The Mechanisms of Molybdate Distribution and Homeostasis with Special Focus on the Model Plant Arabidopsis thaliana
Abstract
:1. Introduction
2. The Importance of Molybdenum for Plants

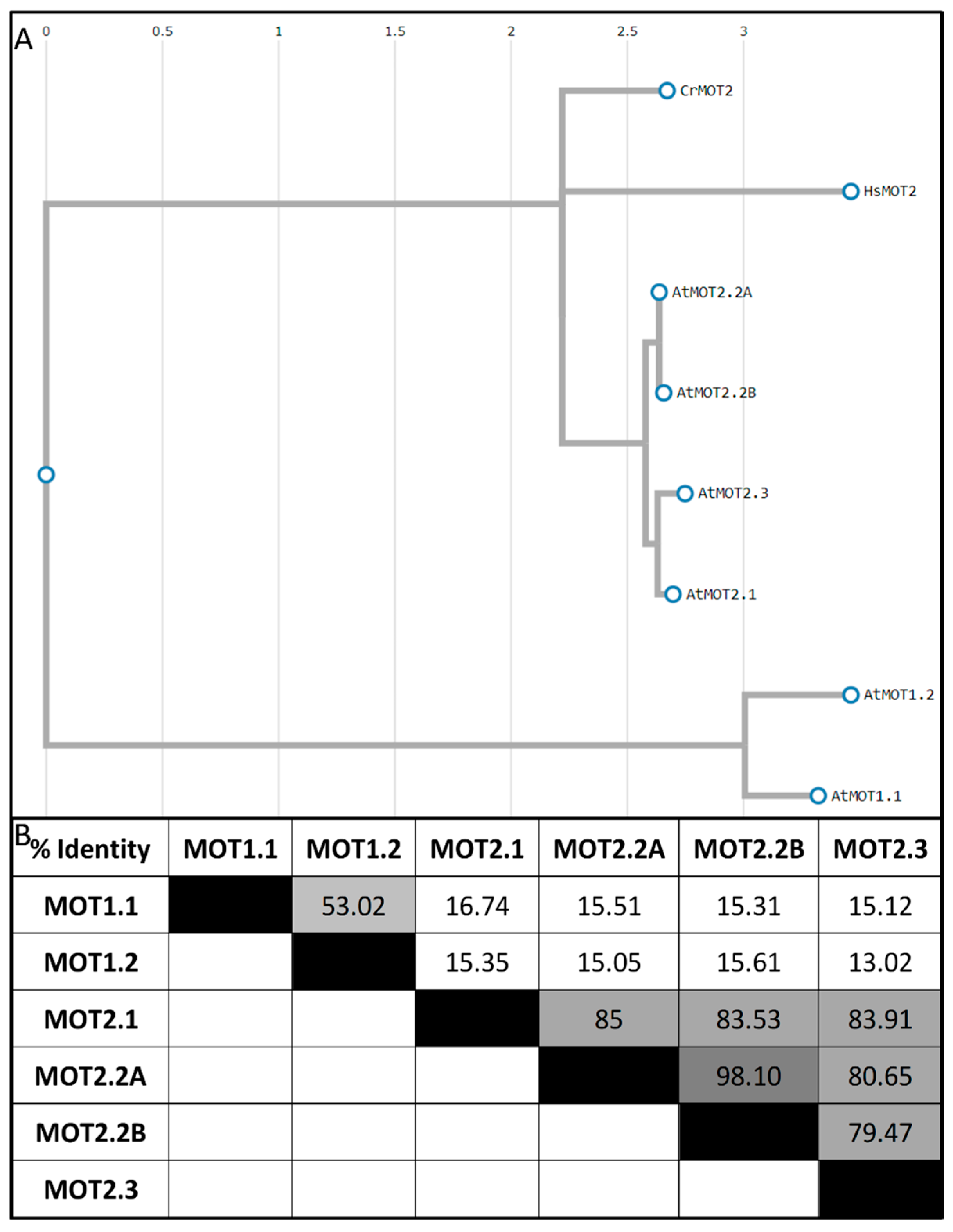
3. Molybdate Transport by Specialized Membrane Transporters
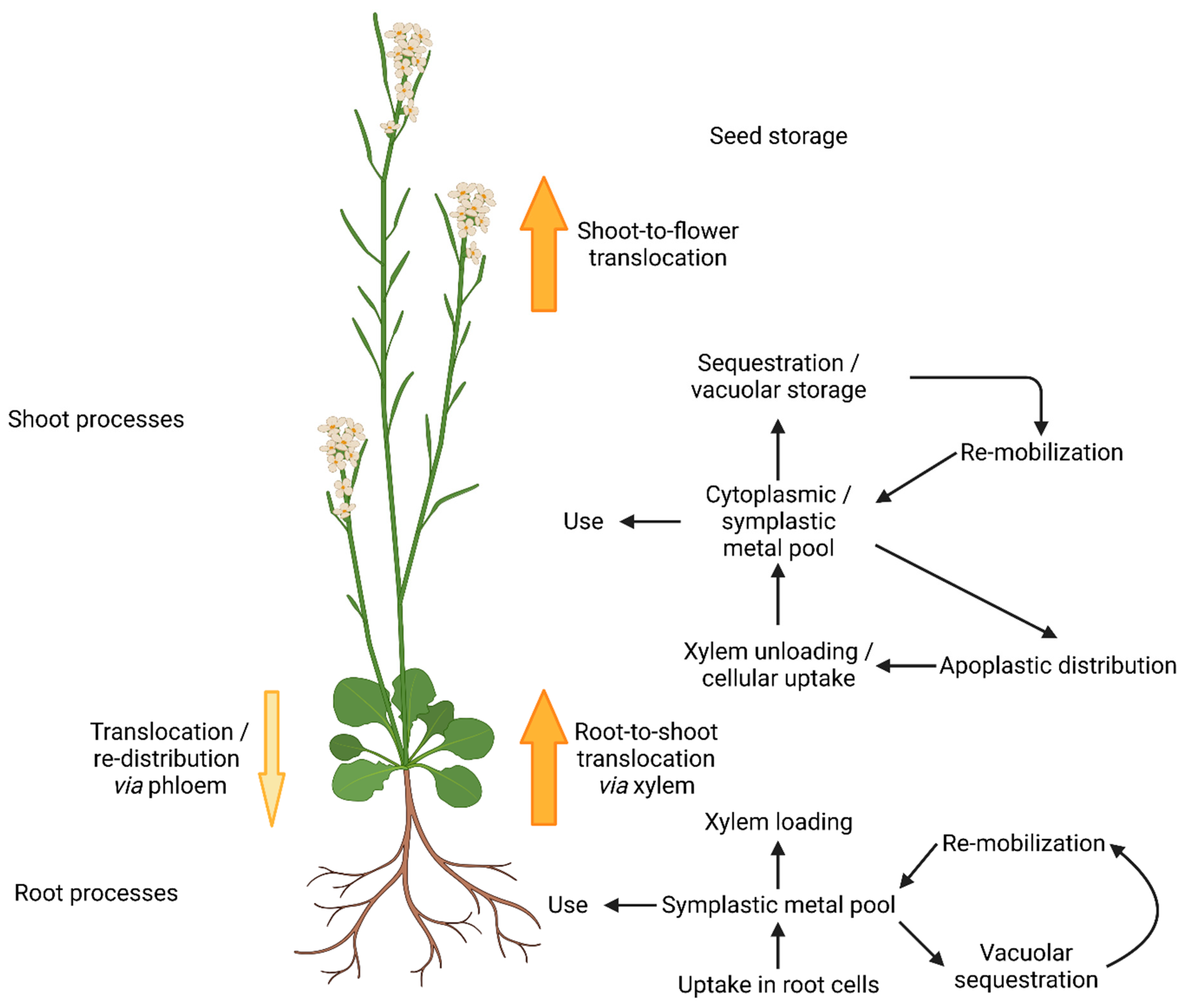
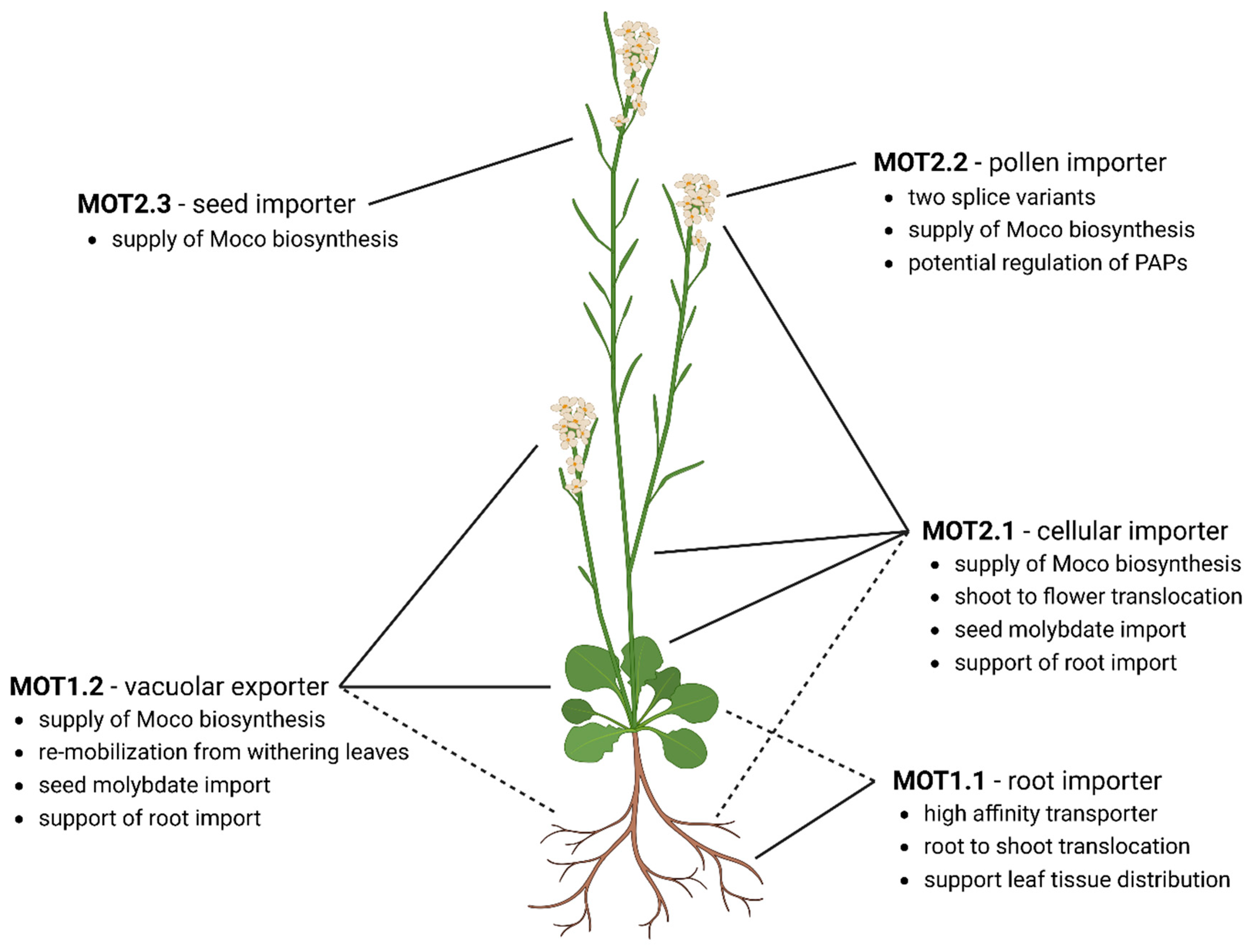
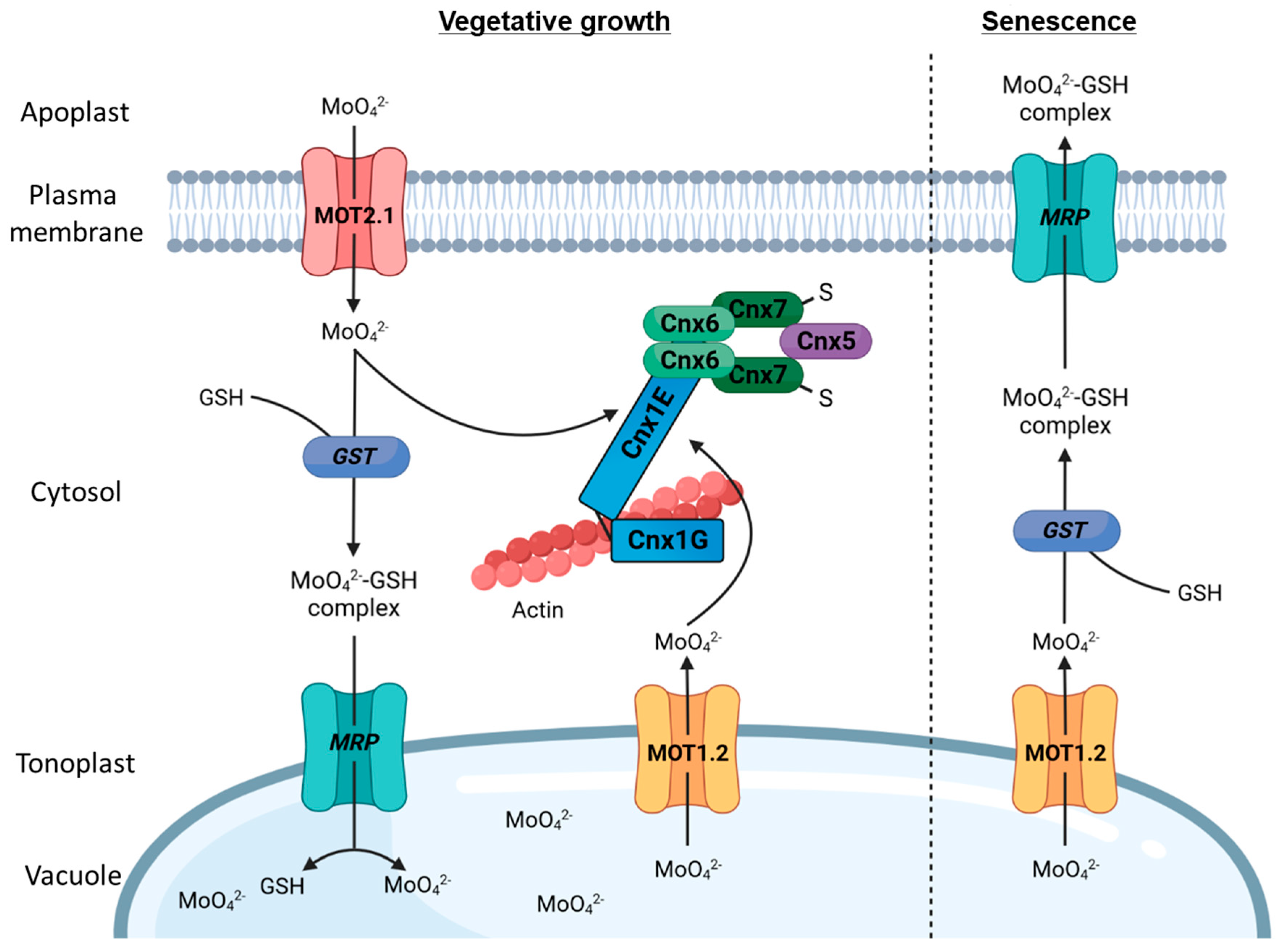
4. Following Molybdate along Its Way through A. thaliana
5. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Angulo-Bejarano, P.I.; Puente-Rivera, J.; Cruz-Ortega, R. Metal and Metalloid Toxicity in Plants: An Overview on Molecular Aspects. Plants 2021, 10, 635. [Google Scholar] [CrossRef] [PubMed]
- Assunção, A.G.L.; Cakmak, I.; Clemens, S.; González-Guerrero, M.; Nawrocki, A.; Thomine, S. Micronutrient Homeostasis in Plants for More Sustainable Agriculture and Healthier Human Nutrition. J. Exp. Bot. 2022, 73, 1789–1799. [Google Scholar] [CrossRef] [PubMed]
- Colangelo, E.P.; Guerinot, M. Lou Put the Metal to the Petal: Metal Uptake and Transport throughout Plants. Curr. Opin. Plant Biol. 2006, 9, 322–330. [Google Scholar] [CrossRef]
- Krämer, U.; Talke, I.N.; Hanikenne, M. Transition Metal Transport. FEBS Lett. 2007, 581, 2263–2272. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S.; Palmgren, M.G.; Krämer, U. A Long Way Ahead: Understanding and Engineering Plant Metal Accumulation. Trends Plant Sci. 2002, 7, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Andresen, E.; Peiter, E.; Küpper, H. Trace Metal Metabolism in Plants. J. Exp. Bot. 2018, 69, 909–954. [Google Scholar] [CrossRef] [PubMed]
