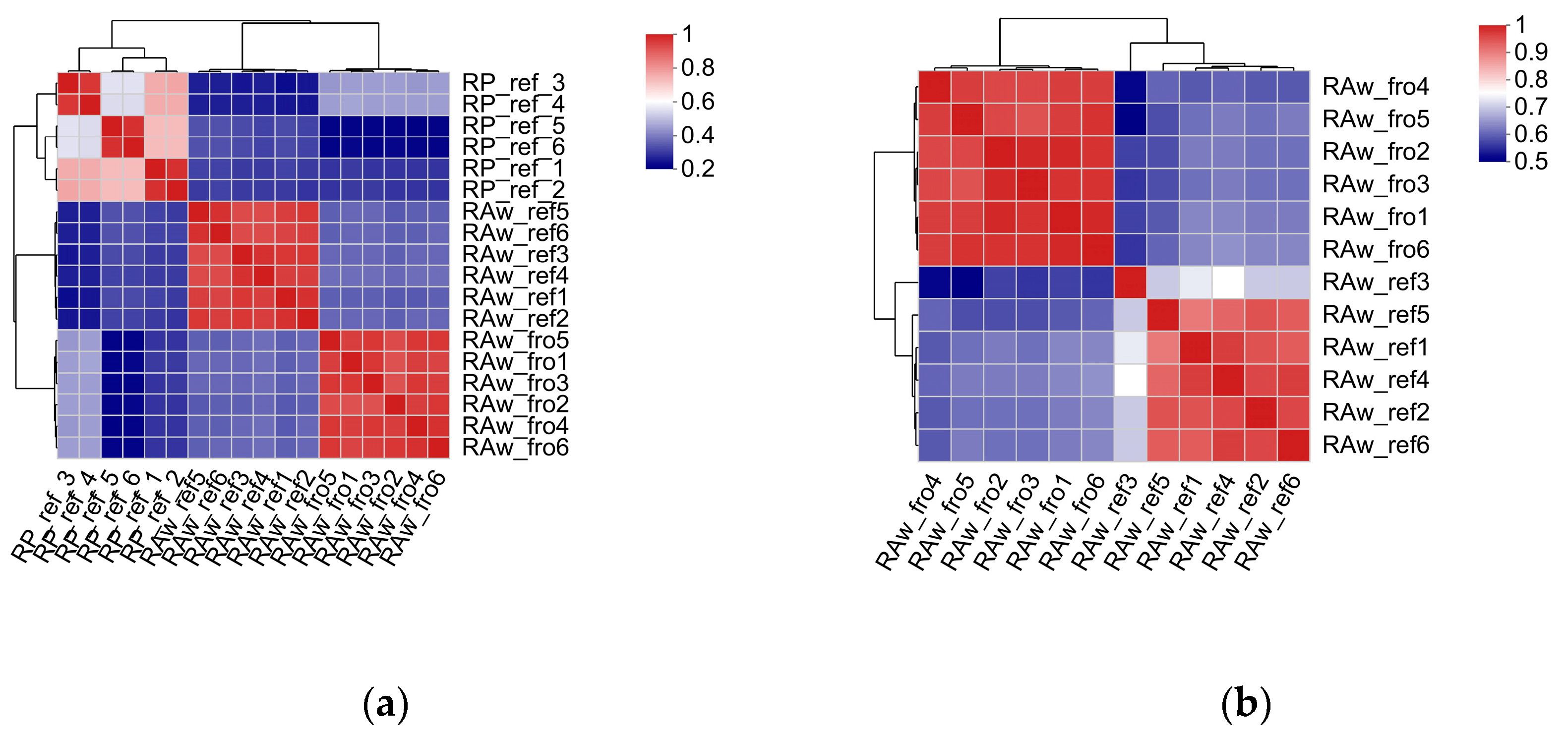
2.2.3. Microbiota Differences and Functional Differences of the Processed Milk Samples
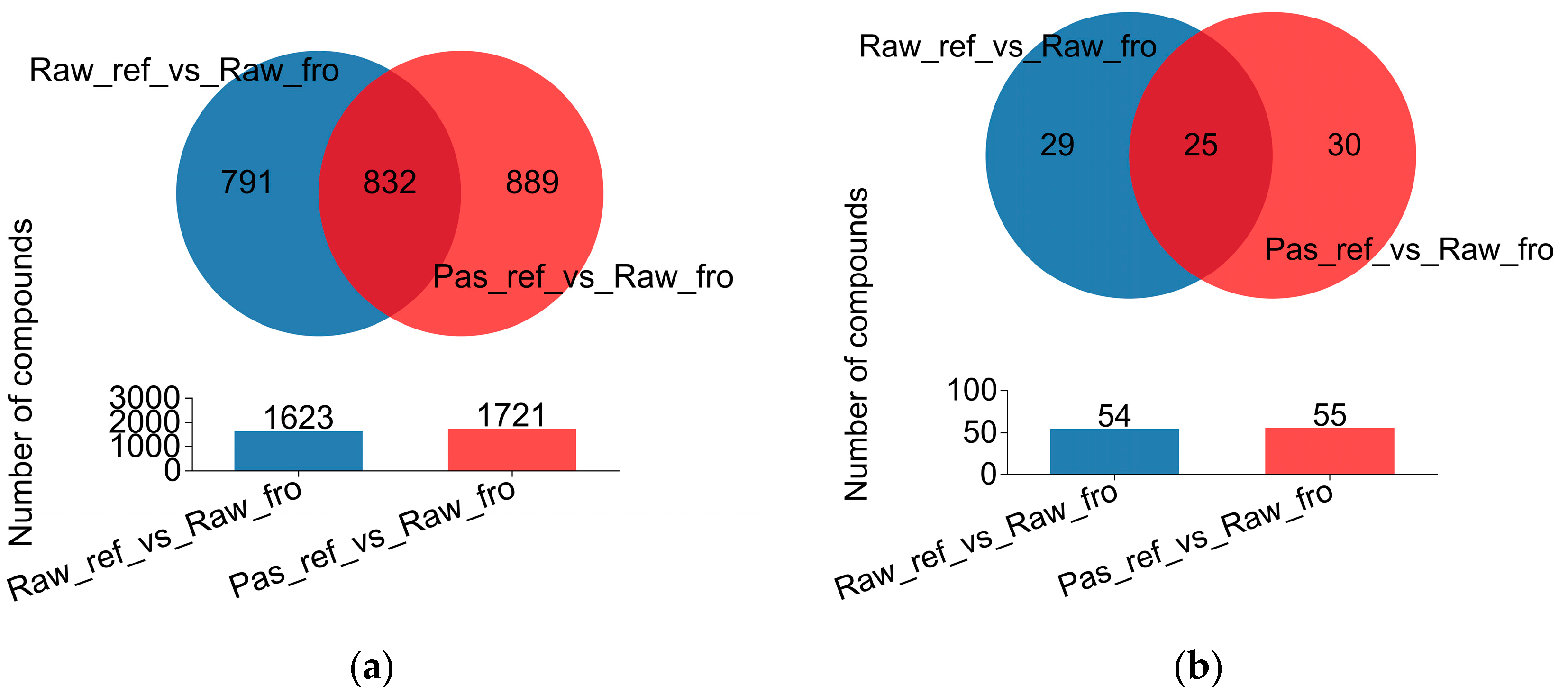
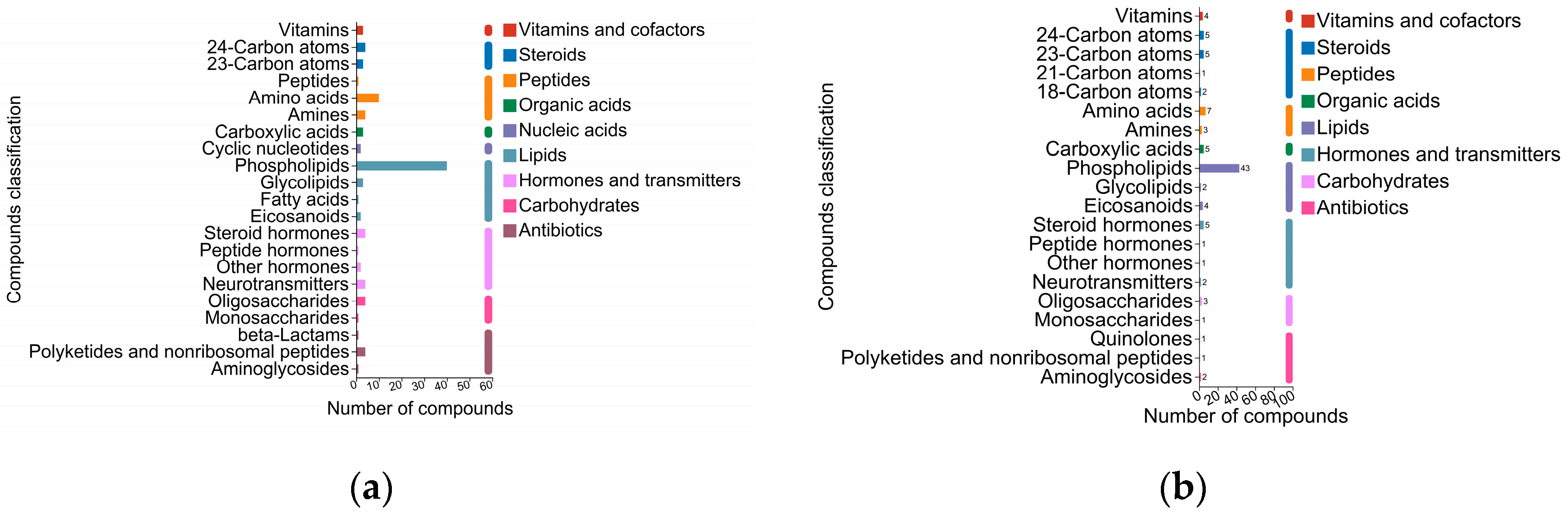
2.3. Identification and Comparison of Milk Metabolites from the Three Groups
2.4. Metabolic Pathways Enrichment
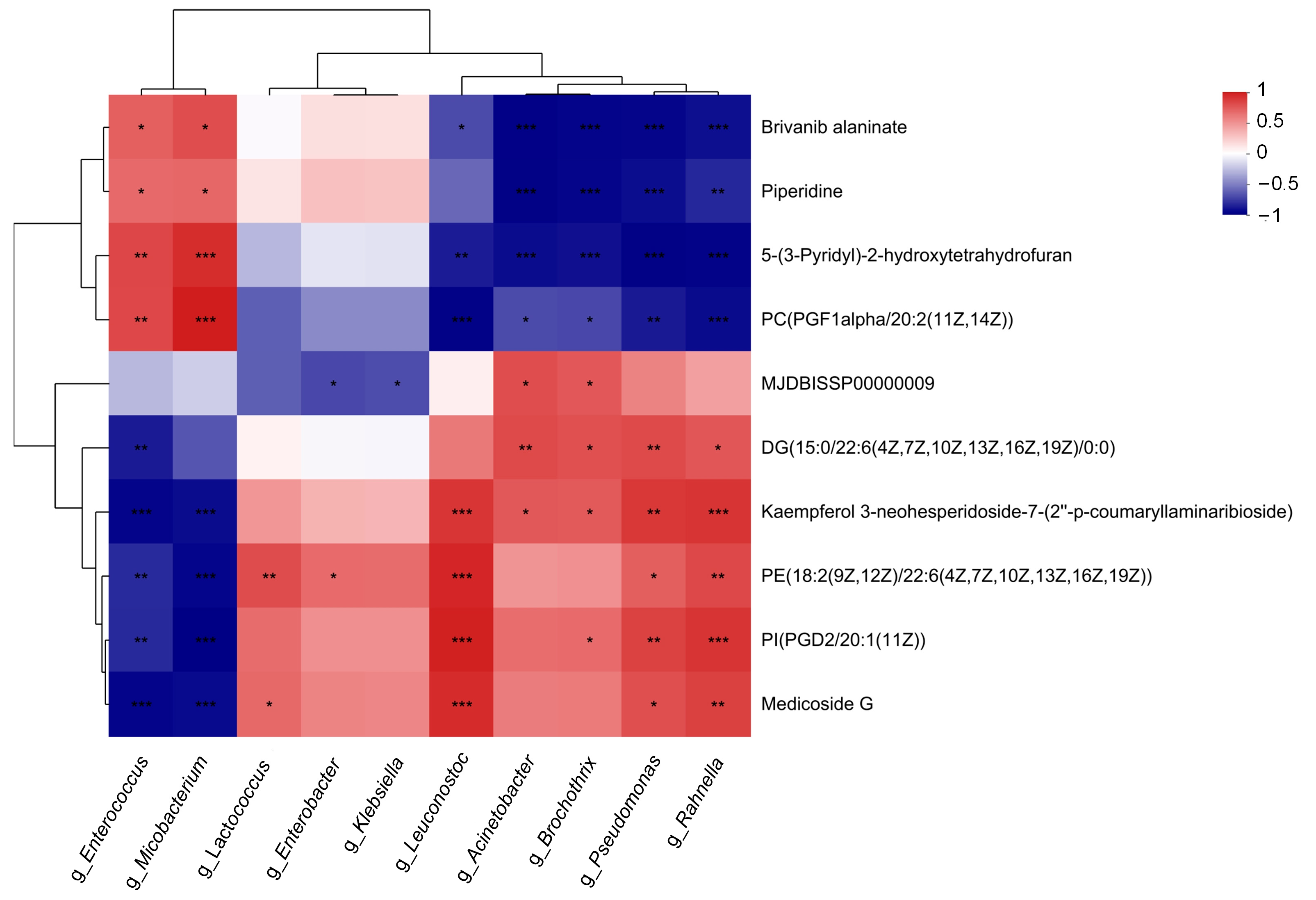
2.5. Correlations between the Refrigerated Pasteurized Milk Microbiome and Differential Metabolites
3. Discussion
4. Materials and Methods
4.1. Samples Collection
4.2. DNA Extraction, Library Construction, and mNGS
4.3. Sequence Quality Control and Genome Assembly
4.4. Gene Prediction, Taxonomy, and Functional Annotation
4.5. Analysis of Microbial Community Diversity
4.6. LC-MS Analysis
4.7. GC-MS Analysis
4.8. MS Data Analysis Processing and Annotation
4.9. Statistical Analysis Methods
4.9.1. Taxa and Metabolite Composition Analysis
4.9.2. Taxa PCoA
4.9.3. Difference Analysis
4.9.4. Correlation Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int. J. Food Microbiol. 2011, 50, 81–94. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed]
- Machado, S.G.; Baglinière, F.; Marchand, S.; Van Coillie, E.; Vanetti, M.C.; De Block, J.; Heyndrickx, M. The biodiversity of the microbiota producing heat-resistant enzymes responsible for spoilage in processed bovine milk and dairy products. Front. Microbiol. 2017, 8, 302. [Google Scholar] [CrossRef]
- O’Connell, A.; Ruegg, P.L.; Jordan, K.; O’Brien, B.; Gleeson, D. The effect of storage temperature and duration on the microbial quality of bulk tank milk. J. Dairy. Sci. 2016, 99, 3367–3374. [Google Scholar] [CrossRef]
- Saha, S.; Majumder, R.; Rout, P.; Hossain, S. Unveiling the significance of psychrotrophic bacteria in milk and milk product spoilage—A review. Microbe 2024, 2, 100034. [Google Scholar] [CrossRef]
- Delbès, C.; Ali-Mandjee, L.; Montel, M.; Montel, M.C. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 2007, 73, 1882–1891. [Google Scholar] [CrossRef]
