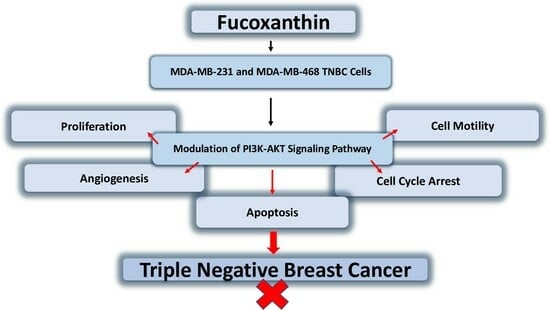
The Anticancer Effects of Marine Carotenoid Fucoxanthin through Phosphatidylinositol 3-Kinase (PI3K)-AKT Signaling on Triple-Negative Breast Cancer Cells
Abstract
1. Introduction
2. Results
2.1. Fucoxanthin Inhibited Cell Viability in TNF-α-Stimulated MDA-MB-231 and MDA-MB-468 Cell Lines
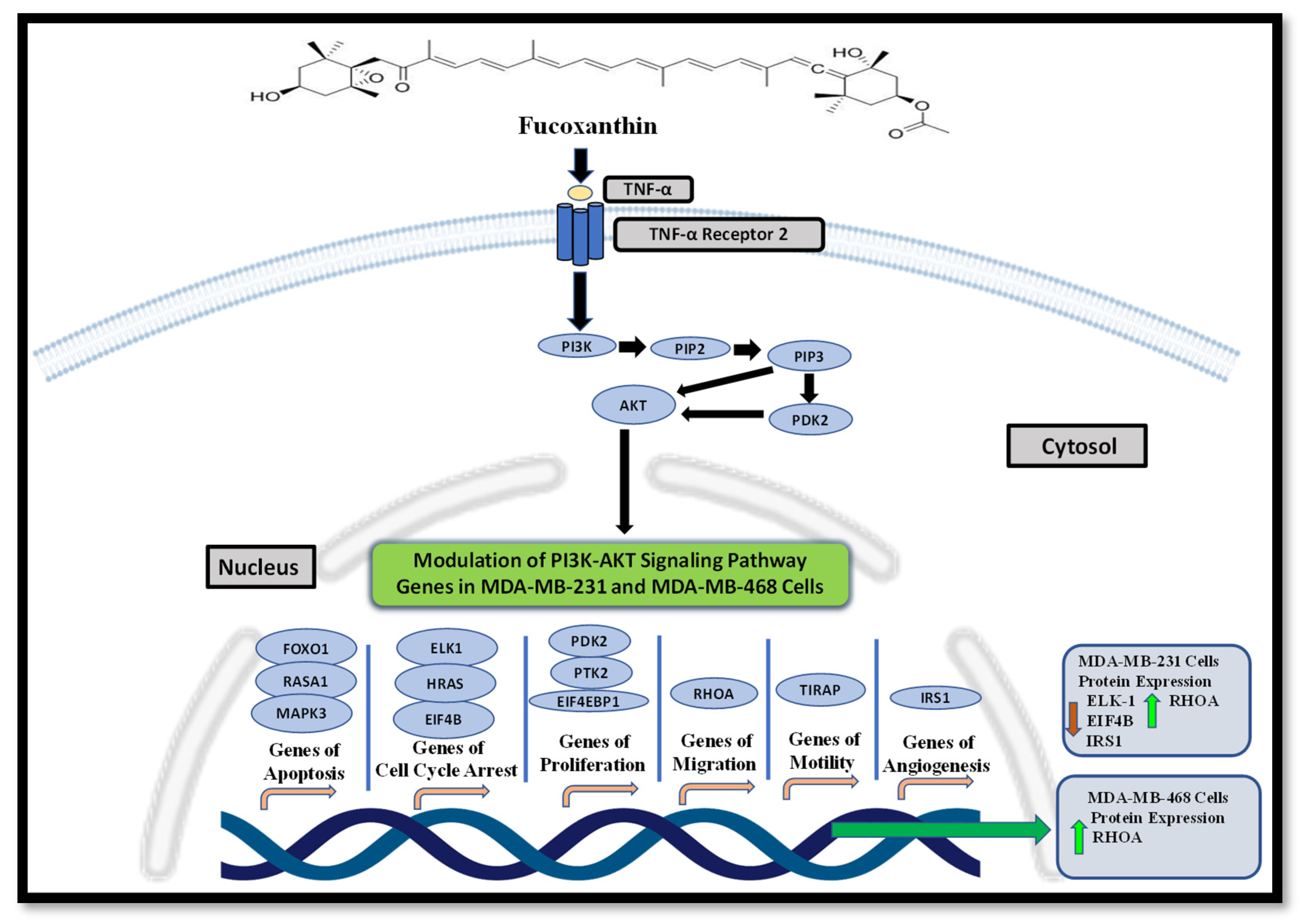
2.2. Fucoxanthin Modulation of the PI3K-AKT Signaling Pathway in MDA-MB-231 and MDA-MB-468 TNBC Cells
2.3. Fucoxanthin Modulated PI3K-AKT Signaling Pathway Protein Expression in MDA-MB-231 and MDA-MB-468 Cells
2.4. Activation of the PI3K-AKT Signaling Pathway through Stimulation of TNF-α Using KEGG Analysis Database
2.5. STRING, Functional Enrichment Analysis of Fucoxanthin’s Effects on Proteins of the PI3K-AKT Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Cell Lines, Chemicals, and Reagents
4.2. Cell Culture
4.3. Cell Viability
4.4. cDNA Synthesis and RT-PCR Array (PI3K-AKT Signaling Pathway)
4.4.1. Extraction of RNA
4.4.2. cDNA Conversion and RT-PCR
4.5. Wes Analysis
4.6. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breast Cancer Research Foundation. Breast Cancer Statistics and Resources; Breast Cancer Research Foundation: New York, NY, USA, 2023; Available online: https://www.bcrf.org/breast-cancer-statistics-and-resources/ (accessed on 18 June 2023).
- National Cancer Institute. Cancer Stat Facts: Female Breast Cancer; National Cancer Institute: Bethesda, MD, USA, 2023. Available online: https://seer.cancer.gov/statfacts/html/breast.html (accessed on 16 June 2023).