- Küpper, H.; Andresen, E. Mechanisms of Metal Toxicity in Plants. Metallomics 2016, 8, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Sitsel, O.; Meloni, G.; Autzen, H.E.; Andersson, M.; Klymchuk, T.; Nielsen, A.M.; Rees, D.C.; Nissen, P.; Gourdon, P. Structure and Mechanism of Zn2+-Transporting P-Type ATPases. Nature 2014, 514, 518–522. [Google Scholar] [CrossRef]
- Murgia, I.; Marzorati, F.; Vigani, G.; Morandini, P. Plant Iron Nutrition: The Long Road from Soil to Seeds. J. Exp. Bot. 2022, 73, 1809–1824. [Google Scholar] [CrossRef]
- Clemens, S. Metal Ligands in Micronutrient Acquisition and Homeostasis. Plant Cell Environ. 2019, 42, 2902–2912. [Google Scholar] [CrossRef]
- Chen, Y.T.; Wang, Y.; Yeh, K.C. Role of Root Exudates in Metal Acquisition and Tolerance. Curr. Opin. Plant Biol. 2017, 39, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, D.; Sun, W.; Wang, T. The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis. Int. J. Mol. Sci. 2019, 20, 2424. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S. The Elements of Plant Micronutrients. Plant Physiol. 2010, 154, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Smedley, P.L.; Kinniburgh, D.G. Molybdenum in Natural Waters: A Review of Occurrence, Distributions and Controls. Appl. Geochem. 2017, 84, 387–432. [Google Scholar] [CrossRef]
- Frascoli, F.; Hudson-Edwards, K.A. Geochemistry, Mineralogy and Microbiology of Molybdenum in Mining-Affected Environments. Minerals 2018, 8, 42. [Google Scholar] [CrossRef]
