Molecular Mechanism of Interaction between DNA Aptamer and Receptor-Binding Domain of Severe Acute Respiratory Syndrome Coronavirus 2 Variants Revealed by Steered Molecular Dynamics Simulations
Abstract
1. Introduction
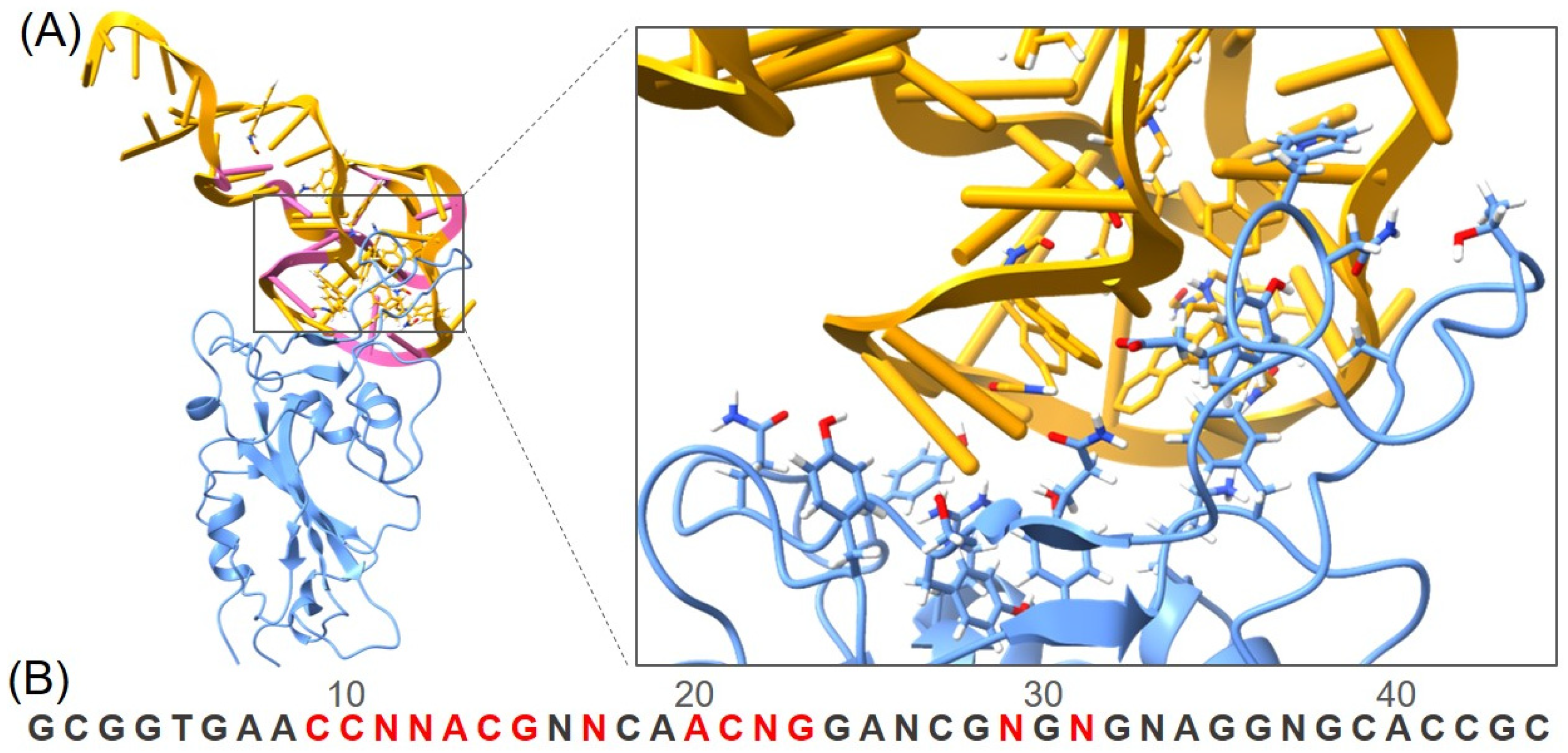
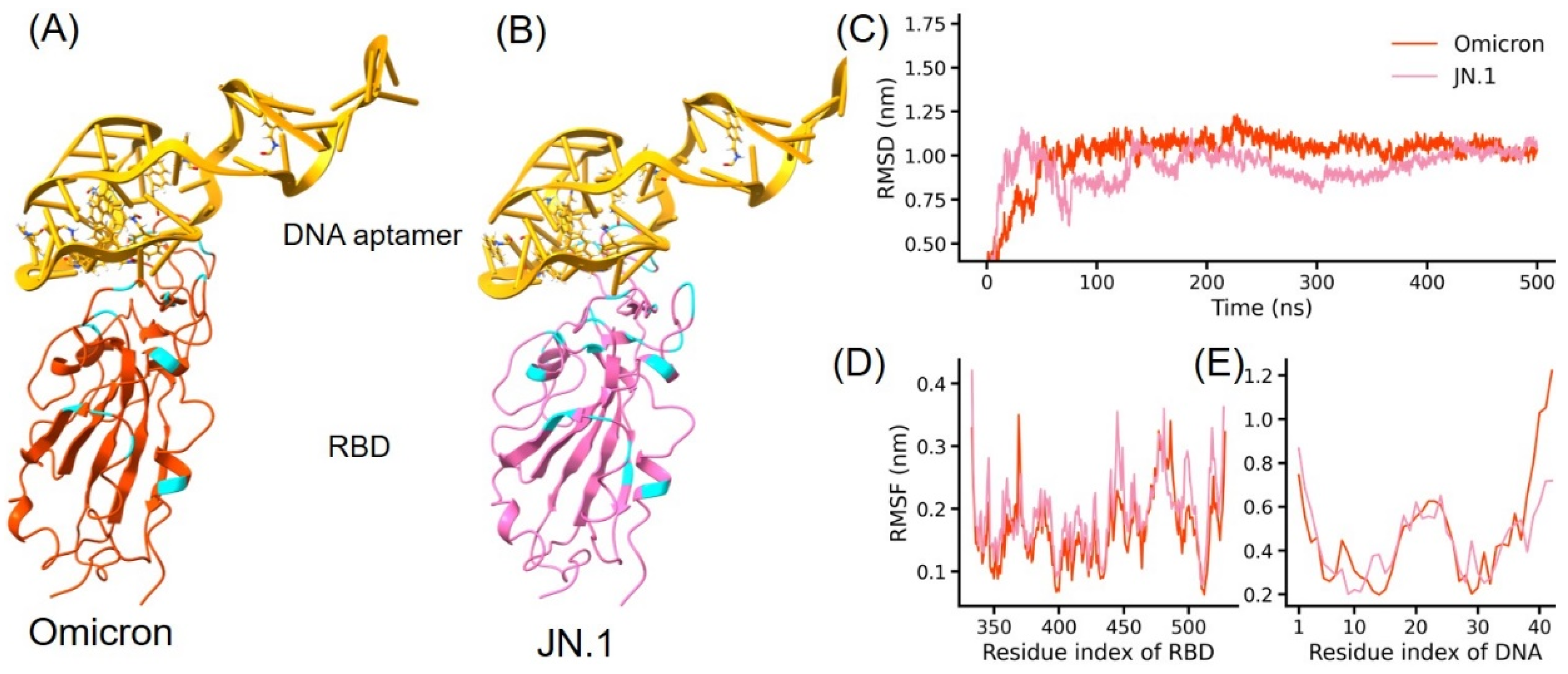
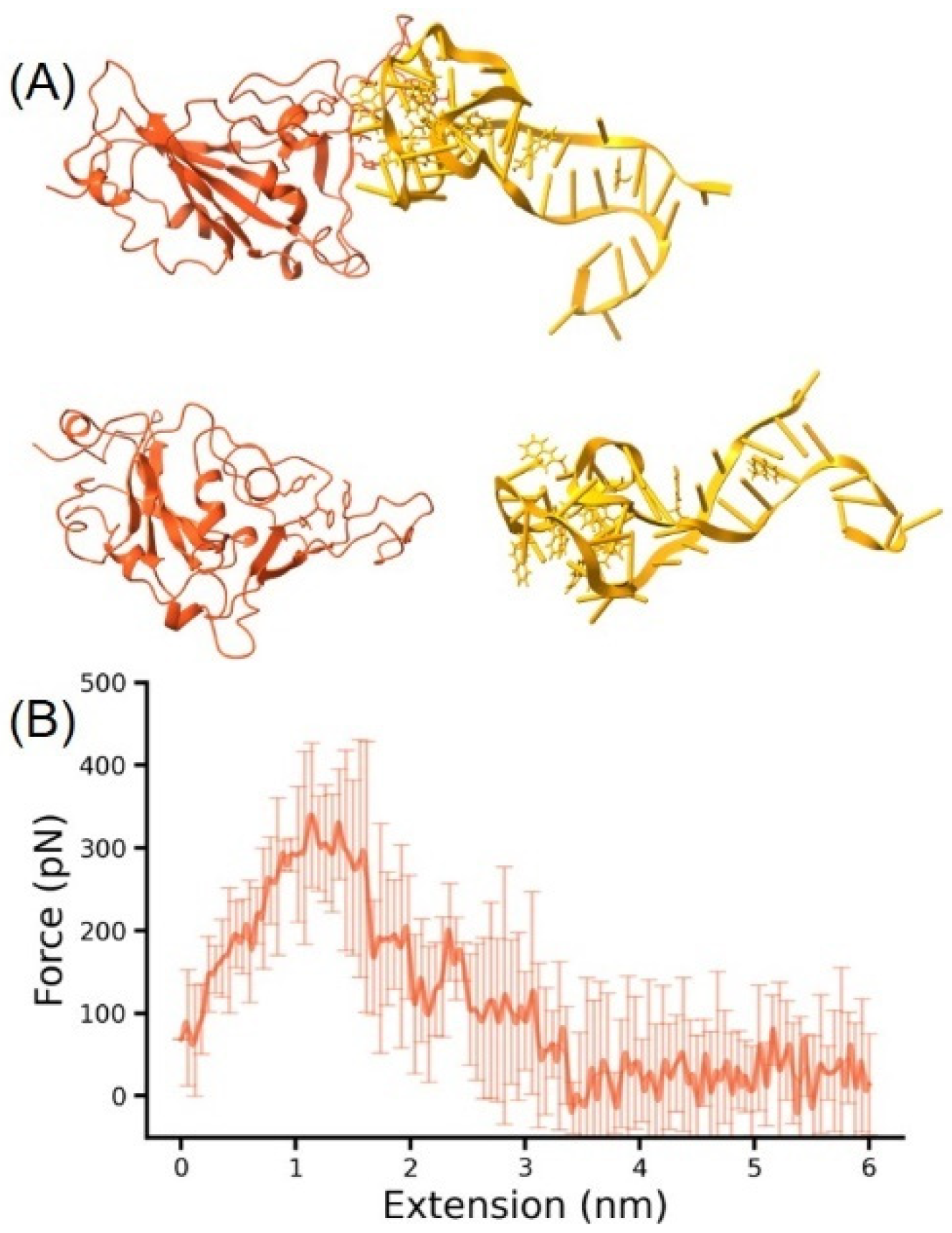
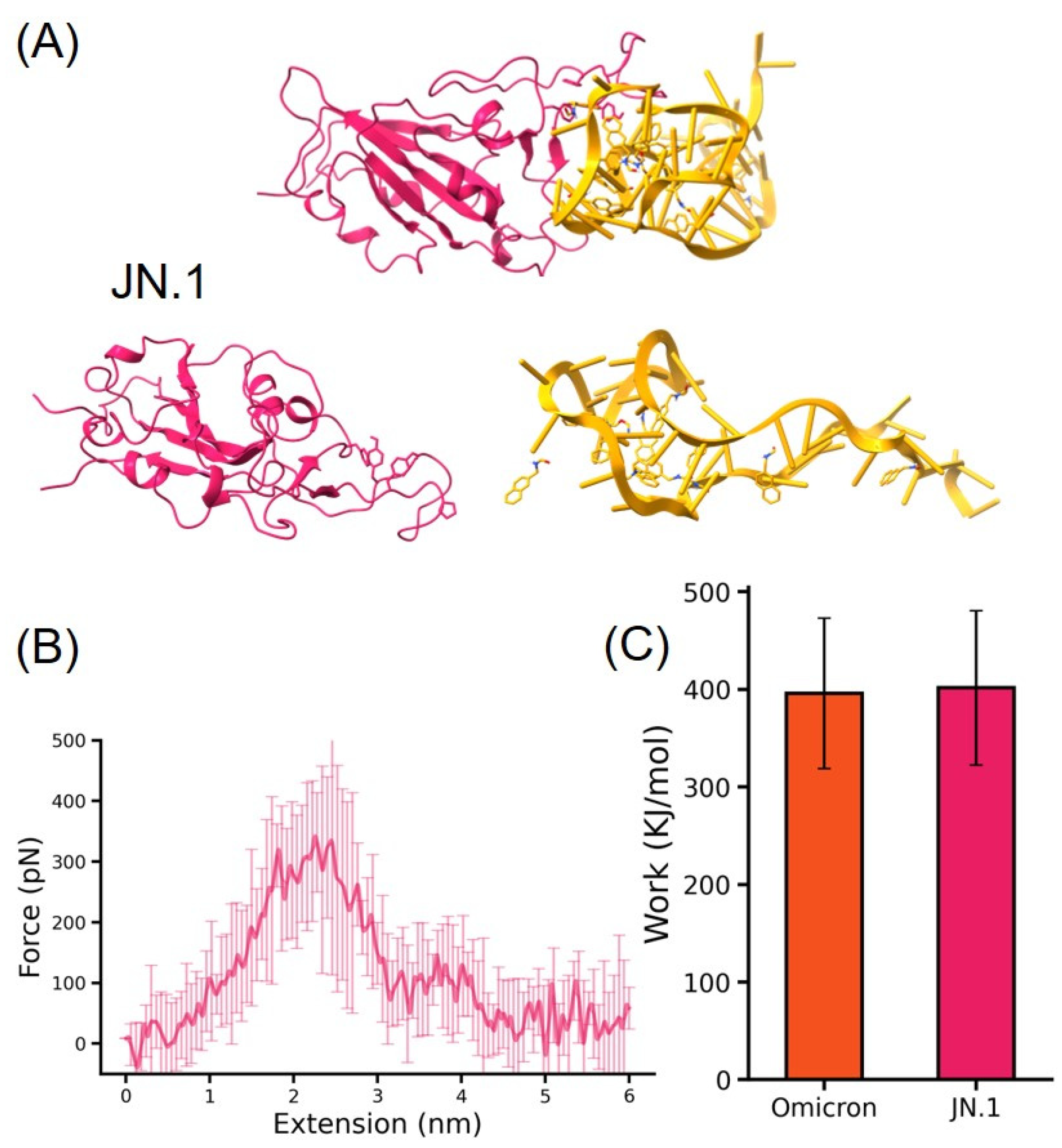
2. Result and Discussion
3. Materials and Methods
3.1. Structure Modeling
3.2. MD Simulation Force Field Preparation
3.3. MD Simulation for RBD-DNA Aptamer Unbinding
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.-M.; et al. COVID-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Elliott, P.; Bodinier, B.; Eales, O.; Wang, H.; Haw, D.; Elliott, J.; Whitaker, M.; Jonnerby, J.; Tang, D.; Walters, C.E.; et al. Rapid increase in Omicron infections in England during December 2021: REACT-1 study. Science 2022, 375, eabn8347. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, Y.; Lavine, C.L.; Peng, H.; Zhu, H.; Anand, K.; Tong, P.; Gautam, A.; Mayer, M.L.; Rits-Volloch, S.; et al. Structural and functional impact by SARS-CoV-2 Omicron spike mutations. Cell Rep. 2022, 39, 110729. [Google Scholar] [CrossRef]
- Yin, W.; Xu, Y.; Xu, P.; Cao, X.; Wu, C.; Gu, C.; He, X.; Wang, X.; Huang, S.; Yuan, Q.; et al. Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science 2022, 375, 1048–1053. [Google Scholar] [CrossRef]
- Kaku, Y.; Okumura, K.; Padilla-Blanco, M.; Kosugi, Y.; Uriu, K.; Hinay, A.A., Jr.; Chen, L.; Plianchaisuk, A.; Kobiyama, K.; Ishii, K.J.; et al. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect. Dis. 2024, 24, e82. [Google Scholar] [CrossRef]
