Response Surface Methodology (RSM) for Optimizing Protein Extraction from Housefly (Musca domestica) Larvae Fed with Toad and Its Structural Characterization
Abstract
1. Introduction
2. Results and Discussion
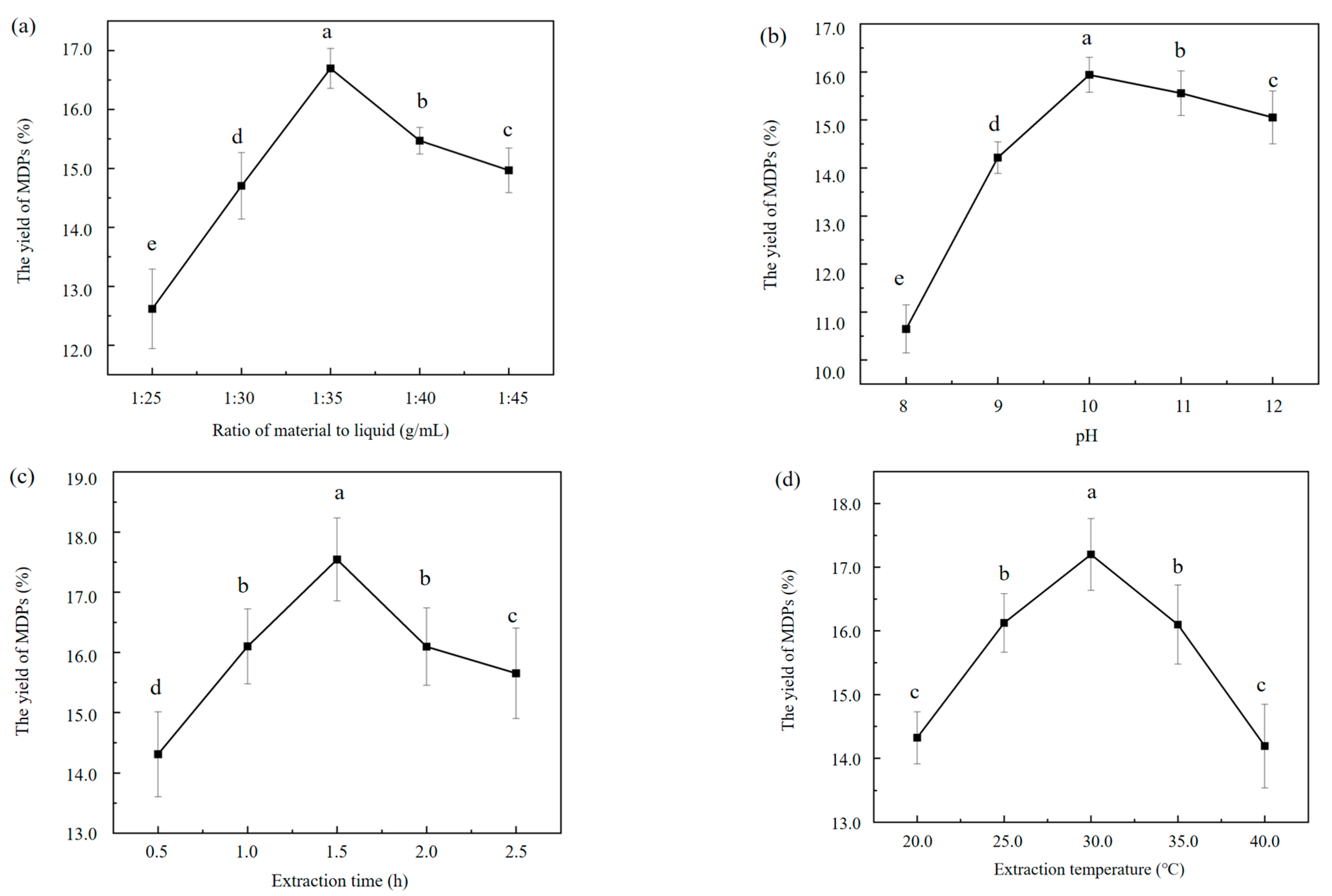
2.1. Influences of Different Extraction Conditions on the Extraction Rate of MDPs
2.2. Optimizing Extraction Parameters of MDPs
2.2.1. Model Fitting
2.2.2. Fitting and Assessing the Adequacy of the Model
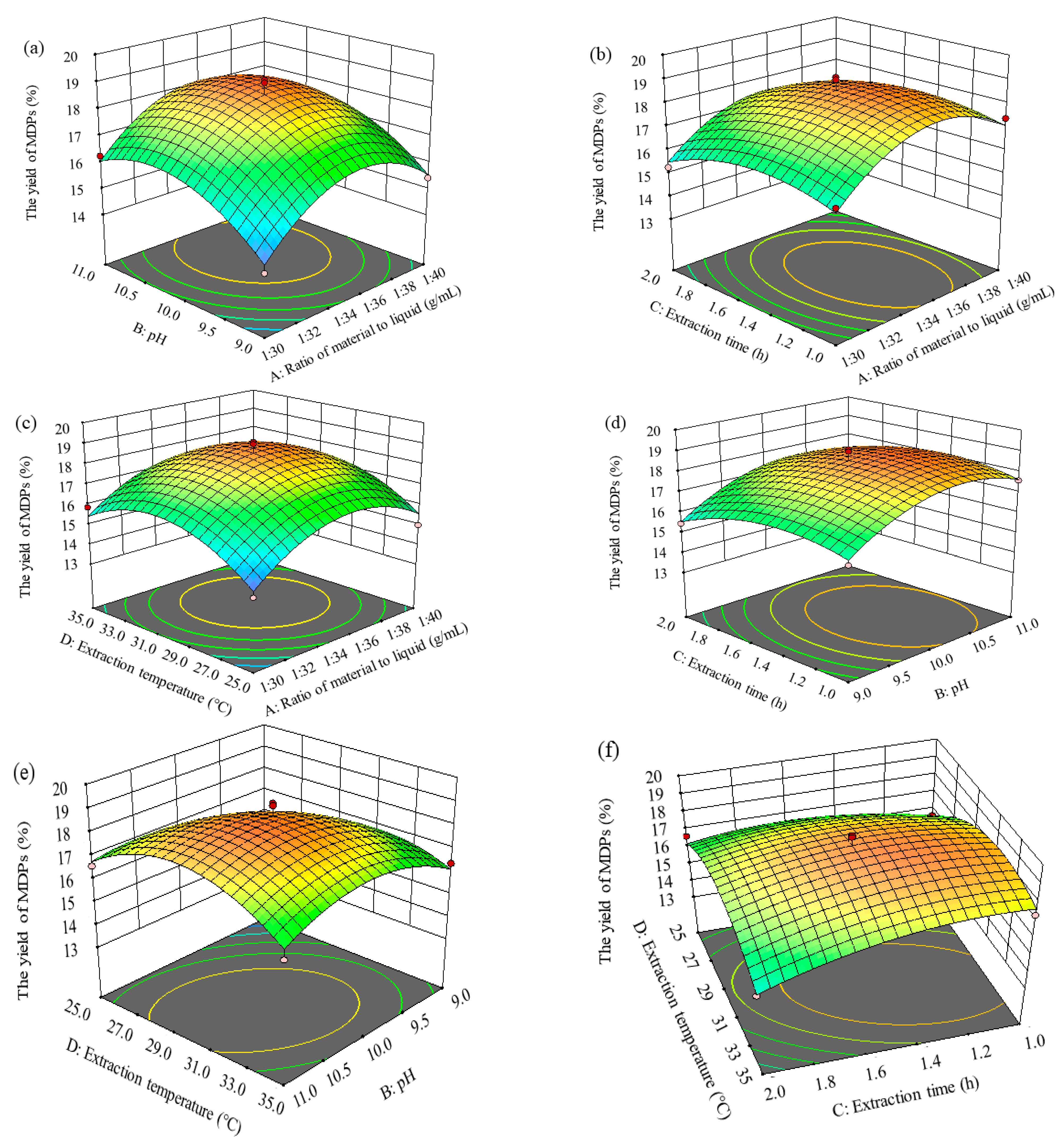
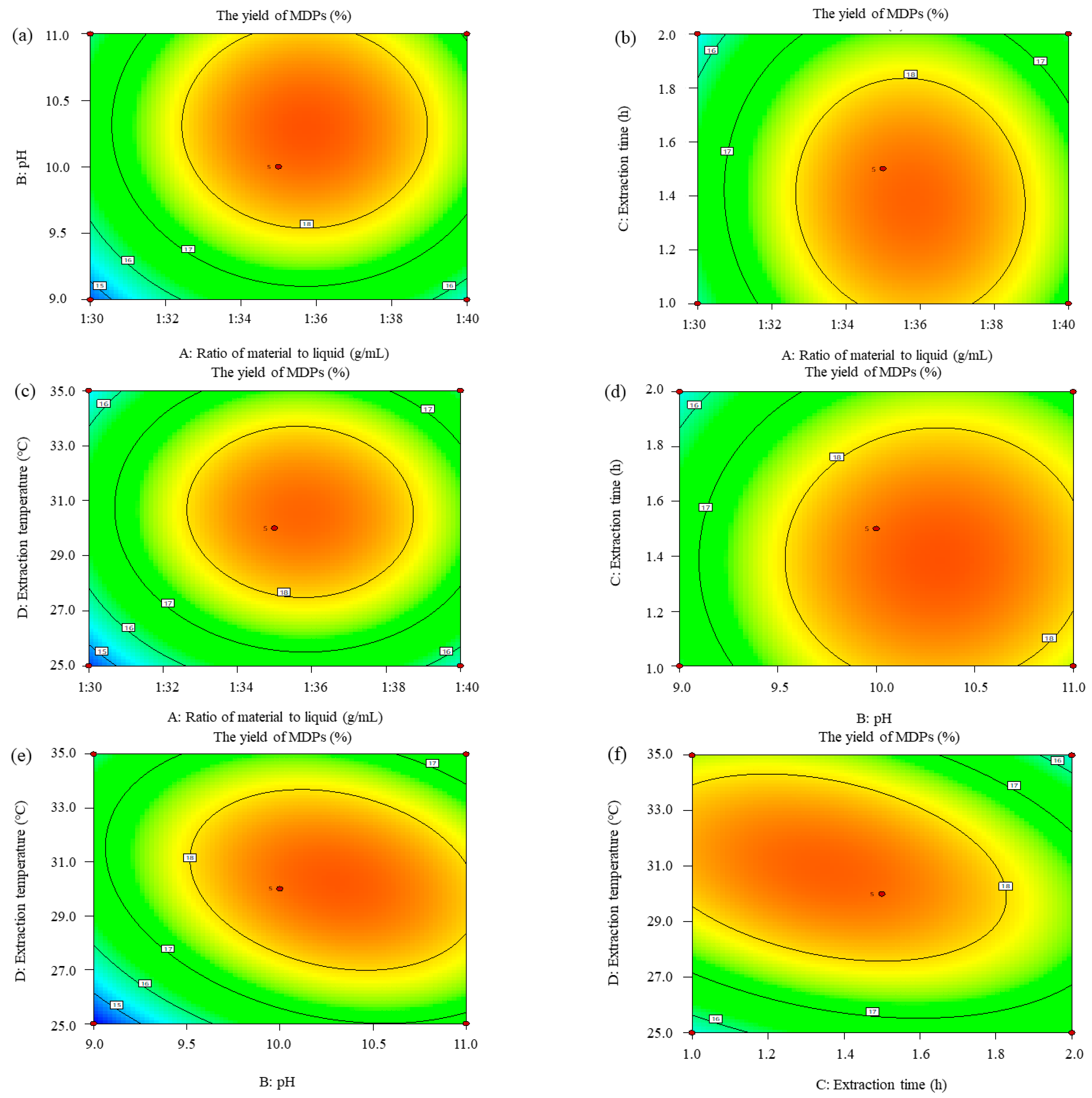
2.2.3. Response Surface Analysis
2.2.4. Model Verification
2.3. Structural Characteristics
2.3.1. Particle Sizing and Zeta Potential
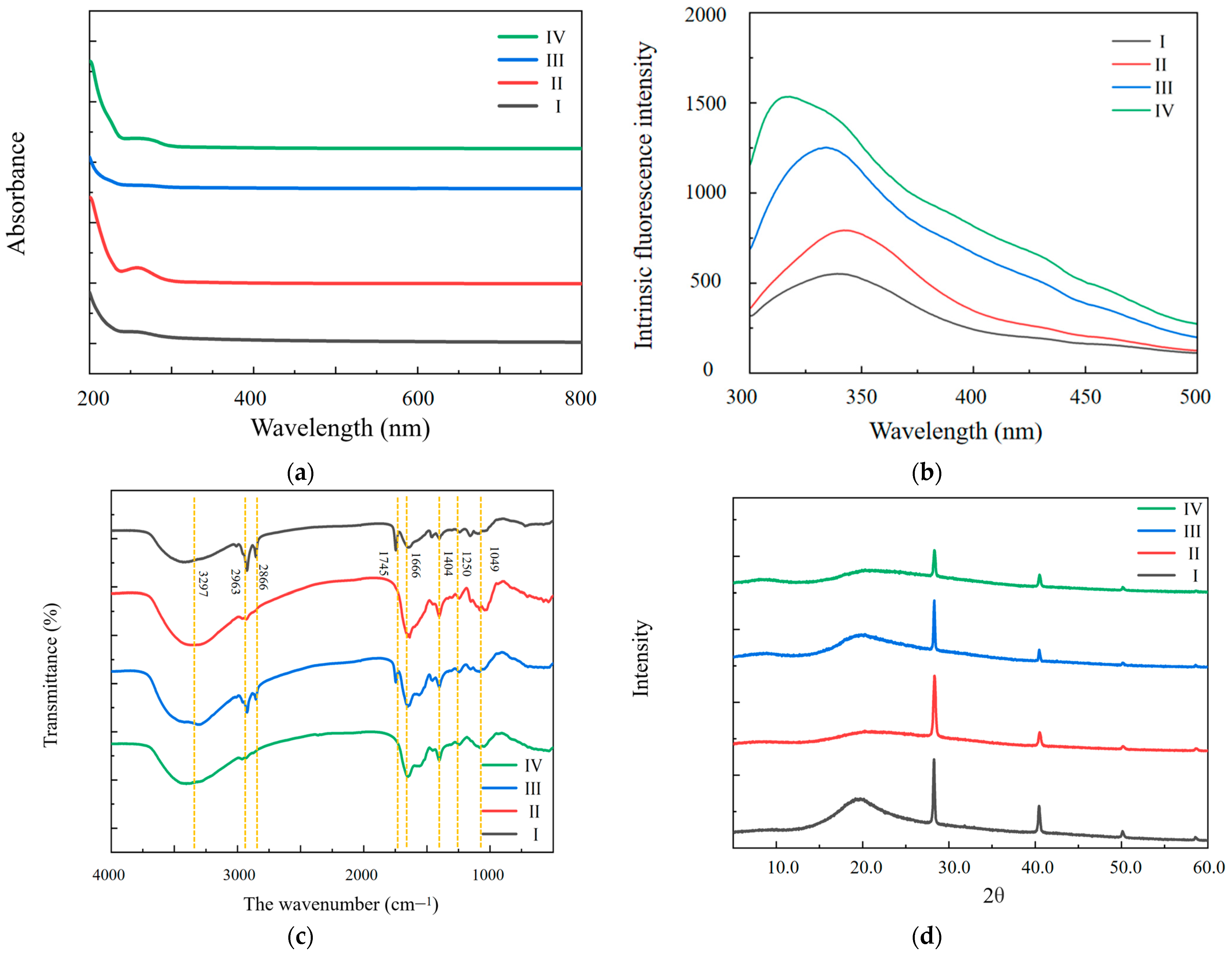
2.3.2. Ultraviolet–Visible (UV–Vis) Spectra and Intrinsic Fluorescence Spectra
2.3.3. FT-IR Spectrometry
2.3.4. Wide-Angle XRD Experiment
2.3.5. Molecular Weight Distribution of MDPs
2.3.6. Amino Acid Analysis of MDPs
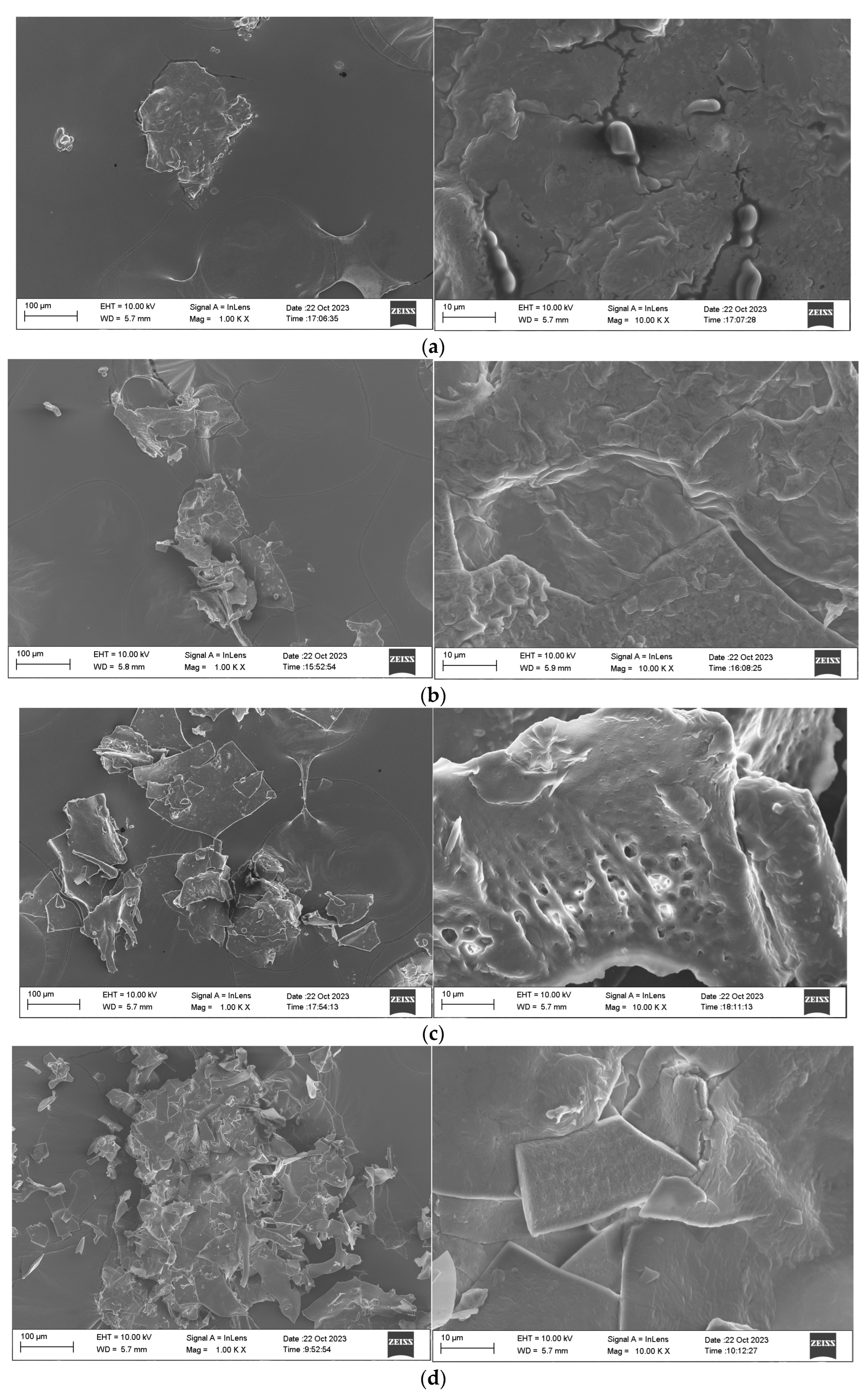
2.3.7. SEM Analysis of MDPs
3. Materials and Methods
3.1. Materials
3.2. Preparing MDPs
3.3. Experimental Design of RSM
3.4. Characterization of MDPs
3.4.1. UV Spectrum and Intrinsic Fluorescence Analysis
3.4.2. Particle Size Distribution and Zeta Potential
3.4.3. FT-IR Spectra
3.4.4. XRD Analysis
3.4.5. Molecular Weight Distribution of MDPs
3.4.6. Analysis of Amino Acid Composition
3.4.7. SEM
3.5. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hou, L.X.; Shi, Y.H.; Zhai, P.; Le, G.W. Antibacterial activity and in vitro anti-tumor activity of the extract of the larvae of the housefly (Musca domestica). J. Ethnopharmacol. 2007, 111, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Hall, H.N.; O’ Nell, H.V.M.; Scholey, D.; Burton, E.; Dickinson, M.; Fitches, E.C. Amino acid digestibility of larval meal (Musca domestica) for broiler chicken. Poultry Sci. 2018, 97, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.; Villamizar-Sarmiento, M.G.; Harmsen, I.; Valdes, F.; Villanueva, V.; Ceballos, R.; Wacyk, J.; Oyarzun-Ampuero, F.; Valenzuela, C. Encapsulation of house fly larvae (Musca domestica) meal by ionic gelation as a strategy to develop a novel nutritive food ingredient with improved aroma and appearance. LWT-Food Sci. Technol. 2022, 163, 113597. [Google Scholar] [CrossRef]
