The Programmable Catalytic Core of 8-17 DNAzymes
Abstract
1. Introduction
2. Results and Discussion
2.1. The Positive Effect of the Internal Stem in 8-17 DNAzymes
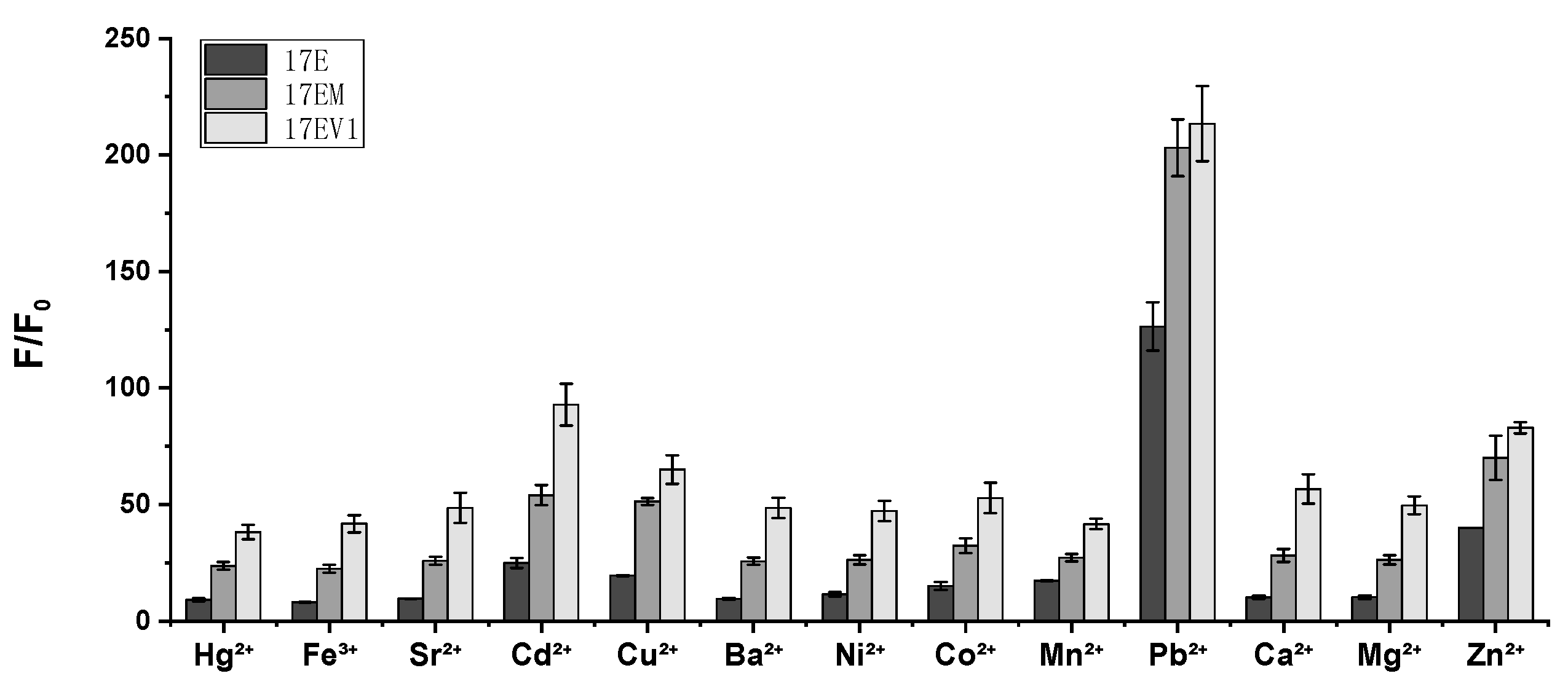
2.2. The Effect of the Extra W12.0 on DNAzymes
2.3. Thermal Stability and CD Spectra of DNAzyme–Substrate Complex System
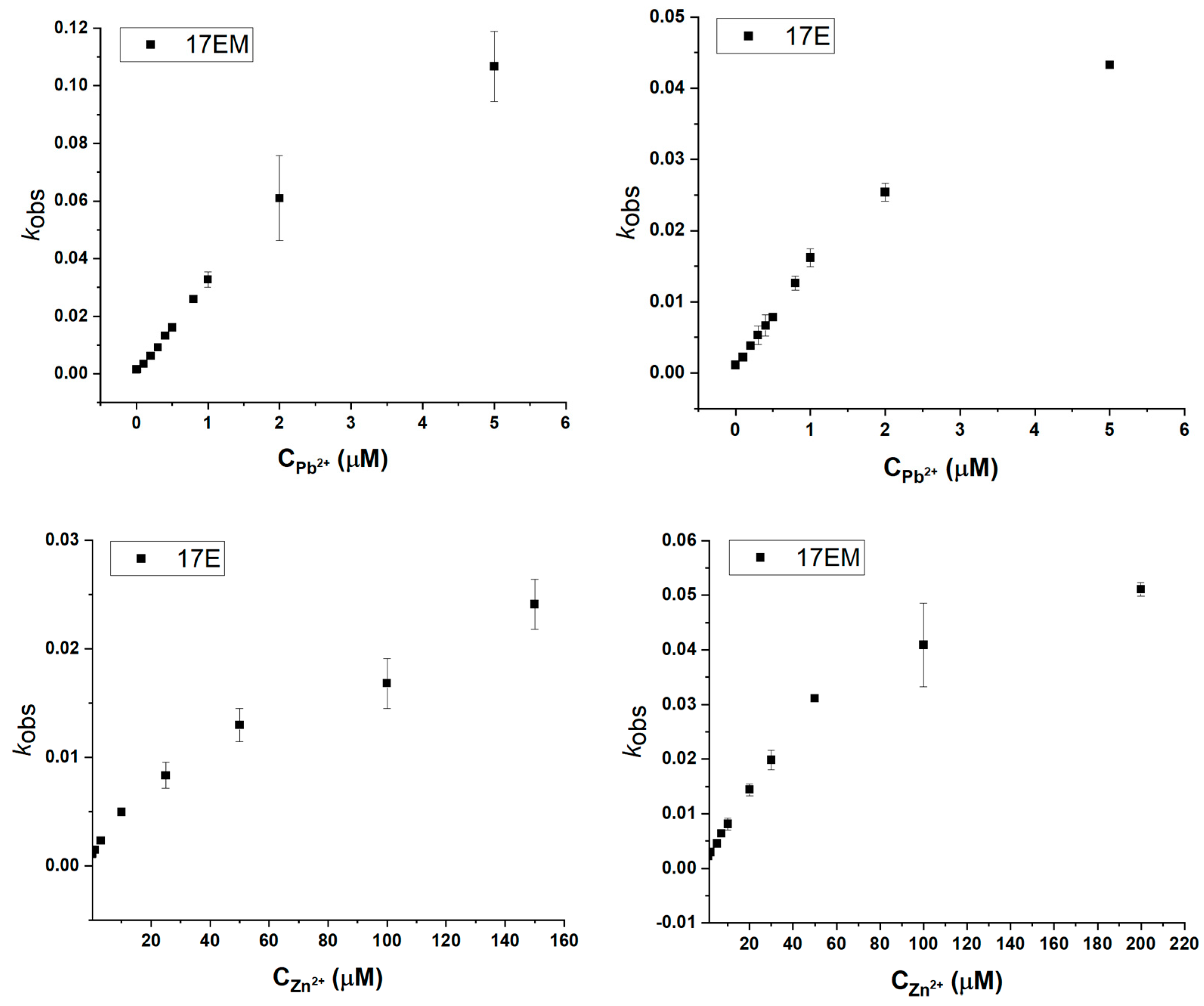
2.4. The Metal Ion Dependence of 17EM
2.5. pH Dependence of Pb2+-Mediated Reaction of 17EM
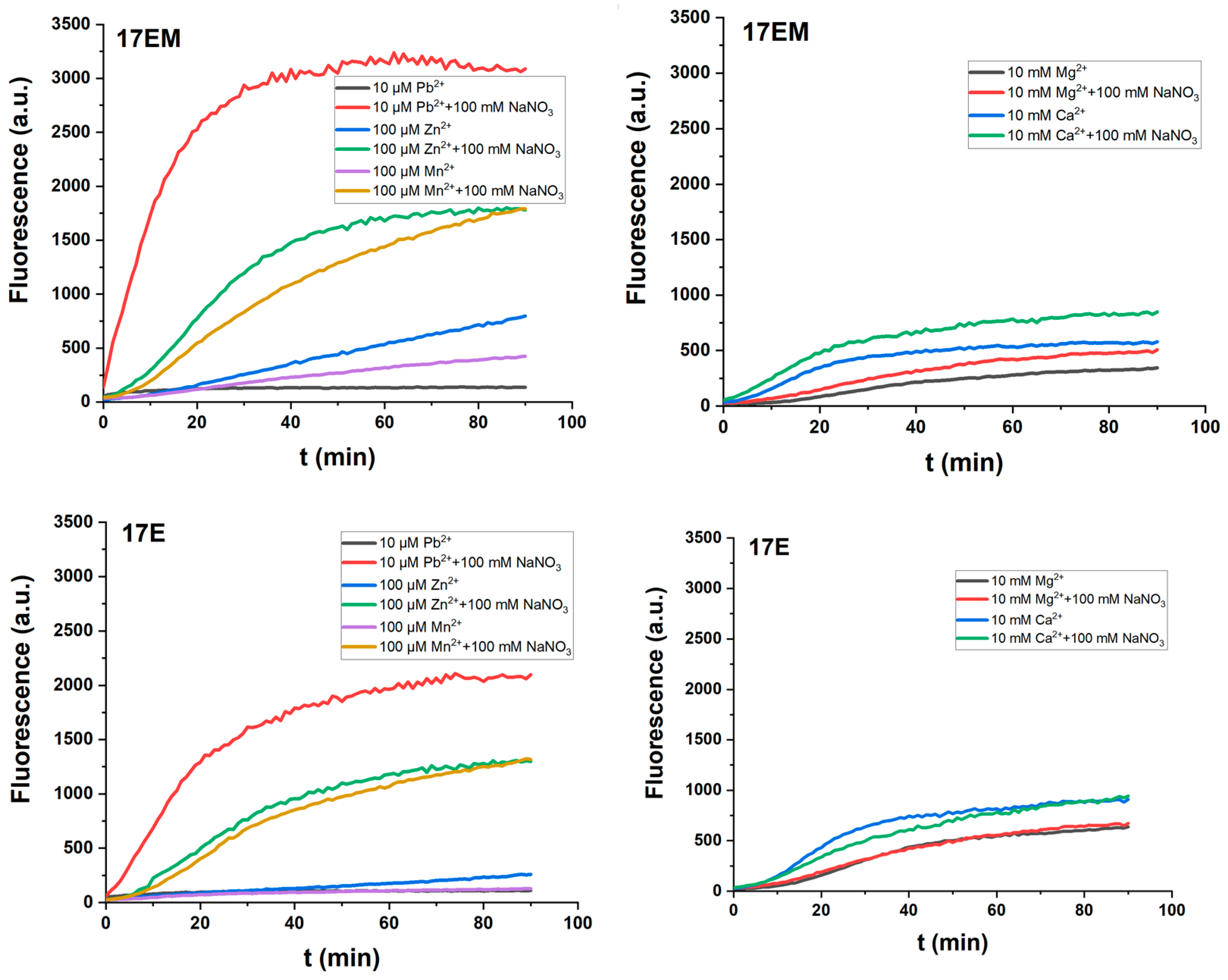
2.6. The Influence of Sodium Ions on the Catalytic Reactions of 17EM and 17E
2.7. The Unique Pb2+-Mediated DNAzyme Reaction
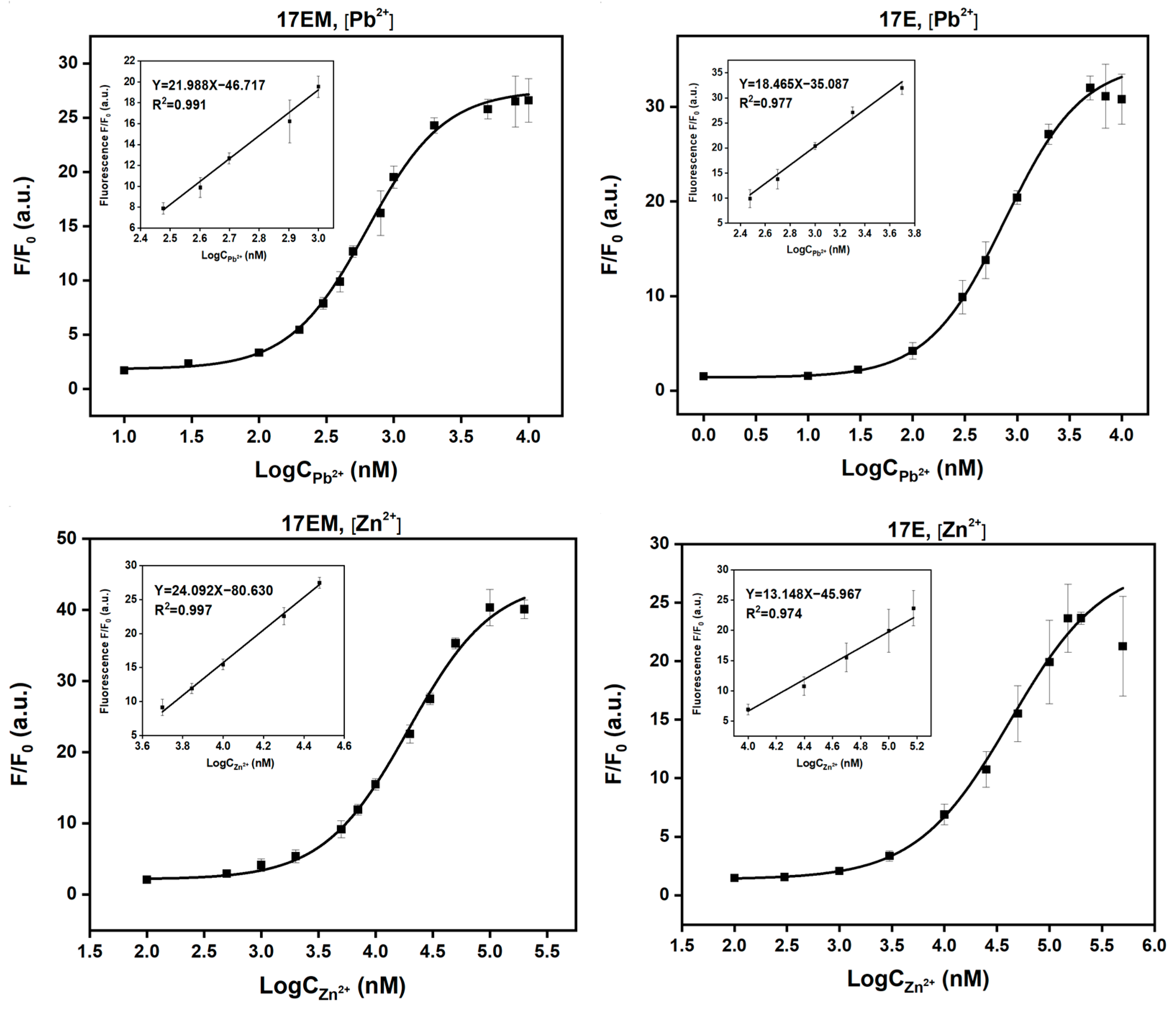
2.8. The Detection Limit of Pb2+ and Zn2+ of 17EM and 17E
3. Materials and Methods
3.1. Materials
3.2. Thermal Stability Measurement
3.3. CD Spectra
3.4. The Catalytic Reaction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Joyce, G.F. Forty years of in vitro evolution. Angew. Chem. Int. Ed. 2007, 46, 6420–6436. [Google Scholar] [CrossRef] [PubMed]
- Silverman, S.K. DNA as a versatile chemical component for catalysis, encoding and stereocontrol. Angew. Chem. Int. Ed. 2010, 49, 7180–7201. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Gordon, C.K.L.; Shin, J.H.; Eisenstein, M.; Soh, H.T. Directed evolution of aptamer discovery technologies. Acc. Chem. Res. 2022, 55, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.-J.J.; Vazin, M.; Liu, J. In Vitro selection of a new lanthanide-dependent dnazyme for ratiometric sensing lanthanides. Anal. Chem. 2014, 86, 9993–9999. [Google Scholar] [CrossRef] [PubMed]