- Amer, I.H.; EI-Aal, S.F.A.A.; Tolba, M.H. Presence and activity of psychrotrophic bacteria in raw cow’s milk and some dairy products. In Proceedings of the 9thVeterinary Medicine Zagazig University Conference, Port-Said, Egypt, 20–22 August 2008; pp. 61–77. [Google Scholar]
- Buehner, K.P.; Anand, S.; Garcia, A. Prevalence of thermoduric bacteria and spores on 10 midwest dairy farms. J. Dairy. Sci. 2014, 97, 6777–6784. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.F.; Tanca, A.; Uzzau, S.; Oikonomou, G.; Moroni, P. The bovine milk microbiota: Insights and perspectives from omics studies. Mol. Biosyst. 2016, 12, 2359–2372. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Júnior, J.C.; De Oliveira, A.M.; Silva, F.D.; Tamanini, R.; De Oliveira, A.L.M.; Beloti, V. The main spoilage-related psychrotrophic bacteria in refrigerated raw milk. J. Dairy. Sci. 2018, 101, 75–83. [Google Scholar] [CrossRef]
- Lo, R.; Turner, M.S.; Weeks, M.; Bansal, N. Culture-independent bacterial community profiling of carbon dioxide. Int. J. Food Microbiol. 2016, 233, 81–89. [Google Scholar] [CrossRef]
- Porcellato, D.; Aspholm, M.; Skeie, S.B.; Monshaugen, M.; Brendehaug, J.; Mellegård, H. Microbial diversity of consumption milk during processing and storage. Int. J. Food Microbiol. 2018, 266, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Renye, J.A.; Feng, L.; Zeng, Q.; Tang, Y.; Huang, L.; Ren, D.; Yang, P. Characterization of the indigenous microflora in raw and pasteurized buffalo milk during storage at refrigeration temperature by high-throughput sequencing. J. Dairy. Sci. 2016, 99, 7016–7024. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Meng, L.; Liu, H.; Zheng, N.; Zhang, Y.; Zhao, S.; Wang, J. Single molecule real-time sequencing and traditional cultivation techniques reveal complex community structures and regional variations of psychrotrophic bacteria in raw milk. Front. Microbiol. 2022, 13, 853263. [Google Scholar] [CrossRef] [PubMed]