- Feng, X.; Cheng, G.; Chen, S.Y.; Yang, H.; Huang, W. Evaluation of the burn healing properties of oil extraction from housefly larva in mice. J. Ethnopharmacol. 2010, 130, 586–592. [Google Scholar] [CrossRef]
- Miron, L.; Montevecchi, G.; Macavei, L.I.; Maistrello, L.; Antonelli, A.; Thomas, M. Effect of black soldier fly larvae protein on the texture of meat analogues. LWT-Food Sci. Technol. 2023, 181, 114745. [Google Scholar] [CrossRef]
- Guo, G.; Tao, R.Y.; Li, Y.; Ma, H.L.; Xiu, J.F.; Fu, P.; Wu, J.W. Identification and characterization of a novel antimicrobial protein from the housefly Musca domestica. Biochem. Biophys. Res. Commun. 2017, 490, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Li, H.J.; Inoue, A.; Taniguchi, S.; Yukutake, T.; Suyama, K.; Nose, T.; Maeda, I. Multifunctional biological activities of water extract of housefly larvae (Musca domestica). Pharma Nutr. 2017, 5, 119–126. [Google Scholar] [CrossRef]
- Sun, H.X.; Chen, L.Q.; Zhang, J.; Chen, F.Y. Anti-tumor and immunomodulatory activity of peptide fraction from the larvae of Musca domestica. J. Ethnopharmacol. 2014, 153, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.J.; Jin, X.B.; Zhu, J.Y. Housefly maggots (Musca domestica) protein-enriched fraction/extracts (PE) inhibit lipo polysaccharide-induced atherosclerosis pro-inflammatory response. J. Atheroscler. Thromb. 2011, 18, 282–290. [Google Scholar] [CrossRef]
- Chu, F.J.; Jin, X.B.; Xu, Y.Y.; Ma, Y.; Li, X.B.; Lu, X.M.; Liu, W.B.; Zhu, J.Y. Inflammatory regulation effect and action mechanism of anti-inflammatory effective parts of housefly (Musca domestica) larvae on atherosclerosis. Evid.-Based Complement. Altern. Med. 2013, 2013, 340267. [Google Scholar] [CrossRef]