- Lake, R.J.; Yang, Z.; Zhang, J.; Lu, Y. DNAzymes as activity-based sensors for metal ions: Recent applications, demonstrated advantages, current challenges, and future directions. Acc. Chem. Res. 2019, 52, 3275–3286. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Vorperian, A.; Shehabat, M.; Chaput, J.C. Evaluating the catalytic potential of a general RNA-cleaving FANA enzyme. ChemBioChem 2020, 21, 1001–1006. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, K.J.; Han, X.; Müller, U.F. A ribozyme that uses lanthanides as cofactor. Nucleic Acids Res. 2023, 51, 7163–7173. [Google Scholar] [CrossRef] [PubMed]
- Santoro, S.W.; Joyce, G.F. A general purpose rna-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef] [PubMed]
- Faulhammer, D.; Famulok, M. The Ca2+ as a cofactor for a novel RNA-cleaving deoxyribozyme. Angew. Chem. Int. Ed. Engl. 1996, 35, 2837–2841. [Google Scholar] [CrossRef]
- Li, J.; Zheng, W.; Kwon, A.H.; Lu, Y. In vitro selection and characterization of a highly efficient Zn (II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000, 28, 481–488. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, Y.; Ding, J.; Liu, J. In vitro selection in serum: RNA-cleaving DNAzymes for measuring Ca2+ and Mg2+. ACS Sens. 2016, 1, 600–606. [Google Scholar] [CrossRef]