- Wang, B.; Guo, C.; Chen, G.; Park, B.; Xu, B. Following aptamer–ricin specific binding by single molecule recognition and force spectroscopy measurements. Chem. Commun. 2012, 48, 1644–1646. [Google Scholar] [CrossRef]
- Sun, M.; Wu, Z.; Zhang, J.; Chen, M.; Lu, Y.; Yang, C.; Song, Y. Spherical neutralizing aptamer suppresses SARS-CoV-2 Omicron escape. Nano Today 2022, 44, 101499. [Google Scholar] [CrossRef]
- Ji, C.; Wei, J.; Zhang, L.; Hou, X.; Tan, J.; Yuan, Q.; Tan, W. Aptamer–Protein Interactions: From Regulation to Biomolecular Detection. Chem. Rev. 2023, 123, 12471–12506. [Google Scholar] [CrossRef]
- Zhang, N.; Chen, Z.; Liu, D.; Jiang, H.; Zhang, Z.-K.; Lu, A.; Zhang, B.-T.; Yu, Y.; Zhang, G. Structural Biology for the Molecular Insight between Aptamers and Target Proteins. Int. J. Mol. Sci. 2021, 22, 4093. [Google Scholar] [CrossRef]
- Adachi, T.; Nakamura, Y. Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application. Molecules 2019, 24, 4229. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.-L.; Wu, J.; Qi, J.; Zeng, Z.; Wan, Q.; Chen, Z.; Manandhar, P.; Cavener, V.S.; Boyle, N.R.; et al. Neutralizing Aptamers Block S/RBD-ACE2 Interactions and Prevent Host Cell Infection. Angew. Chem. Int. Ed. 2021, 60, 10273–10278. [Google Scholar] [CrossRef]
- Vaught, J.D.; Bock, C.; Carter, J.; Fitzwater, T.; Otis, M.; Schneider, D.; Rolando, J.; Waugh, S.; Wilcox, S.K.; Eaton, B.E. Expanding the Chemistry of DNA for in Vitro Selection. J. Am. Chem. Soc. 2010, 132, 4141–4151. [Google Scholar] [CrossRef]