- Ai, H.; Wang, F.; Zhang, N.; Zhang, L.; Lei, C. Antiviral immunomodulatory, and free radical scavenging activities of a protein-enriched fraction from the larvae of the housefly, Musca domestica. J. Insect Sci. 2013, 13, 112. [Google Scholar] [CrossRef]
- Shen, J.; Chen, J.L.; Yang, D.P.; Zhao, Z.M.; Tang, C.L.; Zhang, R.F.; Yang, W.Z.; Niu, Y. Antidiarrheal effects of a thermostable protein fraction obtained from the larvae of Musca domestica. Biomed. Pharmacother. 2019, 115, 108813. [Google Scholar] [CrossRef] [PubMed]
- Chu, F.J.; Jin, X.B.; Ma, H.Y. Anti-diarrhea effects and identification of Musca domestica larvae low molecular weight peptides (LMWP). J. Pharm. Biomed 2019, 173, 162–168. [Google Scholar]
- Wang, F.R.; Ai, H.; Chen, X.M.; Lei, C.L. Hepatoprotective effect of a proteinfraction from the maggots (Musca domestica) against CCl4-induced hepatic damage. Biotechnol. Lett. 2007, 29, 853–858. [Google Scholar] [CrossRef]
- Banerjee, S.; Haldar, S.; Reddy, N.; Reddy, R.; Nagananda, G.S.; Mitra, J. Under-utilized germinated horse gram (Macrotyloma uniflorum) protein-extraction, process optimization, characterization and its use in cookies fortification. LWT-Food Sci. Technol. 2022, 160, 113276. [Google Scholar] [CrossRef]
- Rose, A.; Chiu, Y.C.; Showman, C.; Ku, K.M.; Jaczynski, J.; Matak, K. Characterization of protein concentrates obtained by defatting cricket, locust, and silkworm powders using one-step organic solvent extraction. LWT-Food Sci. Technol. 2023, 182, 114876. [Google Scholar] [CrossRef]
- Garg, D.; Chakraborty, S.; Gokhale, J.S. Optimizing the extraction of protein from Prosopis cineraria seeds using response surface methodology and characterization of seed protein concentrate. LWT-Food Sci. Technol. 2020, 117, 108630. [Google Scholar] [CrossRef]
- Kumar, M.; Potkule, J.; Patil, S.; Saxena, S.; Patil, P.G.; Mageshwaran, V.; Punia, S.; Varghese, E.; Mahapatra, A.; Ashtaputre, N.; et al. Extraction of ultra-low gossypol protein from cottonseed: Characterization based on antioxidant activity, structural morphology and functional group analysis. LWT-Food Sci. Technol. 2021, 140, 110692. [Google Scholar] [CrossRef]