- Liu, Y.; Sen, D. Local rather than global folding enables the lead-dependent activity of the 8–17 deoxyribozyme: Evidence from contact photo-crosslinking. J. Mol. Biol. 2010, 395, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, X.; Chen, Y.; Zhang, J.; Wu, B.; Zheng, L.; Haruehanroengra, P.; Wang, R.; Li, S.; Lin, J.; et al. Crystal structure of an RNA-cleaving DNAzyme. Nat. Commun. 2017, 8, 2006. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, K.; Gu, J.; Sule, L.; Li, Y. Sequence-function relationships provide new insight into the cleavage site selectivity of the 8–17 RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2008, 36, 1472–1481. [Google Scholar] [CrossRef] [PubMed]
- Cruz, R.P.; Withers, J.B.; Li, Y. Dinucleotide junction cleavage versatility of 8-17 deoxyribozyme. Chem. Biol. 2004, 11, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Peracchi, A.; Bonaccio, M.; Clerici, M. A Mutational analysis of the 8–17 deoxyribozyme core. J. Mol. Biol. 2005, 352, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Bonaccio, M.; Credali, A.; Peracchi, A. Kinetic and thermodynamic characterization of the RNA-cleaving 8-17 deoxyribozyme. Nucleic Acids Res. 2004, 32, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Li, Y.; Chai, Z.; Shi, W.; He, J. Functionalization of 8-17 DNAzymes modulates catalytic efficiency and divalent metal ion preference. Bioorg. Chem. 2020, 94, 103401. [Google Scholar] [CrossRef]
- Ekesan, S.; York, D.M. Dynamical ensemble of the active state and transition state mimic for the RNA-cleaving 8–17 DNAzyme in solution. Nucleic Acids Res. 2019, 47, 10282–10295. [Google Scholar] [CrossRef]