- Aysola, K.; Desai, A.; Welch, C.; Xu, J.; Qin, Y.; Reddy, V.; Matthews, R.; Owens, C.; Okoli, J.; Beech, D.J.; et al. Triple Negative Breast Cancer—An Overview. Hered. Genet. 2013, 2013 (Suppl. S2), 1. [Google Scholar] [CrossRef]
- Wahba, H.A.; El-Hadaad, H.A. Current approaches in the treatment of triple-negative breast cancer. Cancer Biol. Med. 2015, 12, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Dewi, C.; Fristiohady, A.; Amalia, R.; Khairul Ikram, N.K.; Ibrahim, S.; Muchtaridi, M. Signaling Pathways and Natural Compounds in Triple-Negative Breast Cancer Cell Line. Molecules 2022, 27, 3661. [Google Scholar] [CrossRef] [PubMed]
- Hashem, S.; Ali, T.A.; Akhtar, S.; Nisar, S.; Sageena, G.; Ali, S.; Al-Mannai, S.; Therachiyil, L.; Mir, R.; Elfaki, I.; et al. Targeting cancer signaling pathways by natural products: Exploring promising anticancer agents. Biomed. Pharmacother. 2022, 150, 113054. [Google Scholar] [CrossRef] [PubMed]
- Zaragozá, M.C.; López, D.; Sáiz, M.P.; Poquet, M.; Pérez, J.; Puig-Parellada, P.; Màrmol, F.; Simonetti, P.; Gardana, C.; Lerat, Y.; et al. Toxicity and antioxidant activity in vitro and in vivo of two Fucus vesiculosus extracts. J. Agric. Food Chem. 2008, 56, 7773–7780. [Google Scholar] [CrossRef] [PubMed]
- Nishino, H. Cancer prevention by carotenoids. Mutat. Res./Fundam. Mol. Mech. Mutagen. 1998, 402, 159–163. [Google Scholar] [CrossRef]
- Airanthi, M.K.; Sasaki, N.; Iwasaki, S.; Baba, N.; Abe, M.; Hosokawa, M.; Miyashita, K. Effect of brown seaweed lipids on fatty acid composition and lipid hydroperoxide levels of mouse liver. J. Agric. Food Chem. 2011, 59, 4156–4163. [Google Scholar] [CrossRef]
- Murakami, C.; Takemura, M.; Sugiyama, Y.; Kamisuki, S.; Asahara, H.; Kawasaki, M.; Ishidoh, T.; Linn, S.; Yoshida, S.; Sugawara, F.; et al. Vitamin A-related compounds, all-trans-retinal and retinoic acids, selectively inhibit the activities of mammalian replicative DNA polymerases. Biochim. Biophys. Acta 2002, 1574, 85–92. [Google Scholar] [CrossRef]
- Ming, J.X.; Wang, Z.C.; Huang, Y.; Ohishi, H.; Wu, R.J.; Shao, Y.; Wang, H.; Qin, M.Y.; Wu, Z.L.; Li, Y.Y.; et al. Fucoxanthin extracted from Laminaria Japonica inhibits metastasis and enhances the sensitivity of lung cancer to Gefitinib. J. Ethnopharmacol. 2021, 265, 113302. [Google Scholar] [CrossRef]
- Mohibbullah, M.; Haque, M.N.; Sohag, A.A.M.; Hossain, M.T.; Zahan, M.S.; Uddin, M.J.; Hannan, M.A.; Moon, I.S.; Choi, J.S. A Systematic Review on Marine Algae-Derived Fucoxanthin: An Update of Pharmacological Insights. Mar. Drugs 2022, 20, 279. [Google Scholar] [CrossRef]
- Zhang, H.; Tang, Y.; Zhang, Y.; Zhang, S.; Qu, J.; Wang, X.; Kong, R.; Han, C.; Liu, Z. Fucoxanthin: A Promising Medicinal and Nutritional Ingredient. Evid. Based Complement. Alternat. Med. 2015, 2015, 723515. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.R.; Hosokawa, M.; Miyashita, K. Fucoxanthin: A marine carotenoid exerting anticancer effects by affecting multiple mechanisms. Mar. Drugs 2013, 11, 5130–5147. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.J. Fucoxanthin and Its Metabolite Fucoxanthinol in Cancer Prevention and Treatment. Mar. Drugs 2015, 13, 4784–4798. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Mendonca, P.; Elhag, R.; Soliman, K.F.A. Anticancer Effects of Fucoxanthin through Cell Cycle Arrest, Apoptosis Induction, Angiogenesis Inhibition, and Autophagy Modulation. Int. J. Mol. Sci. 2022, 23, 16091. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Mendonca, P.; Messeha, S.S.; Soliman, K.F.A. Anticancer Effects of Fucoxanthin through Cell Cycle Arrest, Apoptosis Induction, and Angiogenesis Inhibition in Triple-Negative Breast Cancer Cells. Molecules 2023, 28, 6536. [Google Scholar] [CrossRef]
- Yoshiko, S. Antitumor and Cancer-preventative Function of Fucoxanthin: A Marine Carotenoid. Anticancer Res. 2017, 37, 1557. Available online: http://ar.iiarjournals.org/content/37/4/1557.abstract (accessed on 7 July 2023).