- Tomatsu, H.; Takano, J.; Takahashi, H.; Watanabe-Takahashi, A.; Shibagaki, N.; Fujiwara, T. An Arabidopsis thaliana High-Affinity Molybdate Transporter Required for Efficient Uptake of Molybdate from Soil. Proc. Natl. Acad. Sci. USA 2007, 104, 18807–18812. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, B.N.; Gridley, K.L.; Brady, J.N.; Phillips, T.; Tyerman, S.D. The Role of Molybdenum in Agricultural Plant Production. Ann. Bot. 2005, 96, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Mendel, R.R. The Molybdenum Cofactor. J. Biol. Chem. 2013, 288, 13165–13172. [Google Scholar] [CrossRef]
- Mayr, S.J.; Mendel, R.R.; Schwarz, G. Molybdenum Cofactor Biology, Evolution and Deficiency. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118883. [Google Scholar] [CrossRef]
- Mendel, R.R.; Schwarz, G. Molybdenum Cofactor Biosynthesis in Plants and Humans. Coord. Chem. Rev. 2011, 255, 1145–1158. [Google Scholar] [CrossRef]
- Teschner, J.; Lachmann, N.; Schulze, J.; Geisler, M.; Selbach, K.; Santamaria-Araujo, J.; Balk, J.; Mendel, R.R.; Bittner, F. A Novel Role for Arabidopsis Mitochondrial ABC Transporter ATM3 in Molybdenum Cofactor Biosynthesis. Plant Cell 2010, 22, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Mendel, R.R.; Leimkühler, S. The Biosynthesis of the Molybdenum Cofactors. J. Biol. Inorg. Chem. 2015, 20, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Kuper, J.; Winking, J.; Hecht, H.J.; Mendel, R.R.; Schwarz, G. The Active Site of the Molybdenum Cofactor Biosynthetic Protein Domain Cnx1G. Arch. Biochem. Biophys. 