- Ren, W.; Huang, P.-J.J.; He, M.; Lyu, M.; Wang, S.; Wang, C.; Liu, J. The two classic Pb2+-selective DNAzymes are related: Rational evolution for understanding metal selectivity. ChemBioChem 2020, 21, 1293–1297. [Google Scholar] [CrossRef]
- Fan, H.; McGhee, C.E.; Lake, R.J.; Yang, Z.; Guo, Z.; Zhang, X.-B.; Lu, Y. A highly selective Mn (II)-specific DNAzyme and its application in intracellular sensing. J. Am. Chem. Soc. Au 2023, 3, 1615–1622. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.; Bereiter, R.; Mair, S.; Micura, R. Scaling catalytic contributions of small self-cleaving ribozymes. Angew. Chem. Int. Ed. 2022, 61, e202207590. [Google Scholar] [CrossRef] [PubMed]
- Rong, W.; Xu, L.; Liu, Y.; Yu, J.; Zhou, Y.; Liu, K.; He, J. 8–17 DNAzyme modified with purine analogs in its catalytic core: The conservation of the five-membered moieties of purine residues. Bioorg. Med. Chem. Lett. 2012, 22, 4238–4241. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Satyavolu, N.S.R.; Wu, Z.; Zhang, J.-R.; Zhu, J.-J.; Lu, Y. Near-infrared photothermally activated DNAzyme–gold nanoshells for imaging metal ions in living cells. Angew. Chem. Int. Ed. 2017, 56, 6798–6802. [Google Scholar] [CrossRef]