- Xu, Y.; Guo, J.; Guan, F.C.; Yang, Q.; Zhang, X. Organic solvent extraction and characterization of Antarctic krill protein with high crystallinity and high molecular weight. Process Biochem. 2023, 134, 363–369. [Google Scholar] [CrossRef]
- Xu, J.H.; Xiao, S.; Wang, J.H.; Wang, B.; Cai, Y.X.; Hu, W.F. Comparative study of the effects of ultrasound-assisted alkaline extraction on black soldier fly (Hermetia illucens) larvae protein: Nutritional, structural, and functional properties. Ultrason. Sonochem. 2023, 101, 106662. [Google Scholar] [CrossRef]
- Ma, L.K.; Xu, J.Z.; Yu, Y.S.; Wang, D.P.; Yu, M.; Zhang, X.Y.; Yang, X.Y.; Xu, X.X. Effect of high-intensity ultrasound on the structural and functional properties of proteins in housefly larvae (Musca demestica). Ultrason. Sonochem. 2023, 101, 106673. [Google Scholar] [PubMed]
- Wu, L.L.; Li, J.H.; Wu, W.J.; Wang, L.B.; Qin, F.; Xie, W. Effect of extraction pH on functional properties, structural properties, and in vitro gastrointestinal digestion of tartary buckwheat protein isolates. J. Cereal Sci. 2021, 101, 103314. [Google Scholar] [CrossRef]
- Das, D.; Panesar, P.S.; Saini, C.S. Ultrasonic extraction of soy protein isolate: Characterization and comparison with microwave and enzymatic extraction methods. J. Food Sci. Technol. 2023, 88, 2758–2779. [Google Scholar] [CrossRef]
- Juul, L.; Haue, S.K.; Bruhn, A.; Boderskov, T.; Dalsgaard, T.K. Alkaline pH increases protein extraction yield and solubility of the extracted protein from sugar kelp (Saccharina latissima). Food Bioprod. Process. 2023, 140, 144–150. [Google Scholar] [CrossRef]
- Ortega, M.L.S.; Orellana-Palacios, J.C.; Garcia, S.R.; Rabanal-Ruiz, Y.; Moreno, A.; Hadidi, M. Olive leaf protein: Extraction optimization, in vitro digestibility, structural and techno-functional properties. Int. J. Biol. Macromol. 2024, 256, 128273. [Google Scholar] [CrossRef]
- Wang, Z.J.; Ye, W.F.; Wu, Y.J.; Lin, X.Y.; Luan, C.R.; Xie, X.W.; Peng, Y.; Sun, X.H.; Shi, C.Y.; Lv, Y.C.; et al. Protein extraction from chlorella pyrenoidosa microalgae: Green methodologies, functional assessment, and waste stream valorization for bioenergy production. Bioresour. Technol. 2024, 397, 130508. [Google Scholar] [CrossRef]