- Kacherovsky, N.; Yang, L.F.; Dang, H.V.; Cheng, E.L.; Cardle, I.I.; Walls, A.C.; McCallum, M.; Sellers, D.L.; DiMaio, F.; Salipante, S.J.; et al. Discovery and Characterization of Spike N-Terminal Domain-Binding Aptamers for Rapid SARS-CoV-2 Detection. Angew. Chem. Int. Ed. 2021, 60, 21211–21215. [Google Scholar] [CrossRef]
- Tian, F.; Tong, B.; Sun, L.; Shi, S.; Zheng, B.; Wang, Z.; Dong, X.; Zheng, P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife 2021, 10, e69091. [Google Scholar] [CrossRef]
- Xiao, Y.; Zheng, B.; Ding, X.; Zheng, P. Probing nanomechanical interactions of SARS-CoV-2 variants Omicron and XBB with common surfaces. Chem. Commun. 2023, 59, 11268–11271. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Brooks, C.L.; MacKerell, A.D.; Post, C.B.; Nilsson, L. Biomolecular dynamics in the 21st century. Biochim. Biophys. Acta. Gen. Subj. 2024, 1868, 130534. [Google Scholar] [CrossRef] [PubMed]
- Sang, P.; Chen, Y.-Q.; Liu, M.-T.; Wang, Y.-T.; Yue, T.; Li, Y.; Yin, Y.-R.; Yang, L.-Q. Electrostatic Interactions Are the Primary Determinant of the Binding Affinity of SARS-CoV-2 Spike RBD to ACE2: A Computational Case Study of Omicron Variants. Int. J. Mol. Sci. 2022, 23, 14796. [Google Scholar] [CrossRef] [PubMed]
- Ju, S.-P.; Yang, Y.-C.; Chen, H.-Y. Unraveling the binding mechanisms of SARS-CoV-2 variants through molecular simulations. Heliyon 2024, 10, e27193. [Google Scholar] [CrossRef]
- Pederzoli, M.; Wasif Baig, M.; Kývala, M.; Pittner, J.; Cwiklik, L. Photophysics of BODIPY-Based Photosensitizer for Photodynamic Therapy: Surface Hopping and Classical Molecular Dynamics. J. Chem. Theory Comput. 2019, 15, 5046–5057. [Google Scholar] [CrossRef]