- Ye, G.; Wang, L.; Yang, K.; Wang, C. Fucoxanthin may inhibit cervical cancer cell proliferation via downregulation of HIST1H3D. J. Int. Med. Res. 2020, 48, 0300060520964011. [Google Scholar] [CrossRef]
- Kotake-Nara, E.; Kushiro, M.; Zhang, H.; Sugawara, T.; Miyashita, K.; Nagao, A. Carotenoids affect proliferation of human prostate cancer cells. J. Nutr. 2001, 131, 3303–3306. [Google Scholar] [CrossRef]
- Lau, T.Y.; Kwan, H.Y. Fucoxanthin Is a Potential Therapeutic Agent for the Treatment of Breast Cancer. Mar. Drugs 2022, 20, 370. [Google Scholar] [CrossRef]
- Méresse, S.; Fodil, M.; Fleury, F.; Chénais, B. Fucoxanthin, a Marine-Derived Carotenoid from Brown Seaweeds and Microalgae: A Promising Bioactive Compound for Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 9273. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, J.; Zhang, Y.; Wang, Z.; Yang, Y.; Bai, M.; Dai, Y. Fucoxanthin Activates Apoptosis via Inhibition of PI3K/Akt/mTOR Pathway and Suppresses Invasion and Migration by Restriction of p38-MMP-2/9 Pathway in Human Glioblastoma Cells. Neurochem. Res. 2016, 41, 2728–2751. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Porta, C.; Paglino, C.; Mosca, A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Front. Oncol. 2014, 4, 64. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Feng, C.; Huang, P.; Li, Y.; Liu, R.; Liu, C.; Han, Y.; Chen, L.; Ding, Y.; Shao, C.; et al. TNFα and IFNγ rapidly activate PI3K-AKT signaling to drive glycolysis that confers mesenchymal stem cells enhanced anti-inflammatory properties. Stem Cell Res. Ther. 2022, 13, 491. [Google Scholar] [CrossRef] [PubMed]
- Laha, D.; Grant, R.; Mishra, P.; Nilubol, N. The Role of Tumor Necrosis Factor in Manipulating the Immunological Response of Tumor Microenvironment. Front. Immunol. 2021, 12, 656908. [Google Scholar] [CrossRef]
- Martínez-Reza, I.; Díaz, L.; García-Becerra, R. Preclinical and clinical aspects of TNF-α and its receptors TNFR1 and TNFR2 in breast cancer. J. Biomed. Sci. 2017, 24, 90. [Google Scholar] [CrossRef]
- Mercogliano, M.F.; Bruni, S.; Elizalde, P.V.; Schillaci, R. Tumor Necrosis Factor α Blockade: An Opportunity to Tackle Breast Cancer. Front. Oncol. 2020, 10, 584. [Google Scholar] [CrossRef]
- Wang, Z.; Ao, X.; Shen, Z.; Ao, L.; Wu, X.; Pu, C.; Guo, W.; Xing, W.; He, M.; Yuan, H.; et al. TNF-α augments CXCL10/CXCR3 axis activity to induce Epithelial-Mesenchymal Transition in colon cancer cell. Int. J. Biol. Sci. 2021, 17, 2683–2702. [Google Scholar] [CrossRef]
- Liu, M.; Guo, S.; Stiles, J.K. The emerging role of CXCL10 in cancer (Review). Oncol. Lett. 2011, 2, 583–589. [Google Scholar] [CrossRef]
- Saleh, M.; Chandrashekar, D.S.; Shahin, S.; Agarwal, S.; Kim, H.-G.; Behring, M.; Shaikh, A.J.; Moloo, Z.; Eltoum, I.-E.A.; Yates, C.; et al. Comparative analysis of triple-negative breast cancer transcriptomics of Kenyan, African American and Caucasian Women. Transl. Oncol. 2021, 14, 101086. [Google Scholar] [CrossRef]
- Davis, M.; Martini, R.; Newman, L.; Elemento, O.; White, J.; Verma, A.; Datta, I.; Adrianto, I.; Chen, Y.; Gardner, K.; et al. Identification of Distinct Heterogenic Subtypes and Molecular Signatures Associated with African Ancestry in Triple Negative Breast Cancer Using Quantified Genetic Ancestry Models in Admixed Race Populations. Cancers 2020, 12, 1220. [Google Scholar] [CrossRef] [PubMed]
- Matias, A.T.; Jacinta-Fernandes, A.; Maia, A.-T.; Braga, S.; Jacinto, A.; Cabral, M.G.; Brito, P.H. Differential expression between African-ancestry and White patients diagnosed with Triple-Negative Breast Cancer: EGFR, Myc, Bcl2 and β-Catenin as ancestry-associated markers. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Wang, Q.; Su, B.; Xu, H.; Sun, Y.; Sun, P.; Li, R.; Peng, X.; Cai, J. Role of RASA1 in cancer: A review and update. Oncol. Rep. 2020, 44, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Reagan-Shaw, S.; Ahmad, N. RNA interference-mediated depletion of phosphoinositide 3-kinase activates forkhead box class O transcription factors and induces cell cycle arrest and apoptosis in breast carcinoma cells. Cancer Res. 2006, 66, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Odrowaz, Z.; Sharrocks, A.D. ELK1 Uses Different DNA Binding Modes to Regulate Functionally Distinct Classes of Target Genes. PLoS Genet. 2012, 8, e1002694. [Google Scholar] [CrossRef]
- Chen, B.; Chen, Y.; Rai, K.R.; Wang, X.; Liu, S.; Li, Y.; Xiao, M.; Ma, Y.; Wang, G.; Guo, G.; et al. Deficiency of eIF4B Increases Mouse Mortality and Impairs Antiviral Immunity. Front. Immunol. 2021, 12, 723885. [Google Scholar] [CrossRef]
- Porter, H.A.; Perry, A.; Kingsley, C.; Tran, N.L.; Keegan, A.D. IRS1 is highly expressed in localized breast tumors and regulates the sensitivity of breast cancer cells to chemotherapy, while IRS2 is highly expressed in invasive breast tumors. Cancer Lett. 2013, 338, 239–248. [Google Scholar] [CrossRef]
- Rigiracciolo, D.C.; Santolla, M.F.; Lappano, R.; Vivacqua, A.; Cirillo, F.; Galli, G.R.; Talia, M.; Muglia, L.; Pellegrino, M.; Nohata, N.; et al. Focal adhesion kinase (FAK) activation by estrogens involves GPER in triple-negative breast cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 58. [Google Scholar] [CrossRef]
- Wang, X.; Shen, X.; Yan, Y.; Li, H. Pyruvate dehydrogenase kinases (PDKs): An overview toward clinical applications. Biosci. Rep. 2021, 41, BSR20204402. [Google Scholar] [CrossRef]
- Bin, L.-H.; Xu, L.-G.; Shu, H.-B. TIRP, a Novel Toll/Interleukin-1 receptor (TIR) Domain-containing Adapter Protein Involved in TIR signaling*. J. Biol. Chem. 2003, 278, 24526–24532. [Google Scholar] [CrossRef] [PubMed]
- Murugan, A.K.; Grieco, M.; Tsuchida, N. RAS mutations in human cancers: Roles in precision medicine. Semin. Cancer Biol. 2019, 59, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yang, B.; Nass, N.; Schatz, C.; Haybaeck, J. Impact of Eukaryotic Translation Initiation Factors on Breast Cancer: Still Much to Investigate. Cancers 2020, 12, 1984. Available online: https://www.mdpi.com/2072-6694/12/7/1984 (accessed on 19 September 2023). [CrossRef] [PubMed]
- KEGG TNF Signaling Pathway—Homo Sapiens (Human). 2019. Available online: https://www.genome.jp/pathway/hsa04668 (accessed on 14 November 2022).