- Yang, F.; Chen, W.; Dabbour, M.; Mintah, B.K.; Xu, H.N.; Pan, J.Y.; Dai, C.H.; Ma, H.L.; He, R.H. Preparation of housefly (Musca domestica) larvae protein hydrolysates: Influence of dual-sweeping-frequency ultrasound-assisted enzymatic hydrolysis on yield, antioxidative activity, functional and structural attributes. Food Chem. 2024, 440, 138253. [Google Scholar] [PubMed]
- Mu, J.Y.; Hu, R.S.; Tang, Y.M.; Dong, W.J.; Zhang, Z.Z. Microencapsulation of green coffee oil by complex coacervation of soy protein isolate, sodium casinate and polysaccharides: Physicochemical properties, structural characterisation, and oxidation stability. Int. J. Biol. Macromol. 2024, 256, 128064. [Google Scholar] [CrossRef]
- Xie, M.Y.; Zhou, C.X.; Li, X.; Ma, H.T.; Liu, Q.G.; Hong, P.Z. Preparation and characterization of tilapia protein isolate- Hyaluronic acid complexes using a pH-driven method for improving the stability of tilapia protein isolate emulsion. Food Chem. 2024, 445, 138703. [Google Scholar] [CrossRef]
- Do, D.T.; Ye, A.Q.; Singh, H.; Acevedo-Fani, A. Protein bodies from hemp seeds: Isolation, microstructure and physicochemical characterisation. Food Hydrocoll. 2024, 149, 109597. [Google Scholar] [CrossRef]
- Eckhardt, L.; Fan, B.; Franczyk, A.; Michaels, T.; Ismail, B.P. Hemp (Cannabis sativa L.) protein: Impact of extraction method and cultivar on structure, function, and nutritional quality. Curr. Res. Food Sci. 2024, 8, 100746. [Google Scholar] [CrossRef] [PubMed]
- Puray, J.J.S.; Villaber, R.A.P. Extraction, characterization, and vascular response of proteins from catfish (Clarias batrachus L.) mucus. Food Chem. Adv. 2023, 3, 100444. [Google Scholar] [CrossRef]