- Kim, E.H.; Chin, G.; Rong, G.; Poskanzer, K.E.; Clark, H.A. Optical probes for neurological sensing and imaging. Acc. Chem. Res. 2018, 51, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.; Wu, P.; Kim, T.; Lei, L.; Tian, S.; Wang, Y.; Lu, Y. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew. Chem. Int. Ed. 2014, 53, 13798–13802. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Loh, K.Y.; Chu, Y.-T.; Feng, R.; Satyavolu, N.S.R.; Xiong, M.; Huynh, S.M.N.; Hwang, K.; Li, L.; Xing, H.; et al. Optical control of metal ion probes in cells and zebrafish using highly selective DNAzymes conjugated to upconversion nanoparticles. J. Am. Chem. Soc. 2018, 140, 17656–17665. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.K.; Li, J.; Pavot, C.M.-B.; Lu, Y. A lead-dependent DNAzyme with a two-step mechanism. Biochemistry 2003, 42, 7152–7161. [Google Scholar] [CrossRef]
- Hu, N.; Wang, Y.; Liu, C.; He, M.; Nie, C.; Zhang, J.; Yu, Q.; Zhao, C.; Chen, T.; Chu, X. An enzyme-initiated DNAzyme motor for RNase H activity imaging in living cell. Chem. Commun. 2020, 56, 639–642. [Google Scholar] [CrossRef]
- Wang, X.; Kim, G.; Chu, J.L.; Song, T.; Yang, Z.; Guo, W.; Shao, X.; Oelze, M.L.; Li, K.C.; Lu, Y. Noninvasive and spatiotemporal control of DNAzyme-based imaging of metal ions in vivo using high-intensity focused ultrasound. J. Am. Chem. Soc. 2022, 144, 5812–5819. [Google Scholar] [CrossRef]
- Li, M.; Li, L. Enzyme-triggered DNA sensor technology for spatially-controlled, cell-selective molecular imaging. Acc. Chem. Res. 2023, 56, 1482–1493. [Google Scholar] [CrossRef]