- STRING. STRING CONSORTIUM 2023. 2023. Available online: https://string-db.org/ (accessed on 15 June 2023).
- Asati, V.; Bharti, S.K.; Mahapatra, D.K.; Asati, V.; Budhwani, A.K. Triggering PIK3CA Mutations in PI3K/Akt/mTOR Axis: Exploration of Newer Inhibitors and Rational Preventive Strategies. Curr. Pharm. Des. 2016, 22, 6039–6054. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.; Meng, X.; Gan, R.Y.; Zhang, J.J.; Li, H.B. Dietary Natural Products for Prevention and Treatment of Breast Cancer. Nutrients 2017, 9, 728. [Google Scholar] [CrossRef] [PubMed]
- Metibemu, D.S.; Ogungbe, I.V. Carotenoids in Drug Discovery and Medicine: Pathways and Molecular Targets Implicated in Human Diseases. Molecules 2022, 27, 6005. [Google Scholar] [CrossRef] [PubMed]
- Doolaanea, A.A.; Alfatama, M.; Alkhatib, H.; Mawazi, S.M. Fucoxanthin. In Handbook of Food Bioactive Ingredients: Properties and Applications; Jafari, S.M., Rashidinejad, A., Simal-Gandara, J., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2023; pp. 729–755. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT network at the interface of oncogenic signaling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- Pucci, B.; Kasten, M.; Giordano, A. Cell cycle and apoptosis. Neoplasia 2000, 2, 291–299. [Google Scholar] [CrossRef]
- Alenzi, F.Q. Links between apoptosis, proliferation and the cell cycle. Br. J. Biomed. Sci. 2004, 61, 99–102. [Google Scholar] [CrossRef]
- Yuan, L.; Cai, Y.; Zhang, L.; Liu, S.; Li, P.; Li, X. Promoting Apoptosis, a Promising Way to Treat Breast Cancer with Natural Products: A Comprehensive Review. Front. Pharmacol. 2022, 12, 801662. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, J.; Lu, J.; Ma, R.; Sun, D.; Tang, J. Regulation of the cell cycle and PI3K/Akt/mTOR signaling pathway by tanshinone I in human breast cancer cell lines Retraction in /10.3892/mmr.2021.12080. Mol. Med. Rep. 2015, 11, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Jain, V.K.; Rizwanullah, M.; Ahmad, J.; Jain, K. PI3K/AKT/mTOR pathway inhibitors in triple-negative breast cancer: A review on drug discovery and future challenges. Drug Discov. Today 2019, 24, 2181–2191. [Google Scholar] [CrossRef] [PubMed]
- Nitulescu, G.M.; Van De Venter, M.; Nitulescu, G.; Ungurianu, A.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Grădinaru, D.; Tsatsakis, A.; Tsoukalas, D.; et al. The Akt pathway in oncology therapy and beyond. Int. J. Oncol. 2018, 53, 2319–2331. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.A.; Fraile-Martínez, O.; Asúnsolo, Á.; Buján, J.; García-Honduvilla, N.; Coca, S. Signal Transduction Pathways in Breast Cancer: The Important Role of PI3K/Akt/mTOR. J. Oncol. 2020, 2020, 9258396. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Shiue, C.N.; Zhu, J.; Zhuang, T.; Jonsson, P.; Wright, A.P.; Zhao, C.; Dahlman-Wright, K. AP-1-mediated chromatin looping regulates ZEB2 transcription: New insights into TNFα-induced epithelial-mesenchymal transition in triple-negative breast cancer. Oncotarget 2015, 6, 7804–7814. [Google Scholar] [CrossRef]
- Rivas, M.A.; Carnevale, R.P.; Proietti, C.J.; Rosemblit, C.; Beguelin, W.; Salatino, M.; Charreau, E.H.; Frahm, I.; Sapia, S.; Brouckaert, P.; et al. TNF alpha acting on TNFR1 promotes breast cancer growth via p42/P44 MAPK, JNK, Akt and NF-kappa B-dependent pathways. Exp. Cell Res. 2008, 314, 509–529. [Google Scholar] [CrossRef]
- Catasus, L.; D’Angelo, E.; Pons, C.; Espinosa, I.; Prat, J. Expression profiling of 22 genes involved in the PI3K–AKT pathway identifies two subgroups of high-grade endometrial carcinomas with different molecular alterations. Mod. Pathol. 2010, 23, 694–702. [Google Scholar] [CrossRef]