- Doyle, C.J.; Gleeson, D.; O’Toole, P.W.; Cotter, P.D. High-throughput metataxonomic characterization of the raw milk microbiota identifies changes reflecting lactation stage and storage conditions. Int. J. Food Microbiol. 2017, 255, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kamilaria, E.; Anagnostopoulosa, D.A.; Papademasa, P.; Kamilarisb, A.; Tsaltas, D. Characterizing Halloumi cheese’s bacterial communities through metagenomic analysis. LWT Food Sci. Technol. 2020, 126, 109298. [Google Scholar] [CrossRef]
- Marino, M.; De Wittenau, G.D.; Sacca, E.; Cattonaro, F.; Spadotto, A.; Innocente, N.; Radovic, S.; Piasentier, E.; Marroni, F. Metagenomic profiles of different types of Italian high-moisture Mozzarella cheese. Food Microbiol. 2019, 79, 123–131. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Yang, C.; Jin, H.; Kwok, L.Y.; Sun, Z.; Zhang, H. Metagenomic features of traditional fermented milk products. LWT Food Sci. Technol. 2022, 115, 112945. [Google Scholar] [CrossRef]
- Xia, Y.; Liu, H.; Shuang, Q. Analysis on microbiological diversity and functional genes of koumiss based on metagenomic technology. J. Chin. Inst. Food Sci. Technol. 2022, 02, 301–309. [Google Scholar]
- Wang, Y.; Ju, N.; Gou, M.; Luo, Y.; Li, P. Metagenomics approach to explore the changes of bacterial community in raw milk during refrigerated storage. Trans. Chin. Soc. Agric. Eng. 2020, 36, 333–340. [Google Scholar]
- Nikoloudaki, O.; Lemos Junior, W.J.F.; Campanaro, S.; Di Cagno, R.; Gobbetti, M. Role prediction of Gram-negative species in the resistome of raw cow’s milk. Int. J. Food Microbiol. 2021, 340, 109045. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Zheng, N.; Li, S.; Wen, F.; Zhao, S.; Wang, J. Research progress on metabolomics application in dairy cow nutrition and milk quality and safety. Chin. J. Anim. Nutr. 2017, 29, 3035–3043. [Google Scholar]