- Morriss-Andrews, A.; Shea, J.-E. Simulations of Protein Aggregation: Insights from Atomistic and Coarse-Grained Models. J. Phys. Chem. Lett. 2014, 5, 1899–1908. [Google Scholar] [CrossRef]
- Shaw, D.E.; Maragakis, P.; Lindorff-Larsen, K.; Piana, S.; Dror, R.O.; Eastwood, M.P.; Bank, J.A.; Jumper, J.M.; Salmon, J.K.; Shan, Y.; et al. Atomic-Level Characterization of the Structural Dynamics of Proteins. Science 2010, 330, 341–346. [Google Scholar] [CrossRef]
- Bernardi, R.C.; Durner, E.; Schoeler, C.; Malinowska, K.H.; Carvalho, B.G.; Bayer, E.A.; Luthey-Schulten, Z.; Gaub, H.E.; Nash, M.A. Mechanisms of Nanonewton Mechanostability in a Protein Complex Revealed by Molecular Dynamics Simulations and Single-Molecule Force Spectroscopy. J. Am. Chem. Soc. 2019, 141, 14752–14763. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, D.; Yang, W.T.; Marszalek, P.E. Piecewise All-Atom SMD Simulations Reveal Key Secondary Structures in Luciferase Unfolding Pathway. Biophy. J. 2020, 119, 2251–2261. [Google Scholar] [CrossRef]
- Tian, F.; Li, G.; Zheng, B.; Liu, Y.; Shi, S.; Deng, Y.; Zheng, P. Verification of sortase for protein conjugation by single-molecule force spectroscopy and molecular dynamics simulations. Chem. Commun. 2020, 56, 3943–3946. [Google Scholar] [CrossRef]
- Zheng, B.; Xiao, Y.; Tong, B.; Mao, Y.; Ge, R.; Tian, F.; Dong, X.; Zheng, P. S373P Mutation Stabilizes the Receptor-Binding Domain of the Spike Protein in Omicron and Promotes Binding. JACS Au 2023, 3, 1902–1910. [Google Scholar] [CrossRef]