- Fang, X.; Zhu, Y.; Zhang, T.; Li, Q.; Fan, L.; Li, X.; Jiang, D.; Lin, J.; Zou, L.; Ren, J.; et al. Fucoxanthin Inactivates the PI3K/Akt Signaling Pathway to Mediate Malignant Biological Behaviors of Non-Small Cell Lung Cancer. Nutr. Cancer 2022, 74, 3747–3760. [Google Scholar] [CrossRef]
- Cuesta, C.; Arévalo-Alameda, C.; Castellano, E. The Importance of Being PI3K in the RAS Signaling Network. Genes 2021, 12, 1094. [Google Scholar] [CrossRef]
- Li, Y.; Leisi, T.; Bao, L.; Arunachalam, C.; Alharbi, S.; Cui, J. Fucoxanthin inhibits cell proliferation and stimulates apoptosis through downregulation of PI3K/AKT/mTOR signaling pathway in human ovarian cancer cells. Pharmacogn. Mag. 2020, 16, 311–316. [Google Scholar] [CrossRef]
- Wang, W.; Fu, C.; Lin, M.; Lu, Y.; Lian, S.; Xie, X.; Zhou, G.; Li, W.; Zhang, Y.; Jia, L.; et al. Fucoxanthin prevents breast cancer metastasis by interrupting circulating tumor cells adhesion and transendothelial migration. Front. Pharmacol. 2022, 13, 960375. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ma, Y.; Yang, J.; Jin, L.; Gao, Z.; Xue, L.; Hou, L.; Sui, L.; Liu, J.; Zou, X. Fucoxanthin inhibits tumour-related lymphangiogenesis and growth of breast cancer. J. Cell. Mol. Med. 2019, 23, 2219–2229. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.L.; Fu, X.Y.; Cao, W.Q.; Xiang, W.Z.; Hou, Y.J.; Ma, J.K.; Wang, Y.; Fan, C.D. Induction of Apoptosis in Human Glioma Cells by Fucoxanthin via Triggering of ROS-Mediated Oxidative Damage and Regulation of MAPKs and PI3K-AKT Pathways. J. Agric. Food Chem. 2019, 67, 2212–2219. [Google Scholar] [CrossRef] [PubMed]
- Ismylatifa, D.; Diah, P.; Nita, P.; HerinSetianingsih. The Effect of Brown Seaweed (Sargassum sp.) Extract on Apoptosis Process in Breast Cancer—A Literature Review. Indian J. Forensic Med. Toxicol. 2021, 15, 3247–3251. [Google Scholar] [CrossRef]
- Miricescu, D.; Totan, A.; Stanescu-Spinu, I.-I.; Badoiu, S.C.; Stefani, C.; Greabu, M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int. J. Mol. Sci. 2020, 22, 173. [Google Scholar] [CrossRef]
- Fresno Vara, J.A.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef]
- Guan, B.; Chen, K.; Tong, Z.; Chen, L.; Chen, Q.; Su, J. Advances in Fucoxanthin Research for the Prevention and Treatment of Inflammation-Related Diseases. Nutrients 2022, 14, 4768. [Google Scholar] [CrossRef]
- Li, D.; Zhang, Y.; Zhang, H.; Zhan, C.; Li, X.; Ba, T.; Qiu, Z.; E, F.; Lv, G.; Zou, C.; et al. CADM2, as a new target of miR-10b, promotes tumor metastasis through the FAK/AKT pathway in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 46. [Google Scholar] [CrossRef]
- Bianchi, N.; Brugnoli, F.; Grassilli, S.; Bourgeois, K.; Keillor, J.W.; Bergamini, C.M.; Aguiari, G.; Volinia, S.; Bertagnolo, V. The Motility and Mesenchymal Features of Breast Cancer Cells Correlate with the Levels and Intracellular Localization of Transglutaminase Type 2. Cells 2021, 10, 3059. [Google Scholar] [CrossRef]
- Yoeli-Lerner, M.; Yiu, G.K.; Rabinovitz, I.; Erhardt, P.; Jauliac, S.; Toker, A. Akt Blocks Breast Cancer Cell Motility and Invasion through the Transcription Factor NFAT. Mol. Cell 2005, 20, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Finley, S.D. Mechanistic insight into activation of MAPK signaling by pro-angiogenic factors. BMC Syst. Biol. 2018, 12, 145. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Tocheny, C.E.; Shaw, L.M. The Insulin-like Growth Factor Signaling Pathway in Breast Cancer: An Elusive Therapeutic Target. Life 2022, 12, 1992. [Google Scholar] [CrossRef] [PubMed]
- Timbrell, S.; Aglan, H.; Cramer, A.; Foden, P.; Weaver, D.; Pachter, J.; Kilgallon, A.; Clarke, R.B.; Farnie, G.; Bundred, N.J. FAK inhibition alone or in combination with adjuvant therapies reduces cancer stem cell activity. NPJ Breast Cancer 2021, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Fernald, K.; Kurokawa, M. Evading apoptosis in cancer. Trends Cell Biol. 2013, 23, 620–633. [Google Scholar] [CrossRef]