2003, 411, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Krausze, J.; Hercher, T.W.; Zwerschke, D.; Kirk, M.L.; Blankenfeldt, W.; Mendel, R.R.; Kruse, T. The Functional Principle of Eukaryotic Molybdenum Insertases. Biochem. J. 2018, 475, 1739–1753. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.N.; Minner-Meinen, R.; Behnecke, M.; Biedendieck, R.; Hänsch, V.G.; Hercher, T.W.; Hertweck, C.; van den Hout, L.; Knüppel, L.; Sivov, S.; et al. Moonlighting Arabidopsis Molybdate Transporter 2 Family and GSH-Complex Formation Facilitate Molybdenum Homeostasis. Commun. Biol. 2023, 6, 801. [Google Scholar] [CrossRef] [PubMed]
- Kaufholdt, D.; Gehl, C.; Geisler, M.; Jeske, O.; Voedisch, S.; Ratke, C.; Bollhöner, B.; Mendel, R.R.; Hänsch, R. Visualization and Quantification of Protein Interactions in the Biosynthetic Pathway of Molybdenum Cofactor in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Kaufholdt, D.; Baillie, C.K.; Bikker, R.; Burkart, V.; Dudek, C.A.; von Pein, L.; Rothkegel, M.; Mendel, R.R.; Hänsch, R. The Molybdenum Cofactor Biosynthesis Complex Interacts with Actin Filaments via Molybdenum Insertase Cnx1 as Anchor Protein in Arabidopsis thaliana. Plant Sci. 2016, 244, 8–18. [Google Scholar] [CrossRef]
- Kaufholdt, D.; Baillie, C.K.; Meyer, M.H.; Schwich, O.D.; Timmerer, U.L.; Tobias, L.; van Thiel, D.; Hänsch, R.; Mendel, R.R. Identification of a Protein-Protein Interaction Network Downstream of Molybdenum Cofactor Biosynthesis in Arabidopsis thaliana. J. Plant Physiol. 2016, 207, 42–50. [Google Scholar] [CrossRef]