- Gunnoo, M.; Cazade, P.-A.; Orlowski, A.; Chwastyk, M.; Liu, H.; Ta, D.T.; Cieplak, M.; Nash, M.; Thompson, D. Steered molecular dynamics simulations reveal the role of Ca2+ in regulating mechanostability of cellulose-binding proteins. Phys. Chem. Chem. Phys. 2018, 20, 22674–22680. [Google Scholar] [CrossRef] [PubMed]
- Rico, F.; Gonzalez, L.; Casuso, I.; Puig-Vidal, M.; Scheuring, S. High-Speed Force Spectroscopy Unfolds Titin at the Velocity of Molecular Dynamics Simulations. Science 2013, 342, 741–743. [Google Scholar] [CrossRef]
- Zhao, P.; Xu, C.-Q.; Sun, C.; Xia, J.; Sun, L.; Li, J.; Xu, H. Exploring the difference of bonding strength between silver(i) and chalcogenides in block copolymer systems. Polym. Chem. 2020, 11, 7087–7093. [Google Scholar] [CrossRef]
- Chen, G.; Wang, H.; Bumba, L.; Masin, J.; Sebo, P.; Li, H. The adenylate cyclase toxin RTX domain follows a series templated folding mechanism with implications for toxin activity. J. Biol. Chem. 2023, 299, 105150. [Google Scholar] [CrossRef] [PubMed]
- Baier, D.; Müller, T.; Mohr, T.; Windberger, U. Red Blood Cell Stiffness and Adhesion Are Species-Specific Properties Strongly Affected by Temperature and Medium Changes in Single Cell Force Spectroscopy. Molecules 2021, 26, 2771. [Google Scholar] [CrossRef] [PubMed]
- Sen Mojumdar, S.; Scholl, Z.N.; Dee, D.R.; Rouleau, L.; Anand, U.; Garen, C.; Woodside, M.T. Partially native intermediates mediate misfolding of SOD1 in single-molecule folding trajectories. Nat. Commun. 2017, 8, 1881. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Caballero, A.; Echelman, D.J.; Tapia-Rojo, R.; Haldar, S.; Eckels, E.C.; Fernandez, J.M. Protein folding modulates the chemical reactivity of a Gram-positive adhesin. Nat. Chem. 2021, 13, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.S.; Gruber, S.; Hausch, A.; Gomes, P.; Milles, L.F.; Nicolaus, T.; Schendel, L.C.; Navajas, P.L.; Procko, E.; Lietha, D.; et al. A tethered ligand assay to probe SARS-CoV-2:ACE2 interactions. Proc. Natl. Acad. Sci. USA 2022, 119, e2114397119. [Google Scholar] [CrossRef]
- Bauer, M.S.; Gruber, S.; Hausch, A.; Melo, M.C.R.; Gomes, P.S.F.C.; Nicolaus, T.; Milles, L.F.; Gaub, H.E.; Bernardi, R.C.; Lipfert, J. Single-molecule force stability of the SARS-CoV-2–ACE2 interface in variants-of-concern. Nat. Nanotechnol. 2024, 19, 399–405. [Google Scholar] [CrossRef]
- Zhu, R.; Canena, D.; Sikora, M.; Klausberger, M.; Seferovic, H.; Mehdipour, A.R.; Hain, L.; Laurent, E.; Monteil, V.; Wirnsberger, G.; et al. Force-tuned avidity of spike variant-ACE2 interactions viewed on the single-molecule level. Nat. Commun. 2022, 13, 7926. [Google Scholar] [CrossRef]
- Köhler, M.; Karner, A.; Leitner, M.; Hytönen, V.P.; Kulomaa, M.; Hinterdorfer, P.; Ebner, A. pH-Dependent Deformations of the Energy Landscape of Avidin-like Proteins Investigated by Single Molecule Force Spectroscopy. Molecules 2014, 19, 12531–12546. [Google Scholar] [CrossRef] [PubMed]
- Tong, B.; Tian, F.; Zheng, P. Interdomain Linker Effect on the Mechanical Stability of Ig Domains in Titin. Int. J. Mol. Sci. 2022, 23, 9836. [Google Scholar] [CrossRef]
- Suay-Corredera, C.; Pricolo, M.R.; Velazquez-Carreras, D.; Pathak, D.; Nandwani, N.; Pimenta-Lopes, C.; Sanchez-Ortiz, D.; Urrutia-Irazabal, I.; Vilches, S.; Dominguez, F.; et al. Nanomechanical Phenotypes in Cardiac Myosin-Binding Protein C Mutants That Cause Hypertrophic Cardiomyopathy. Acs Nano 2021, 15, 10203–10216. [Google Scholar] [CrossRef]