- Zhu, D.; Kebede, B.; Chen, G.; McComb, K.; Frew, R. Effects of the vat pasteurization process and refrigerated storage on the bovine milk metabolome. J. Dairy. Sci. 2020, 103, 2077–2088. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, M.L.; Huck, J.R.; Sonnen, M.; Barbano, D.M.; Boor, K.J. High temperature, short time pasteurization temperatures inversely affect bacterial numbers during refrigerated storage of pasteurized fluid milk. J. Dairy. Sci. 2009, 92, 4823–4832. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; McCarthy, R.; O’Sullivan, O.; Beresford, T.P.; Fitzgerald, G.F.; Ross, R.P.; Stanton, C.; Cotter, P.D. The microbial content of raw and pasteurized cow milk as determined by molecular approaches. J. Dairy. Sci. 2013, 96, 4928–4937. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.G. Ropiness in pasteurized milk due to a heat resistant micrococcus. J. Appl. Microbiol. 2010, 9, 91–93. [Google Scholar] [CrossRef]
- Fromm, H.I.; Boor, K.J. Characterization of pasteurized fluid milk shelf-life attributes. J. Food Sci. 2004, 69, M207–M214. [Google Scholar] [CrossRef]
- Alothman, M.; Bremer, P.J.; Lusk, K.; Silcock, P. When does milk spoil? The use of rejection threshold methodology to investigate the influence of total microbial numbers on the acceptability of fresh chilled pasteurised milk. Beverages 2023, 9, 53. [Google Scholar] [CrossRef]
- Ziyaina, M.; Rasco, B.; Coffey, T.; Mattinson, D.S.; Sablani, S. Correlation of volatile compound concentrations with bacterial counts in whole pasteurised milk under various storage conditions. Int. J. Dairy. Technol. 2019, 72, 36–46. [Google Scholar] [CrossRef]
- Hahne, J.; Isele, D.; Berning, J.; Lipski, A. The contribution of fast growing, psychrotrophic microorganisms on biodiversity of refrigerated raw cow’s milk with high bacterial counts and their food spoilage potential. Food Microbiol. 2019, 79, 11–19. [Google Scholar] [CrossRef]