- Lehrke, M.; Rump, S.; Heidenreich, T.; Wissing, J.; Mendel, R.R.; Bittner, F. Identification of Persulfide-Binding and Disulfide-Forming Cysteine Residues in the NifS-like Domain of the Molybdenum Cofactor Sulfurase ABA3 by Cysteine-Scanning Mutagenesis. Biochem. J. 2012, 441, 823–832. [Google Scholar] [CrossRef]
- Kaufholdt, D.; Baillie, C.K.; Meinen, R.; Mendel, R.R.; Hänsch, R. The Molybdenum Cofactor Biosynthesis Network: In Vivo Protein-Protein Interactions of an Actin Associated Multi-Protein Complex. Front. Plant Sci. 2017, 8, 1946. [Google Scholar] [CrossRef]
- Mendel, R.R. The History of the Molybdenum Cofactor—A Personal View. Molecules 2022, 27, 4934. [Google Scholar] [CrossRef]
- Magalon, A.; Mendel, R.R. Biosynthesis and Insertion of the Molybdenum Cofactor. EcoSal Plus 2015, 6, 10-1128. [Google Scholar] [CrossRef]
- Mendel, R.R.; Hänsch, R. Molybdoenzymes and Molybdenum Cofactor in Plants. J. Exp. Bot. 2002, 53, 1689–1698. [Google Scholar] [CrossRef]
- Sano, N.; Marion-Poll, A. Aba Metabolism and Homeostasis in Seed Dormancy and Germination. Int. J. Mol. Sci. 2021, 22, 5069. [Google Scholar] [CrossRef]
- Soltabayeva, A.; Srivastava, S.; Kurmanbayeva, A.; Bekturova, A.; Fluhr, R.; Sagi, M. Early Senescence in Older Leaves of Low Nitrate-Grown Atxdh1 Uncovers a Role for Purine Catabolism in n Supply. Plant Physiol. 2018, 178, 1027–1044. [Google Scholar] [CrossRef]
- Yesbergenova, Z.; Yang, G.; Oron, E.; Soffer, D.; Fluhr, R.; Sagi, M. The Plant Mo-Hydroxylases Aldehyde Oxidase and Xanthine Dehydrogenase Have Distinct Reactive Oxygen Species Signatures and Are Induced by Drought and Abscisic Acid. Plant J. 2005, 42, 862–876. [Google Scholar] [CrossRef]