- Lei, H.; Ma, Q.; Li, W.; Wen, J.; Ma, H.; Qin, M.; Wang, W.; Cao, Y. An ester bond underlies the mechanical strength of a pathogen surface protein. Nat. Commun. 2021, 12, 5082. [Google Scholar] [CrossRef]
- Zhang, S.; Qian, H.-J.; Liu, Z.; Ju, H.; Lu, Z.-Y.; Zhang, H.; Chi, L.; Cui, S. Towards Unveiling the Exact Molecular Structure of Amorphous Red Phosphorus by Single-Molecule Studies. Angew. Chem. Int. Ed. 2019, 58, 1659–1663. [Google Scholar] [CrossRef]
- Shi, S.; Wang, Z.; Deng, Y.; Tian, F.; Wu, Q.; Zheng, P. Combination of Click Chemistry and Enzymatic Ligation for Stable and Efficient Protein Immobilization for Single-Molecule Force Spectroscopy. CCS Chem. 2022, 4, 598–604. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Z.; Yang, Z.; Li, G.; Zheng, P. pH-Dependent PdS3 Site in α3DIV Revealed by Single-Molecule Force Spectroscopy. J. Phys. Chem. B 2023, 127, 2934–2940. [Google Scholar] [CrossRef]
- Yuan, G.; Ma, Q.; Wu, T.; Wang, M.; Li, X.; Zuo, J.; Zheng, P. Multistep Protein Unfolding Scenarios from the Rupture of a Complex Metal Cluster Cd3S9. Sci. Rep. 2019, 9, 10518–10525. [Google Scholar] [CrossRef]
- Song, G.; Tian, F.; Liu, H.; Li, G.; Zheng, P. Pioglitazone Inhibits Metal Cluster Transfer of mitoNEET by Stabilizing the Labile Fe–N Bond Revealed at Single-Bond Level. J. Phys. Chem. Lett. 2021, 12, 3860–3867. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.; Mittal, J.; Feig, M.; Mackerell, A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Van der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2015, 12, 405–413. [Google Scholar] [CrossRef]