- Von Neubeck, M.; Baur, C.; Krewinkel, M.; Stoeckel, M.; Kranz, B.; Stressler, T.; Fischer, L.; Hinrichs, J.; Scherer, S.; Wenning, M. Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int. J. Food Microbiol. 2015, 211, 57–65. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, Z.; You, C. Biodiversity of psychrotrophic bacteria isolated from raw milk and their enzymatic profiles. Chin. J. Dairy. Sci. Technol. 2017, 40, 1–8. [Google Scholar]
- Zhao, C.C.; Eun, J.B. Shotgun metagenomics approach reveals the bacterial community and metabolic pathways in commercial hongeo product, a traditional Korean fermented skate product. Food Res. Int. 2020, 131, 109030. [Google Scholar] [CrossRef] [PubMed]
- Glück, C.; Rentschler, E.; Krewinkel, M.; Merz, M.; Von Neubeck, M.; Wenning, M.; Scherer, S.; Stoeckel, M.; Hinrichs, J.; Stressler, T.; et al. Thermostability of peptidases secreted by microorganisms associated with raw milk. Int. Dairy. J. 2016, 56, 186–197. [Google Scholar] [CrossRef]
- Chen, T.R.; Wei, Q.K.; Chen, Y.J. Pseudomonas spp. and Hafnia alvei growth in UHT milk at cold storage. Food Control 2011, 22, 697–701. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, J.; Zhang, H. Detect microorganism in UHT milk by metagenome sequencing. China Dairy. Ind. 2015, 43, 55–58. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Park, J.; Takagi, T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006, 34, 5623–5630. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Guo, Z.; Yao, Y.; Xia, L.; Liu, R.; Song, H.; Zhang, S. Acetic acid alters rhizosphere microbes and metabolic composition to improve willows drought resistance. Sci. Total Environ. 2022, 844, 157132. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, J. Characterization of metabolite, genome and volatile organic compound changes provides insights into the spoilage and cold adaptive markers of Acinetobacter johnsonii XY27. LWT 2022, 162, 113453. [Google Scholar] [CrossRef]