- Ma, X.; Wang, W.; Bittner, F.; Schmidt, N.; Berkey, R.; Zhang, L.; King, H.; Zhang, Y.; Feng, J.; Wen, Y.; et al. Dual and Opposing Roles of Xanthine Dehydrogenase in Defense-Associated Reactive Oxygen Species Metabolism in Arabidopsis. Plant Cell 2016, 28, 1108–1126. [Google Scholar] [CrossRef]
- Krompholz, N.; Krischkowski, C.; Reichmann, D.; Garbe-Schönberg, D.; Mendel, R.R.; Bittner, F.; Clement, B.; Havemeyer, A. The Mitochondrial Amidoxime Reducing Component (MARC) Is Involved in Detoxification of N-Hydroxylated Base Analogues. Chem. Res. Toxicol. 2012, 25, 2443–2450. [Google Scholar] [CrossRef]
- Maiber, L.; Koprivova, A.; Bender, D.; Kopriva, S.; Fischer-Schrader, K. Characterization of the Amidoxime Reducing Components ARC1 and ARC2 from Arabidopsis thaliana. FEBS J. 2022, 289, 5656–5669. [Google Scholar] [CrossRef]
- Chamizo-Ampudia, A.; Galvan, A.; Fernandez, E.; Llamas, A. The Chlamydomonas reinhardtii Molybdenum Cofactor Enzyme CrARC Has a Zn-Dependent Activity and Protein Partners Similar to Those of Its Human Homologue. Eukaryot. Cell 2011, 10, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, Á.; Ocaña-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galván, A.; Fernández, E. A Dual System Formed by the ARC and NR Molybdoenzymes Mediates Nitrite-Dependent NO Production in Chlamydomonas. Plant Cell Environ. 2016, 39, 2097–2107. [Google Scholar] [CrossRef]