- Banys-Paluchowski, M.; Milde-Langosch, K.; Fehm, T.; Witzel, I.; Oliveira-Ferrer, L.; Schmalfeldt, B.; Müller, V. Clinical relevance of H-RAS, K-RAS, and N-RAS mRNA expression in primary breast cancer patients. Breast Cancer Res. Treat. 2020, 179, 403–414. [Google Scholar] [CrossRef]
- Pązik, M.; Michalska, K.; Żebrowska-Nawrocka, M.; Zawadzka, I.; Łochowski, M.; Balcerczak, E. Clinical significance of HRAS and KRAS genes expression in patients with non–small-cell lung cancer—Preliminary findings. BMC Cancer 2021, 21, 130. [Google Scholar] [CrossRef]
- Galiè, M. RAS as Supporting Actor in Breast Cancer. Front. Oncol. 2019, 9, 1199. [Google Scholar] [CrossRef]
- Wu, C.-C.; Pan, M.-R.; Shih, S.-L.; Shiau, J.-P.; Wu, C.-C.; Chang, S.-J.; Kao, C.-N.; Chen, F.-M.; Hou, M.-F.; Luo, C.-W. Combination of FAK inhibitor and cytokine-induced killer cell therapy: An alternative therapeutic strategy for patients with triple-negative breast cancer. Biomed. Pharmacother. 2023, 163, 114732. [Google Scholar] [CrossRef]
- Atas, E.; Oberhuber, M.; Kenner, L. The Implications of PDK1–4 on Tumor Energy Metabolism, Aggressiveness and Therapy Resistance. Front. Oncol. 2020, 10, 583217. [Google Scholar] [CrossRef]
- Chan, T.O.; Tsichlis, P.N. PDK2: A complex tail in one Akt. Sci. STKE 2001, 2001, pe1. [Google Scholar] [CrossRef] [PubMed]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar] [CrossRef] [PubMed]
- Sophie, P.; Nicolas, C.; Jérôme, T.; Valérie, B.; Pascale, C.-L.; Lise, W.; Alexa, G.; Patrick, M.; Catherine, L.; Didier, B. Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia. Haematologica 2010, 95, 819–828. [Google Scholar] [CrossRef]
- Shahbazian, D.; Parsyan, A.; Petroulakis, E.; Hershey, J.; Sonenberg, N. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle 2010, 9, 4106–4109. [Google Scholar] [CrossRef] [PubMed]
- Lazaris-Karatzas, A.; Montine, K.S.; Sonenberg, N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 1990, 345, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Madden, J.M.; Mueller, K.L.; Bollig-Fischer, A.; Stemmer, P.; Mattingly, R.R.; Boerner, J.L. Abrogating phosphorylation of eIF4B is required for EGFR and mTOR inhibitor synergy in triple-negative breast cancer. Breast Cancer Res. Treat. 2014, 147, 283–293. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fitzgerald, K.A.; Rowe, D.C.; Barnes, B.J.; Caffrey, D.R.; Visintin, A.; Latz, E.; Monks, B.; Pitha, P.M.; Golenbock, D.T. LPS-TLR4 Signaling to IRF-3/7 and NF-κB Involves the Toll Adapters TRAM and TRIF. J. Exp. Med. 2003, 198, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Bumpass, D.C.; Perkins, N.D.; Sharrocks, A.D. The ETS domain transcription factor Elk-1 contains a novel class of repression domain. Mol. Cell. Biol. 2002, 22, 5036–5046. [Google Scholar] [CrossRef]
- Yan, Q.; Ni, C.; Lin, Y.; Sun, X.; Shen, Z.; Zhang, M.; Han, S.; Shi, J.; Mao, J.; Yang, Z.; et al. ELK1 Enhances Pancreatic Cancer Progression Via LGMN and Correlates with Poor Prognosis. Front. Mol. Biosci. 2021, 8, 764900. [Google Scholar] [CrossRef]
- Chen, B.H.; Tzen, J.T.; Bresnick, A.R.; Chen, H.C. Roles of Rho-associated kinase and myosin light chain kinase in morphological and migratory defects of focal adhesion kinase-null cells. J. Biol. Chem. 2002, 277, 33857–33863. [Google Scholar] [CrossRef]
- Phuyal, S.; Farhan, H. Multifaceted Rho GTPase Signaling at the Endomembranes. Front. Cell Dev. Biol. 2019, 7, 127. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Tindall, D.J. FOXOs, cancer and regulation of apoptosis. Oncogene 2008, 27, 2312–2319. [Google Scholar] [CrossRef] [PubMed]