- Jiang, J.; Li, W.; Yu, S. The effect of inoculation Leuconostoc mesenteroides and Lactiplantibacillus planetarium on the quality of Pleurotus eryngii Jiaosu. LWT 2022, 163, 113445. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multi-omics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]



| Groups | Samples | Optimized Reads | Optimized Bases (bp) | Contigs | ORFs |
|---|---|---|---|---|---|
| Raw_fro | RAw_fro_1 | 53,867,630 | 8,116,741,291 | 70,515 | 142,253 |
| RAw_fro_2 | 55,008,670 | 8,286,484,529 | 64,916 | 135,320 | |
| RAw_fro_3 | 63,800,856 | 9,608,917,943 | 67,471 | 143,276 | |
| Raw_ref | RAw_ref_1 | 62,946,434 | 9,490,863,131 | 70,997 | 154,713 |
| RAw_ref_2 | 71,983,366 | 10,849,612,040 | 72,332 | 160,636 | |
| RAw_ref_3 | 63,740,996 | 9,604,086,372 | 71,333 | 153,063 | |
| Pas_ref | RP_ref_1 | 61,152,560 | 9,214,137,144 | 21,165 | 39,291 |
| RP_ref_2 | 58,224,872 | 8,774,437,192 | 23,589 | 41,496 | |
| RP_ref_3 | 91,038,714 | 13,715,234,644 | 24,320 | 46,970 | |
| UHT_ref 1 | RU_ref_2 | 4,349,358 | 654,056,765 | 14,633 | 53,055 |
| Comparison Groups | Total Number of Differential Metabolites | Top Five Differential Metabolites | VIP 1 | Relative Quantity of Raw_ref (RP_ref) | Relative Quantity of Raw_fro | Regulate | HMDB Subclass |
|---|---|---|---|---|---|---|---|
| Raw_ref vs. Raw_fro | 462 (LC-MS) | Valylmethionine | 2.4517 | 7.35 ± 0.06 | 1.41 ± 0.03 | up | Amino acids, peptides, and analogues |
| Biocytin | 2.2861 | 7.12 ± 0.05 | 1.96 ± 0.03 | up | Amino acids, peptides, and analogues | ||
| PA(8:0/8:0) | 2.1459 | 6.57 ± 0.09 | 2.03 ± 0.03 | up | Glycerophosphates | ||
| Pyridinoline | 2.128 | 6.87 ± 0.17 | 2.21 ± 0.73 | up | Amino acids, peptides, and analogues | ||
| Hydroxytetradecenoylcarnitine | 2.0905 | 6.24 ± 0.16 | 1.92 ± 0.03 | up | Fatty acids and conjugates | ||
| 17 (GC-MS) | Citric Acid | 2.889 | 4.17 ± 0.22 | 7.66 ± 0.04 | down | Tricarboxylic acids and derivatives | |