- Li, R.M.; Wang, Q.T.; Shen, Y.Y.; Li, M.B.; Sun, L.L. Integrated extraction, structural characterization, and activity assessment of squid pen protein hydrolysates and β-chitin with different protease hydrolysis. Int. J. Biol. Macromol. 2024, 262, 130069. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.K.; Xu, J.Q.; Zhang, J.L.; Wei, Y.N.; Cai, X.; Tan, J.Q. Modeling, optimization, purification, and characterization of polysaccharides from Lilium lancifolium Thunb. LWT-Food Sci. Technol. 2022, 162, 113491. [Google Scholar] [CrossRef]
- Tian, S.Q.; Du, K.; Yan, F.; Li, Y.H. Microwave-assisted enzymatic hydrolysis of wheat germ albumin to prepare polypeptides and influence on physical and chemical properties. Food Chem. 2022, 374, 131707. [Google Scholar] [CrossRef]


| Run | Ratio of Material to Liquid (g/mL) X1 | pH X2 | Extraction Time (h) X3 | Extraction Temperature (°C) X4 | MDPs Yield (%) |
|---|---|---|---|---|---|
| 1 | 1:35 | 9 | 1.5 | 25 | 14.6 |
| 2 | 1:30 | 9 | 1.5 | 30 | 14.3 |
| 3 | 1:35 | 10 | 1.5 | 30 | 18.5 |
| 4 | 1:35 | 10 | 1.5 | 30 | 17.9 |
| 5 | 1:35 | 11 | 1.5 | 25 | 16.6 |
| 6 | 1:35 | 10 | 2.0 | 35 | 15.5 |
| 7 | 1:40 | 10 | 1.5 | 35 | 16.3 |
| 8 | 1:40 | 9 | 1.5 | 30 | 15.4 |
| 9 | 1:40 | 11 | 1.5 | 30 | 17.3 |
| 10 | 1:40 | 10 | 1.5 | 25 | 15.0 |
| 11 | 1:35 | 10 | 1.0 | 25 | 15.2 |
| 12 | 1:30 | 10 | 2.0 | 30 | 15.3 |
| 13 | 1:40 | 10 | 2.0 | 30 | 16.1 |
| 14 | 1:35 | 9 | 1.0 | 30 | 15.6 |
| 15 | 1:40 | 10 | 1.0 | 30 | 17.0 |
| 16 | 1:35 | 10 | 2.0 | 25 | 16.6 |
| 17 | 1:30 | 11 | 1.5 | 30 | 16.3 |
| 18 | 1:35 | 10 | 1.0 | 35 | 16.9 |
| 19 | 1:35 | 9 | 2.0 | 30 | 15.5 |
| 20 | 1:30 | 10 | 1.5 | 35 | 15.9 |
| 21 | 1:35 | 10 | 1.5 | 30 | 18.2 |
| 22 | 1:35 | 10 | 1.5 | 30 | 18.3 |
| 23 | 1:35 | 9 | 1.5 | 35 | 16.4 |
| 24 | 1:35 | 10 | 1.5 | 30 | 18.1 |
| 25 | 1:30 | 10 | 1.5 | 25 | 14.2 |
| 26 | 1:35 | 11 | 1.0 | 30 | 17.2 |
| 27 | 1:30 | 10 | 1.0 | 30 | 15.7 |
| 28 | 1:35 | 11 | 1.5 | 35 | 15.9 |
| 29 | 1:35 | 11 | 2.0 | 30 | 17.1 |
| Parameter | Sum of Square | df | Mean Square | F-Value | p-Value | Significance |
|---|---|---|---|---|---|---|
| Model | 45.06 | 14 | 3.22 | 14.93 | <0.0001 | ** |
| X1 | 2.44 | 1 | 2.44 | 11.31 | 0.0046 | ** |
| X2 | 6.06 | 1 | 6.06 | 28.12 | 0.0001 | ** |