- Ravikumar, Y.; Koonyosying, P.; Srichairatanakool, S.; Ponpandian, L.N.; Kumaravelu, J.; Srichairatanakool, S. In Silico Molecular Docking and Dynamics Simulation Analysis of Potential Histone Lysine Methyl Transferase Inhibitors for Managing β-Thalassemia. Molecules 2023, 28, 7266. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, X.; Xu, C.; Zheng, B.; Yu, H.; Zheng, P. Molecular Mechanism of Interaction between DNA Aptamer and Receptor-Binding Domain of Severe Acute Respiratory Syndrome Coronavirus 2 Variants Revealed by Steered Molecular Dynamics Simulations. Molecules 2024, 29, 2215. https://doi.org/10.3390/molecules29102215
Ding X, Xu C, Zheng B, Yu H, Zheng P. Molecular Mechanism of Interaction between DNA Aptamer and Receptor-Binding Domain of Severe Acute Respiratory Syndrome Coronavirus 2 Variants Revealed by Steered Molecular Dynamics Simulations. Molecules. 2024; 29(10):2215. https://doi.org/10.3390/molecules29102215
Chicago/Turabian StyleDing, Xuan, Chao Xu, Bin Zheng, Hanyang Yu, and Peng Zheng. 2024. "Molecular Mechanism of Interaction between DNA Aptamer and Receptor-Binding Domain of Severe Acute Respiratory Syndrome Coronavirus 2 Variants Revealed by Steered Molecular Dynamics Simulations" Molecules 29, no. 10: 2215. https://doi.org/10.3390/molecules29102215
APA StyleDing, X., Xu, C., Zheng, B., Yu, H., & Zheng, P. (2024). Molecular Mechanism of Interaction between DNA Aptamer and Receptor-Binding Domain of Severe Acute Respiratory Syndrome Coronavirus 2 Variants Revealed by Steered Molecular Dynamics Simulations. Molecules, 29(10), 2215. https://doi.org/10.3390/molecules29102215