| 2,3-Butanediol | 2.005 | 7.27 ± 0.14 | 5.58 ± 0.14 | up | Alcohols and polyols | ||
| Butane-2,3-diol | 1.9093 | 7.29 ± 0.09 | 5.76 ± 0.11 | up | Alcohols and polyols | ||
| 2-hydroxy-4-methylpentanoic acid | 1.808 | 5.98 ± 0.10 | 4.61 ± 0.13 | up | Fatty acids and conjugates | ||
| L-Serine | 1.7551 | 5.86 ± 0.86 | 4.28 ± 0.06 | up | Amino acids, peptides, and analogues | ||
| RP_ref vs. Raw_fro | 519 (LC-MS) | 3b,6a-Dihydroxy-alpha-ionol 9-[apiosyl-(1->6)-glucoside] | 2.6584 | 7.31 ± 0.39 | 0.64 ± 0.40 | up | Fatty acyl glycosides |
| PS(22:4(7Z,10Z,13Z,16Z)/22:5(7Z,10Z,13Z,16Z,19Z)) | 2.5301 | 5.93 ± 0.52 | 0.31 ± 0.71 | up | Glycerophosphoserines | ||
| 11-Maleimidoundecanoic acid | 2.5084 | 6.61 ± 0.34 | 1.16 ± 0.01 | up | Fatty acids and conjugates | ||
| Prolyl-Alanine | 2.4519 | 6.09 ± 0.67 | 0.36 ± 0.02 | up | Amino acids, peptides, and analogues | ||
| 3-[(2R)-2-Hydroxy-3-methyl-3-[(phosphonooxy)methyl]butanamido]propanoylcarnitine | 2.449 | 6.37 ± 1.07 | 0.58 ± 0.02 | up | Fatty acid esters | ||
| 22 (GC-MS) | D-Glucose | 2.0015 | 7.34 ± 0.24 | 6.02 ± 0.08 | up | Carbohydrates and carbohydrate conjugates | |
| Ethanamine | 1.9081 | 5.41 ± 1.09 | 7.00 ± 0.06 | down | Amines | ||
| Butanoic acid | 1.6228 | 5.38 ± 0.05 | 4.53 ± 0.07 | up | Fatty acids and conjugates | ||
| Gamma-Aminobutyric Acid | 1.5961 | 6.21 ± 0.50 | 5.23 ± 0.13 | up | Amino acids, peptides, and analogues | ||
| 2-hydroxyhexanoic acid | 1.5905 | 5.32 ± 0.10 | 4.44 ± 0.29 | up | Fatty acids and conjugates |
| Highly Correlated Metabolites | HMDB Subclass | Correlation | |||
|---|---|---|---|---|---|
| Mriobacterium | Micrococcus | Acinetobacter | Pseudomonas | ||
| Gln Leu Leu | – | 0.8743 | 0.8456 | −0.3182 | −0.6086 |
| Arg Leu | – | 0.9686 | 0.9883 | −0.8071 | −0.939 |
| 13-Demethyl tacrolimus | Pyrimidines and pyrimidine derivatives | 0.9683 | 0.9854 | −0.8193 | −0.9456 |
| Levonorgestrel | Estrane steroids | 0.9712 | 0.9938 | −0.7845 | −0.9236 |