- Zanella, F.; Link, W.; Carnero, A. Understanding FOXO, new views on old transcription factors. Curr. Cancer Drug Targets 2010, 10, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Fan, L.Y.; Lam, E.W. The FOXO3-FOXM1 axis: A key cancer drug target and a modulator of cancer drug resistance. Semin. Cancer Biol. 2018, 50, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Chau, J.E.; Vish, K.J.; Boggon, T.J.; Stiegler, A.L. SH3 domain regulation of RhoGAP activity: Crosstalk between p120RasGAP and DLC1 RhoGAP. Nat. Commun. 2022, 13, 4788. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Zheng, S.; Zou, Y.; Yang, A.; Xie, X.; Tang, H.; Xie, X. CircAHNAK1 inhibits proliferation and metastasis of triple-negative breast cancer by modulating miR-421 and RASA1. Aging 2019, 11, 12043–12056. [Google Scholar] [CrossRef] [PubMed]
- Rutkovsky, A.C.; Yeh, E.S.; Guest, S.T.; Findlay, V.J.; Muise-Helmericks, R.C.; Armeson, K.; Ethier, S.P. Eukaryotic initiation factor 4E-binding protein as an oncogene in breast cancer. BMC Cancer 2019, 19, 491. [Google Scholar] [CrossRef] [PubMed]
- Assoian, R.K.; Zhu, X. Cell anchorage and the cytoskeleton as partners in growth factor-dependent cell cycle progression. Curr. Opin. Cell Biol. 1997, 9, 93–98. [Google Scholar] [CrossRef]
- O’Meara, T.; Safonov, A.; Casadevall, D.; Qing, T.; Silber, A.; Killelea, B.; Hatzis, C.; Pusztai, L. Immune microenvironment of triple-negative breast cancer in African-American and Caucasian women. Breast Cancer Res. Treat. 2019, 175, 247–259. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. BCR 2020, 22, 61. [Google Scholar] [CrossRef]







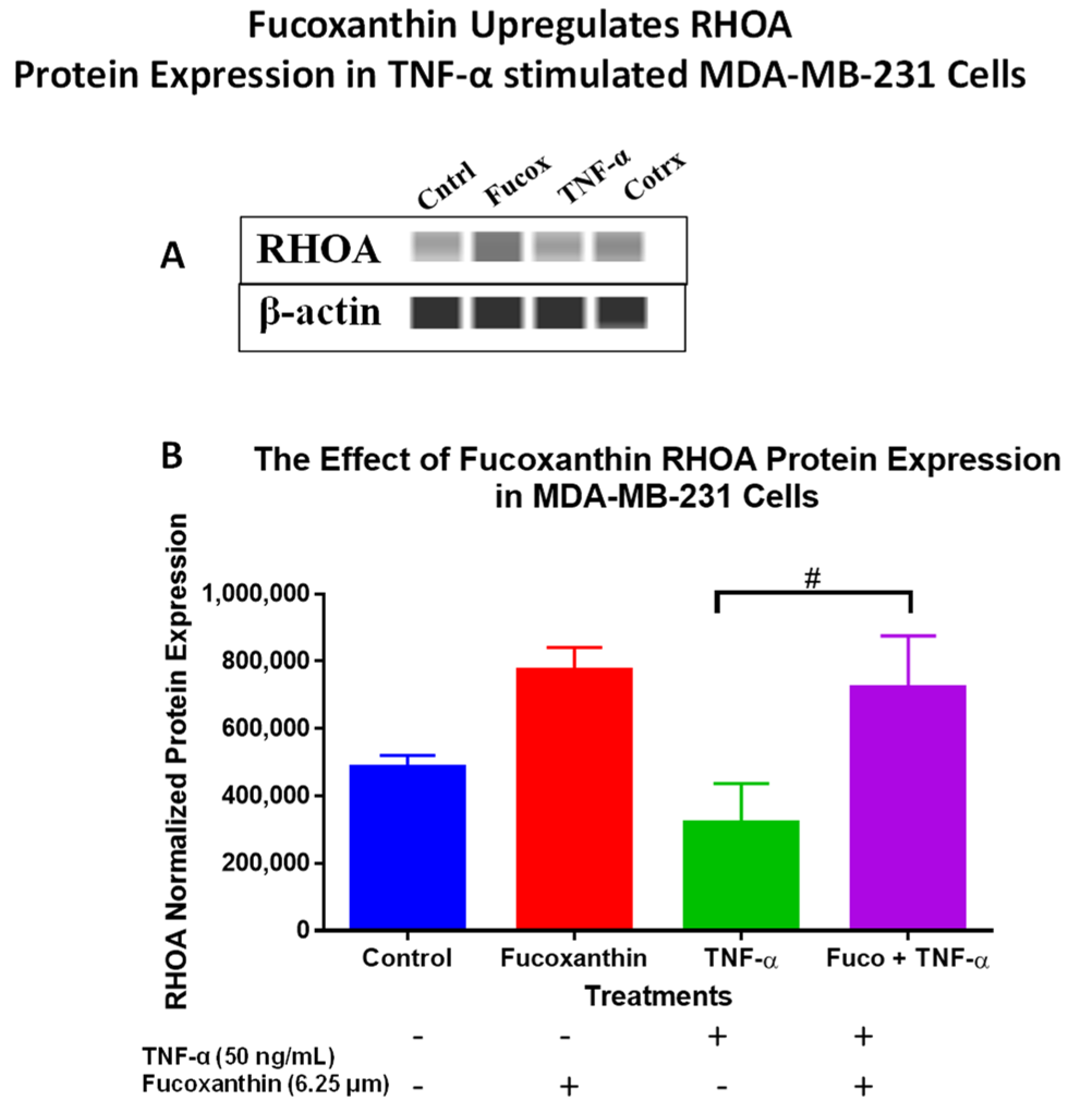

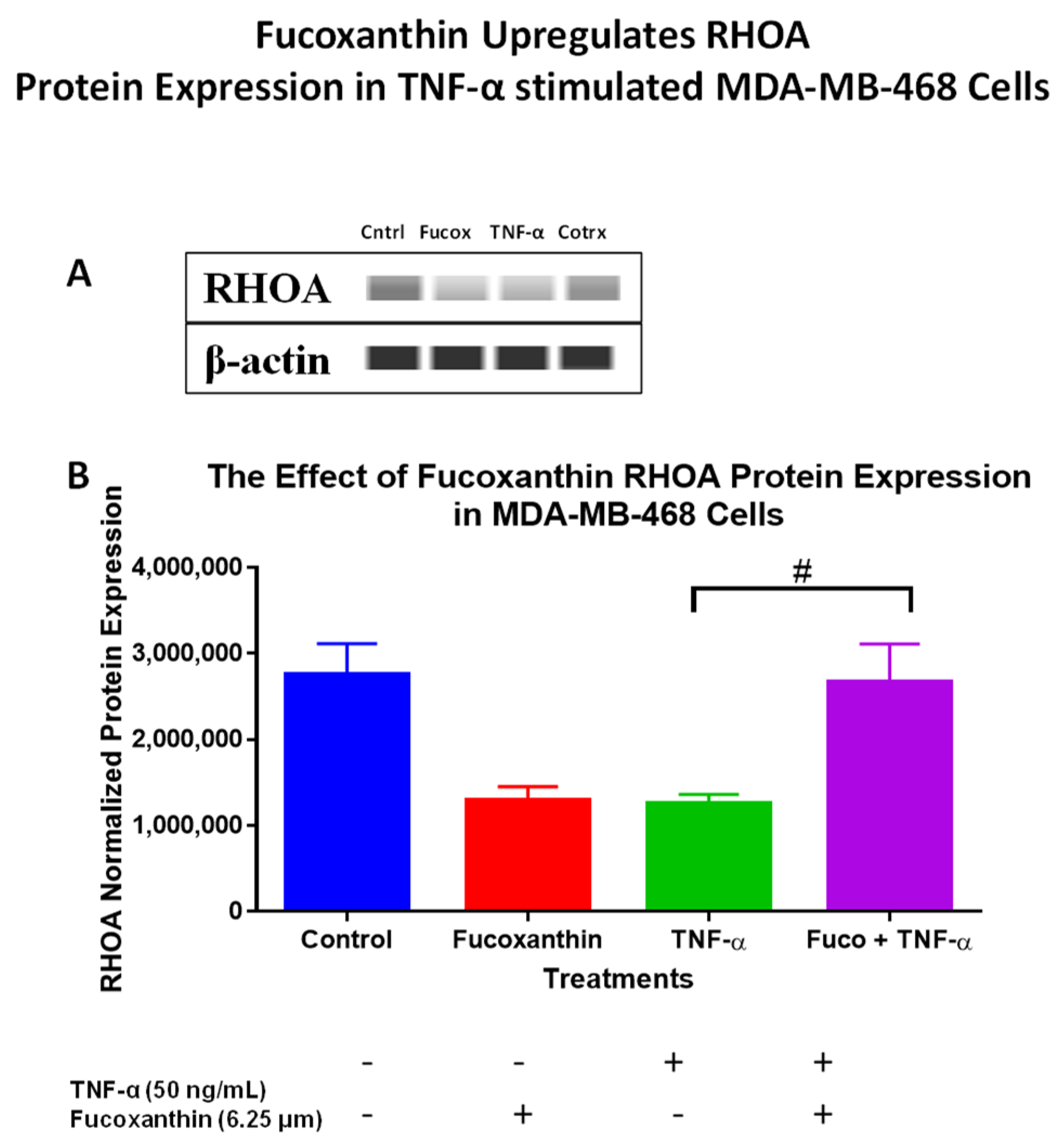


| Gene Name | Description | Fold Change (Downregulated or Upregulated) | Function |
|---|---|---|---|
| RASA1 | Ras P21 Protein Activator 1 | +3.24 | Provides instruction for protein p120-RasGap and helps regulate Ras-MAPK [35]. |
| FOXO1 | Forkhead Box 1 | +2.66 | Tumor suppressor that induces cell cycle arrest and apoptosis [36]. |
| RHOA | Rho Family Protein A | +1.89 | Contributes to cell cycle progression, adhesion, and microtubule and actin cytoskeleton organization [37]. |
| ELK1 | ETS Transcription Factor 1 | −3.91 | Regulates a network of genes related to actin/migration [38]. |
| EIF4B | Eukaryotic Translation Initiation Factor 4B | −3.83 | Regulated by both Ras-MAPK and PI3K/mTOR and plays a role in cell viability, cell growth, and survival [39]. |
| IRS1 | Insulin Receptor Substrate 1 | −3.51 | Regulates cell motility, proliferation, and cell survival [40]. |
| PTK2 | Protein Tyrosine Kinase 2 | −2.64 | Stimulates tumor cell invasiveness and adhesion [41]. |
| PDK2 | Pyruvate Dehydrogenase Kinase 2 | −2.46 | Associated with proliferation, anti-apoptosis, tumor aggressiveness, and therapy resistance [42]. |
| TIRAP | TIR Domain-Containing Adaptor Protein | −2.14 | Activates NF-κB, MAPK3, and JNK signaling, resulting in the inflammatory response and cytokine release [43]. |
| HRAS | HRas Proto-Oncogene, GTPase | −1.97 | Regulates adhesion, cytoskeleton rearrangements, proliferation, cell survival, and cell motility [44]. |
| MAPK3 | Mitogen-Activated Protein Kinase 3 | −1.90 | Regulates inflammation, proliferation, apoptosis, and differentiation [45]. |
| EIF4EBP1 | Eukaryotic Transition Initiation Factor 4E-Binding Protein 1 | −1.23 | Increases metastasis, invasion, and tumor formation [46]. |
| Gene Name | Description | Fold Change (Downregulated or Upregulated) | Function |
|---|---|---|---|
| EIF4EBP1 | Eukaryotic Transition Initiation Factor 4E-Binding Protein 1 | +2.16 | Increases metastasis, invasion, and tumor formation [46]. |
| TIRAP | TIR Domain-Containing Adaptor Protein | −1.61 | Activates NF-κB, MAPK3, and JNK signaling, resulting in the inflammatory response and cytokine release [43]. |
| EIF4B | Eukaryotic Translation Initiation Factor 4B | −1.51 | Regulated by both Ras-MAPK and PI3K/mTOR, it plays a role in cell viability, growth, and survival [39]. |