| X3 | 0.129 | 1 | 0.129 | 6.00 | 0.0280 | * |
| X4 | 1.72 | 1 | 1.72 | 8.00 | 0.0134 | * |
| X1X2 | 0.0021 | 1 | 0.0021 | 0.0096 | 0.9233 | - |
| X1X3 | 0.0415 | 1 | 0.0415 | 0.1926 | 0.6675 | - |
| X1X4 | 0.0309 | 1 | 0.0309 | 0.1433 | 0.7107 | - |
| X2X3 | 0.0034 | 1 | 0.0034 | 0.0158 | 0.9019 | - |
| X2X4 | 1.43 | 1 | 1.43 | 6.62 | 0.0221 | * |
| X3X4 | 1.93 | 1 | 1.93 | 8.97 | 0.0097 | ** |
| X12 | 16.64 | 1 | 16.64 | 77.20 | <0.0001 | ** |
| X22 | 8.71 | 1 | 8.71 | 40.42 | <0.0001 | ** |
| X32 | 3.78 | 1 | 3.78 | 17.56 | 0.0009 | ** |
| X42 | 15.33 | 1 | 15.33 | 71.73 | <0.0001 | ** |
| Residual | 3.02 | 14 | 0.2155 | |||
| Lack of fit | 2.04 | 10 | 0.2040 | 0.8341 | 0.6304 | - |
| Pure error | 0.9781 | 4 | 0.2445 | CV% = 2.82 | ||
| R-Squared | 0.9372 | Adj.R-squared | 0.8745 |
| Samples | Particle Size Distribution | Zeta Potential |
|---|---|---|
| MDPs-ND | 363.2 ± 16.6 | −37.0 ± 1.7 |
| Defatted MDPs-ND | 135.7 ± 3.3 | −46.0 ± 3.4 |
| MDPs-T | 246.4 ± 5.6 **## | −39.0 ## ± 1.5 |
| Defatted MDPs-T | 219.3 ± 31.1 | −8.3 ± 1.1 |
| Content of Each Component in the Amide I Band/% | ||||
|---|---|---|---|---|
| α-Helix | β-Fold | β-Turn | Random Coil | |
| MDPs-ND | 18.40 | 33.34 | 31.40 | 16.86 |
| Defatted MDPs-ND | 15.25 | 30.02 | 40.80 | 13.93 |
| MDPs-T | 43.55 | 28.67 | 27.78 | - |
| Defatted MDPs-T | 43.05 | 28.34 | 28.60 | - |
| Samples | MDPs-ND | Defatted MDPs-ND | MDPs-T | Defatted MDPs-T |
|---|---|---|---|---|
| Peak 1 (9.12 × 103 kDa) | 3.28% | 2.24% | 6.24% | - |
| Peak 2 (3.96 × 103 kDa) | 38.68% | 28.78% | 10.30% | 6.31% |
| Peak 3 (1.70 × 103 kDa) | 20.22% | 24.41% | 12.34% | 10.37% |
| Peak 4 (1.23 × 103 kDa) | 10.31% | 11.54% | 11.44% | 11.14% |
| Peak 5 (8.68 kDa) | 4.92% | 6.04% | 9.46% | 10.31% |
| Peak 6 (4.26 kDa) | 18.62% | 23.42% | 31.65% | 40.05% |
| Peak 7 (1.02 kDa) | 3.96% | 3.57% | 18.57% | 21.83% |
| Amino Acid | MDPs-ND (mg/mL) | Defatted MDPs-ND (mg/mL) | MDPs-T (mg/mL) | Defatted MDPs-T (mg/mL) |
|---|---|---|---|---|
| Aspartic acid | 0.595 | 0.389 | 1.055 | 0.745 |
| Glutamic acid | 1.609 | 1.098 | 1.322 | 1.197 |