| S-(PGA1)-glutathione | Amino acids, peptides, and analogues | 0.9782 | 0.9957 | −0.7563 | −0.9144 |
| PC(PGF1alpha/20:2(11Z,14Z)) | Not Available | 0.9986 | 0.9609 | −0.6973 | −0.8746 |
| Cucurbitacin I 2-glucoside | Steroidal glycosides | 0.9751 | 0.9907 | −0.6389 | −0.8406 |
| APC | Carbonyl compounds | 0.8287 | 0.8517 | −0.9142 | −0.9413 |
| 2-Isopropylmalic Acid | Fatty acids and conjugates | 0.8969 | 0.8692 | −0.9454 | −0.9729 |
| Hydroxysepiapterin | Pterins and derivatives | 0.892 | 0.9071 | −0.9093 | −0.9447 |
| M-Coumaric acid | Phenylpropanoids and polyketides | 0.9021 | 0.9043 | −0.9253 | −0.9873 |
| PS(20:5(5Z,8Z,11Z,14Z,17Z)/16:0) | Glycerophosphoserines | 0.8241 | 0.9162 | −0.8303 | −0.8884 |
| Ppack | Amino acids, peptides, and analogues | 0.9534 | 0.9183 | −0.8924 | −0.9792 |
| 5-(3-Pyridyl)-2-hydroxytetrahydrofuran | Not available | 0.9316 | 0.9244 | −0.9179 | −0.9804 |
| L-Lysine | Amino acids, peptides, and analogues | 0.9223 | 0.9413 | −0.901 | −0.9792 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Yu, P.; Tao, F. Dynamic Interplay between Microbiota Shifts and Differential Metabolites during Dairy Processing and Storage. Molecules 2024, 29, 2745. https://doi.org/10.3390/molecules29122745
Zhang Y, Yu P, Tao F. Dynamic Interplay between Microbiota Shifts and Differential Metabolites during Dairy Processing and Storage. Molecules. 2024; 29(12):2745. https://doi.org/10.3390/molecules29122745
Chicago/Turabian StyleZhang, Yinan, Peng Yu, and Fei Tao. 2024. "Dynamic Interplay between Microbiota Shifts and Differential Metabolites during Dairy Processing and Storage" Molecules 29, no. 12: 2745. https://doi.org/10.3390/molecules29122745
APA StyleZhang, Y., Yu, P., & Tao, F. (2024). Dynamic Interplay between Microbiota Shifts and Differential Metabolites during Dairy Processing and Storage. Molecules, 29(12), 2745. https://doi.org/10.3390/molecules29122745