| PTK2 | Protein Tyrosine Kinase 2 | −1.51 | Stimulates tumor cell invasiveness and adhesion [41]. |
| IRS1 | Insulin Receptor Substrate 1 | −1.49 | Regulates cell motility, proliferation, and cell survival [40]. |
| ELK1 | ETS Transcription Factor 1 | −1.44 | Regulates a network of genes related to actin/migration [38]. |
| RHOA | Rho Family Protein A | −1.38 | Contributes to cell cycle progression, adhesion, and microtubule and actin cytoskeleton organization [37]. |
| HRAS | HRas Proto-Oncogene, GTPase | −1.34 | Regulates adhesion, cytoskeleton rearrangements, proliferation, cell survival, and cell motility [44]. |
| MAPK3 | Mitogen-Activated Protein Kinase 3 | −1.22 | Regulates inflammation, proliferation, apoptosis, and differentiation [45]. |
| FOXO1 | Forkhead Box 1 | −1.21 | Tumor suppressor that induces cell cycle arrest and apoptosis [36]. |
| RASA1 | Ras P21 Protein Activator 1 | −1.19 | Provides instruction for protein p120-RasGap and helps regulate Ras-MAPK [35]. |
| PDK2 | Pyruvate Dehydrogenase Kinase 2 | −1.14 | Associated with proliferation, anti-apoptosis, tumor aggressiveness, and therapy resistance [42]. |
| Node 1 | Node 2 | Node l Accession | Node 2 Accession | Score |
|---|---|---|---|---|
| RHOA | PDK2 | ENSPO0000400175 | ENSP00000420927 | 0.184 |
| RHOA | IRS1 | ENSPO0000400175 | ENSP0000030489S | 0.386 |
| RHOA | FOXO1 | ENSP00000400175 | ENSP00000368880 | 0.364 |
| RHOA | EIF4B | ENSP00000400175 | ENSP00000388806 | 0.324 |
| PDK2 | RHOA | ENSP00000420927 | ENSPO0000400175 | 0.184 |
| PDK2 | IRS1 | ENSPO0000420927 | ENSPO0000304895 | 0.436 |
| PDK2 | FOXO1 | ENSP00000420927 | ENSP00000368880 | 0.413 |
| PDK2 | EIF4B | ENSP00000420927 | ENSP00000388806 | 0.179 |
| IRS1 | RHOA | ENSP00000304895 | ENSPO0000400175 | 0.386 |
| IRS1 | PDK2 | ENSP00000304895 | ENSPO0000420927 | 0.436 |
| IRS1 | FOXO1 | ENSP00000304895 | ENSP00000368880 | 0.854 |
| IRS1 | EIF4B | ENSPO0000304895 | ENSP00000388806 | 0.357 |
| FOXO1 | RHOA | ENSP00000368880 | ENSPO0000400175 | 0.364 |
| FOXO1 | PDK2 | ENSP00000368880 | ENSP00000420927 | 0.413 |
| FOXO1 | IRS1 | ENSP00000368880 | ENSP00000304895 | 0.854 |
| FOXO1 | EIF4B | ENSP00000368880 | ENSPO0000388806 | 0.244 |
| EIF4B | RHOA | ENSP00000388806 | ENSP00000400175 | 0.324 |
| EIF4B | PDK2 | ENSPO0000388806 | ENSPO0000420927 | 0.179 |
| EIF4B | IRS1 | ENSPO0000388806 | ENSPO0000304895 | 0.357 |
| EIF4B | FOXO1 | ENSPO0000388806 | ENSPO0000368880 | 0.244 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, S.A.; Mendonca, P.; Messeha, S.S.; Oriaku, E.T.; Soliman, K.F.A. The Anticancer Effects of Marine Carotenoid Fucoxanthin through Phosphatidylinositol 3-Kinase (PI3K)-AKT Signaling on Triple-Negative Breast Cancer Cells. Molecules 2024, 29, 61. https://doi.org/10.3390/molecules29010061
Ahmed SA, Mendonca P, Messeha SS, Oriaku ET, Soliman KFA. The Anticancer Effects of Marine Carotenoid Fucoxanthin through Phosphatidylinositol 3-Kinase (PI3K)-AKT Signaling on Triple-Negative Breast Cancer Cells. Molecules. 2024; 29(1):61. https://doi.org/10.3390/molecules29010061
Chicago/Turabian StyleAhmed, Shade’ A., Patricia Mendonca, Samia S. Messeha, Ebenezer T. Oriaku, and Karam F. A. Soliman. 2024. "The Anticancer Effects of Marine Carotenoid Fucoxanthin through Phosphatidylinositol 3-Kinase (PI3K)-AKT Signaling on Triple-Negative Breast Cancer Cells" Molecules 29, no. 1: 61. https://doi.org/10.3390/molecules29010061
APA StyleAhmed, S. A., Mendonca, P., Messeha, S. S., Oriaku, E. T., & Soliman, K. F. A. (2024). The Anticancer Effects of Marine Carotenoid Fucoxanthin through Phosphatidylinositol 3-Kinase (PI3K)-AKT Signaling on Triple-Negative Breast Cancer Cells. Molecules, 29(1), 61. https://doi.org/10.3390/molecules29010061