| Serine | 0.283 | 0.191 | 0.373 | 0.316 |
| Glycine | 0.331 | 0.197 | 0.337 | 0.318 |
| Histidine | 0.801 | 0.486 | 0.708 | 0.720 |
| Arginine | 0.387 | 0.240 | 0.452 | 0.413 |
| Threonine | 0.228 | 0.157 | 0.369 | 0.301 |
| Alanine | 0.337 | 0.221 | 0.410 | 0.491 |
| Proline | 0.318 | 0.192 | 0.320 | 0.294 |
| Tyrptophan | 0.491 | 0.314 | 0.993 | 0.725 |
| Valine | 0.323 | 0.209 | 0.510 | 0.398 |
| Methionine | 0.014 | 0.024 | 0.276 | 0.142 |
| Leucine | 0.259 | 0.147 | 0.600 | 0.404 |
| Isoleucine | 0.166 | 0.102 | 0.382 | 0.280 |
| Phenylalanine | 0.561 | 0.376 | 0.801 | 0.514 |
| Lysine | 0.441 | 0.270 | 0.753 | 0.524 |
| Cystine | 0.023 | 0.017 | 0.040 | 0.037 |
| Trptophan | 0.091 | 0.036 | 0.117 | 0.108 |
| Essential amino acids | 2.793 | 1.771 | 4.399 | 3.283 |
| Total amino acids | 7.258 | 4.666 | 9.818 | 7.927 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, J.; Yu, C.; Cheng, X.; Qiu, J.; Liu, S. Response Surface Methodology (RSM) for Optimizing Protein Extraction from Housefly (Musca domestica) Larvae Fed with Toad and Its Structural Characterization. Molecules 2024, 29, 2595. https://doi.org/10.3390/molecules29112595
Miao J, Yu C, Cheng X, Qiu J, Liu S. Response Surface Methodology (RSM) for Optimizing Protein Extraction from Housefly (Musca domestica) Larvae Fed with Toad and Its Structural Characterization. Molecules. 2024; 29(11):2595. https://doi.org/10.3390/molecules29112595
Chicago/Turabian StyleMiao, Jingnan, Chenglu Yu, Xianhe Cheng, Junqiang Qiu, and Shumin Liu. 2024. "Response Surface Methodology (RSM) for Optimizing Protein Extraction from Housefly (Musca domestica) Larvae Fed with Toad and Its Structural Characterization" Molecules 29, no. 11: 2595. https://doi.org/10.3390/molecules29112595
APA StyleMiao, J., Yu, C., Cheng, X., Qiu, J., & Liu, S. (2024). Response Surface Methodology (RSM) for Optimizing Protein Extraction from Housefly (Musca domestica) Larvae Fed with Toad and Its Structural Characterization. Molecules, 29(11), 2595. https://doi.org/10.3390/molecules29112595