- Cieslak, M.; Szymanski, J.; Adamiak, R.W.; Cierniewski, C.S. Structural rearrangements of the 10–23 DNAzyme to 3 integrin subunit mRNA induced by cations and their relations to the catalytic activity. J. Biol. Chem. 2003, 278, 47987–47996. [Google Scholar] [CrossRef]
- Cortés-Guajardo, C.; Rojas-Hernández, F.; Paillao-Bustos, R.; Cepeda-Plaza, M. Hydrated metal ion as a general acid in the catalytic mechanism of the 8–17 DNAzyme. Org. Biomol. Chem. 2021, 19, 5395–5402. [Google Scholar] [CrossRef]
- Breaker, R.R.; Emilsson, G.M.; Lazarev, D.; Nakamura, S.; Puskarz, I.J.; Roth, A.; Sudarsan, N. A common speed limit for RNA cleaving ribozymes and deoxyribozymes. RNA 2003, 9, 949–957. [Google Scholar] [CrossRef]
- Parra-Meneses, V.; Rojas-Hernández, F.; Cepeda-Plaza, M. The role of Na+ in catalysis by the 8–17 DNAzyme. Org. Biomol. Chem. 2022, 20, 6356–6362. [Google Scholar] [CrossRef]
- Moon, W.J.; Huang, P.-J.J.; Liu, J. Probing metal-dependent phosphate binding for the catalysis of the 17E DNAzyme. Biochemistry 2021, 60, 1909–1918. [Google Scholar] [CrossRef]
- Leung, E.K.Y.; Sen, D. Electron hole flow patterns through the RNA-cleaving 8-17 deoxyribozyme yield unusual information about its structure and folding. Chem. Biol. 2007, 14, 41–51. [Google Scholar] [CrossRef]
- Liu, Y.; Sen, D. A contact photo-cross-linking investigation of the active site of the 8–17 Deoxyribozyme. J. Mol. Biol. 2008, 381, 845–859. [Google Scholar] [CrossRef]
- Kim, H.-K.; Rasnik, I.; Liu, J.; Ha, T.; Lu, Y. Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat. Chem. Biol. 2007, 3, 763–768. [Google Scholar] [CrossRef]
- Saito, K.; Shimada, N.; Maruyama, A. Cooperative enhancement of deoxyribozyme activity by chemical modification and added cationic copolymer. Sci. Technol. Adv. Mater. 2016, 17, 437–442. [Google Scholar] [CrossRef]
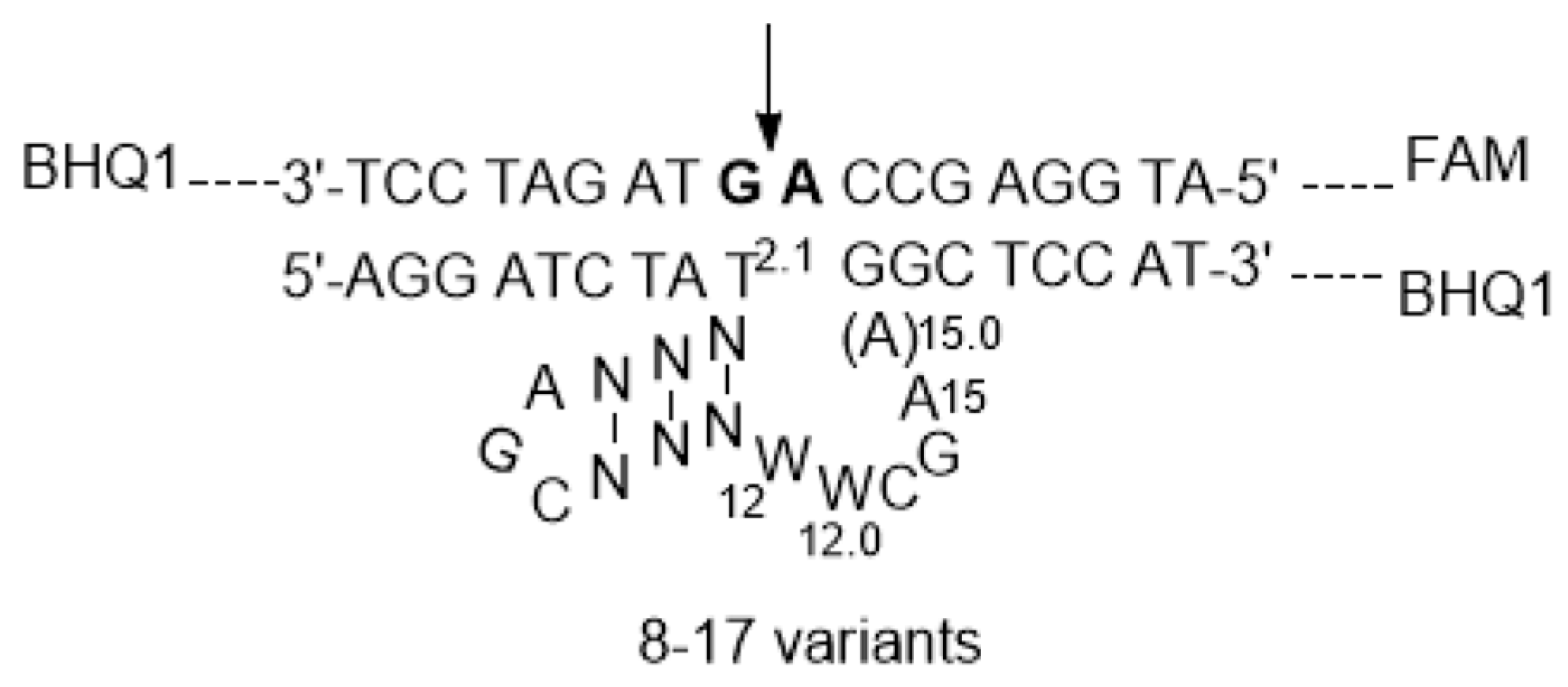
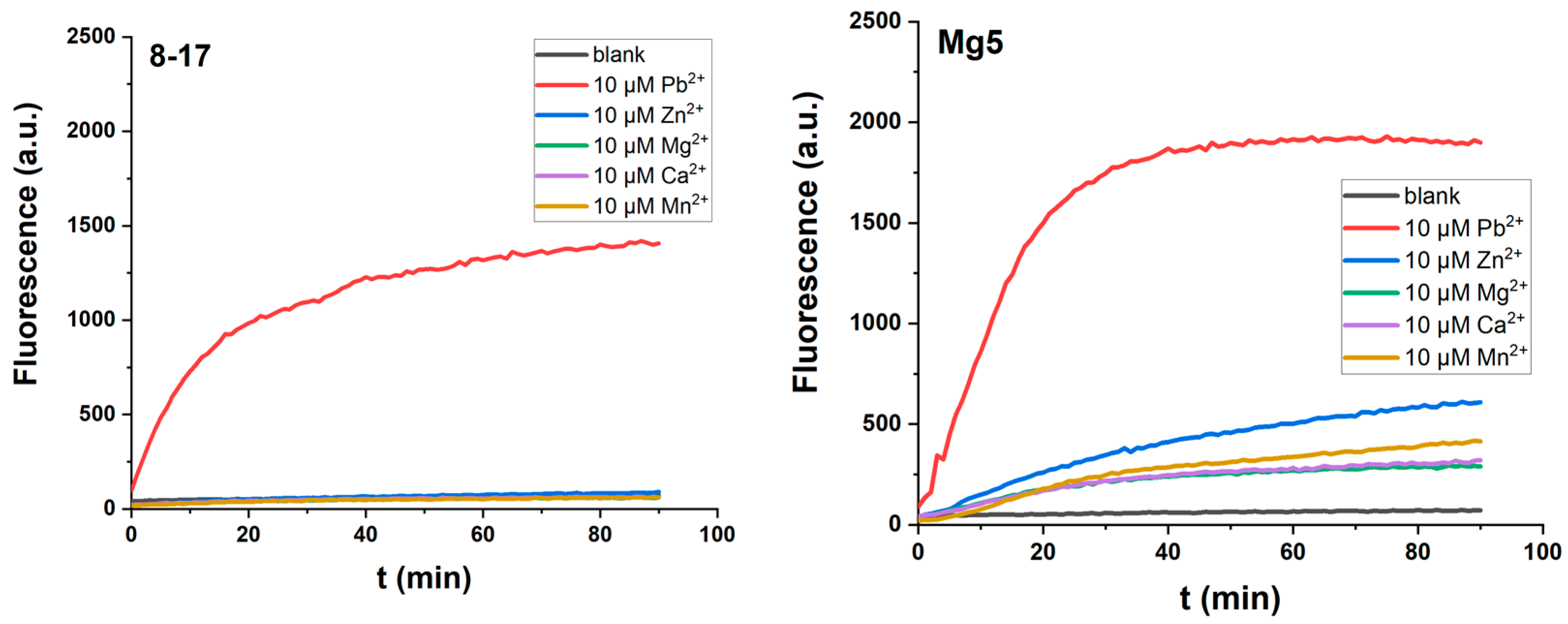

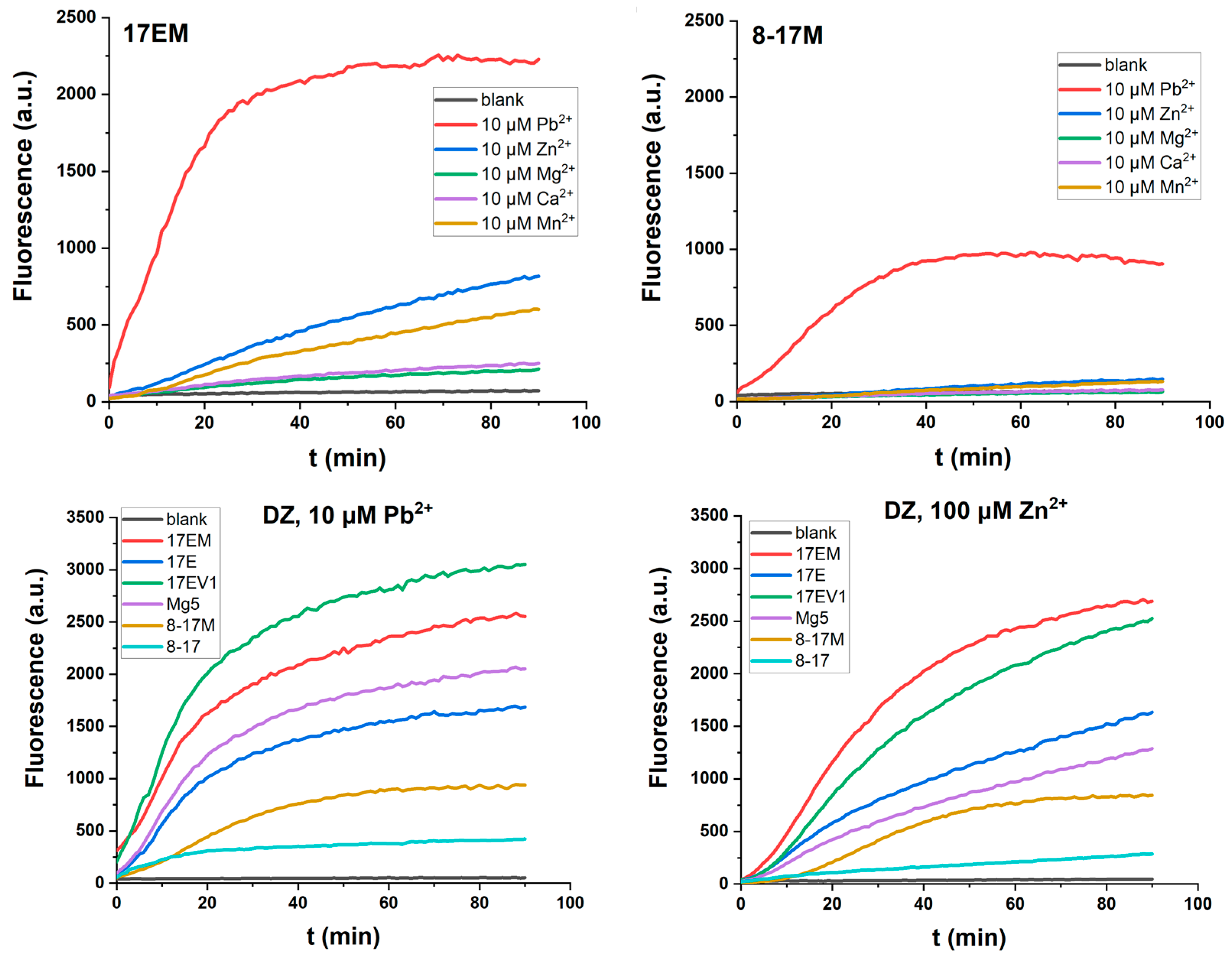
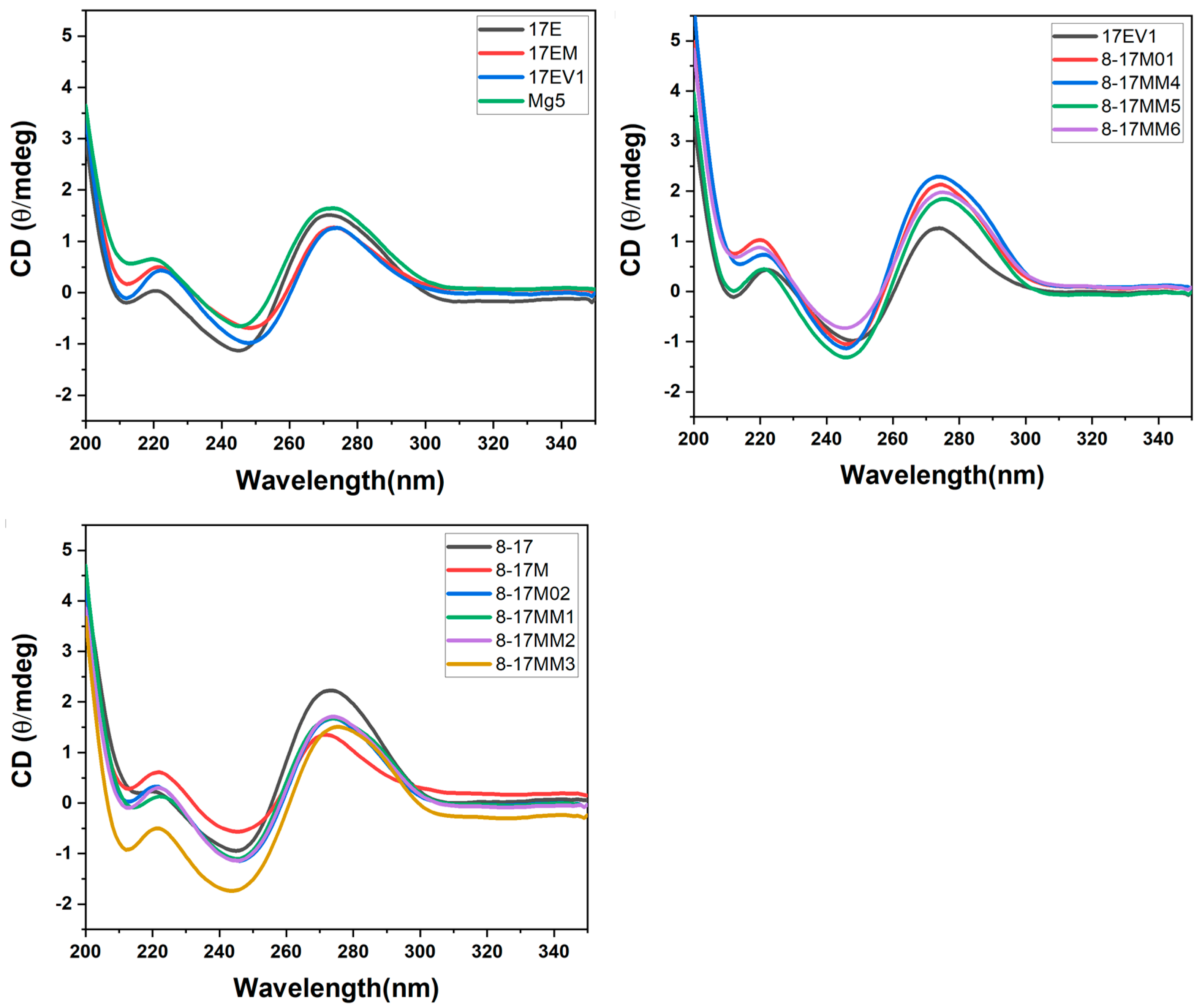

| DNAzyme | Selection Conditions | End Loop | Internal Stem | Bulge Loop | Metal Ion Dependence | Ref. |
|---|---|---|---|---|---|---|
| 8-17 | 10 mM MgCl2/1 M NaCl, 50 mM Tris-HCl, pH 7.5, 37 °C | AGC | CCG GGC | ACGA | Pb2+ >> Mg2+, Ca2+ | [8] |
| Mg5 | 0.5 mM Mg2+/50 mM histidine, 50 mM Na3PO4, pH 7.0, 125 mM NaCl, 125 mM KCl, 37 °C | AGC | CCG GGC | ACGAA | Pb2+ >> Zn2+, Ca2+ | [9] |
| 17E | 100 μM Zn2+, 500 mM NaCl, 50 mM HEPES, pH 7.0, 25 °C | AGC | CCG GGC | TCGAA | Pb2+ >> Zn2+ >> Mn2+ > Mg2+~Ca2+ | [10] |
| 17EV1 | 50 mM MES, pH 6.0, 25 mM NaCl, human serum | AGC | CTC GAG | ACGAA | Pb2+ >> Zn2+, Mn2+ > Ca2+, Mg2+ | [11] |
| 17EM | - | AGC | CTC GAG | TCGAA | Pb2+ >> Zn2+, Mn2+ > Ca2+, Mg2+ | |
| 8-17M | - | AGC | CTC GAG | ACGA | Pb2+ >> Mg2+, Ca2+ |
| DNAzyme | Sequence (5′-3′) | Tm 1 |
|---|---|---|
| 17EV1 | agg atc tat CTC AGC GAG ACGAA ggc tcc at-BHQ1 | 39.8 |
| Mg5 | agg atc tat CCG AGC CGG ACGAA ggc tcc at-BHQ1 | 40.0 |
| 17E | agg atc tat CCG AGC CGG TCGAA ggc tcc at-BHQ1 | 41.6 |
| 17EM | agg atc tat CTC AGC GAG TCGAA ggc tcc at-BHQ1 | 42.2 |
| 8-17 | agg atc tat CCG AGC CGG ACGA ggc tcc at-BHQ1 | 39.8 |
| 8-17M | agg atc tat CTC AGC GAG ACGA ggc tcc at-BHQ1 | 40.0 |
| 8-17M01 | agg atc tat CTC AGC GAG AACGAA ggc tcc at-BHQ1 | 41.3 |
| 8-17MM4 | agg atc tat CTC AGC GAG ATCGAA ggc tcc at-BHQ1 | 41.9 |
| 8-17MM5 | agg atc tat CTC AGC GAG TACGAA ggc tcc at-BHQ1 | 41.8 |
| 8-17MM6 | agg atc tat CTC AGC GAG TTCGAA ggc tcc at-BHQ1 | 40.2 |
| 8-17M02 | agg atc tat CTC AGC GAG AACGA ggc tcc at-BHQ1 | 40.5 |
| 8-17MM1 | agg atc tat CTC AGC GAG ATCGA ggc tcc at-BHQ1 | 42.9 |
| 8-17MM2 | agg atc tat CTC AGC GAG TACGA ggc tcc at-BHQ1 | 40.0 |
| 8-17MM3 | agg atc tat CTC AGC GAG TTCGA ggc tcc at-BHQ1 | 43.9 |
| 8-17S | FAM-d(ATGGAGCC)-r(AG)-d(TAGATCCT)-BHQ1 | |
| D18 | ATGGAGCCAGTAGATCCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Shi, W.; Guo, L.; Liu, S.; He, J. The Programmable Catalytic Core of 8-17 DNAzymes. Molecules 2024, 29, 2420. https://doi.org/10.3390/molecules29112420
Zhang F, Shi W, Guo L, Liu S, He J. The Programmable Catalytic Core of 8-17 DNAzymes. Molecules. 2024; 29(11):2420. https://doi.org/10.3390/molecules29112420
Chicago/Turabian StyleZhang, Fumei, Weiguo Shi, Lei Guo, Shihui Liu, and Junlin He. 2024. "The Programmable Catalytic Core of 8-17 DNAzymes" Molecules 29, no. 11: 2420. https://doi.org/10.3390/molecules29112420
APA StyleZhang, F., Shi, W., Guo, L., Liu, S., & He, J. (2024). The Programmable Catalytic Core of 8-17 DNAzymes. Molecules, 29(11), 2420. https://doi.org/10.3390/molecules29112420