- Mohn, M.A.; Thaqi, B.; Fischer-Schrader, K. Isoform-Specific NO Synthesis by Arabidopsis thaliana Nitrate Reductase. Plants 2019, 8, 67. [Google Scholar] [CrossRef]
- Campbell, W.H. Nitrate Reductase Structure, Function and Regulation: Bridging the Gap between Biochemistry and Physiology. Annu. Rev. Plant Biol. 1999, 50, 277–303. [Google Scholar] [CrossRef]
- Nowak, K.; Luniak, N.; Witt, C.; Wüstefeld, Y.; Wachter, A.; Mendel, R.R.; Hänsch, R. Peroxisomal Localization of Sulfite Oxidase Separates It from Chloroplast-Based Sulfur Assimilation. Plant Cell Physiol. 2004, 45, 1889–1894. [Google Scholar] [CrossRef]
- Baillie, C.K.; Kaufholdt, D.; Karpinski, L.H.; Schreiber, V.; Hänsch, S.; Evers, C.; Bloem, E.; Schnug, E.; Kreuzwieser, J.; Herschbach, C.; et al. Detoxification of Volcanic Sulfur Surplus in Planta: Three Different Strategies of Survival. Environ. Exp. Bot. 2016, 126, 44–54. [Google Scholar] [CrossRef]
- Weber, J.N.; Kaufholdt, D.; Minner-Meinen, R.; Bloem, E.; Shahid, A.; Rennenberg, H.; Hänsch, R. Impact of Wildfires on SO2 Detoxification Mechanisms in Leaves of Oak and Beech Trees. Environ. Pollut. 2021, 272, 116389. [Google Scholar] [CrossRef]
- Hu, Y.; Ribbe, M.W. Biosynthesis of the Iron-Molybdenum Cofactor of Nitrogenase. J. Biol. Chem. 2013, 288, 13173–13177. [Google Scholar] [CrossRef]
- Bellenger, J.P.; Wichard, T.; Kustka, A.B.; Kraepiel, A.M.L. Uptake of Molybdenum and Vanadium by Nitrogen-Fixing Soil Bacterium Using Siderophores. Nat. Geosci. 2008, 1, 243–246. [Google Scholar] [CrossRef]
- Lindström, K.; Mousavi, S.A. Effectiveness of Nitrogen Fixation in Rhizobia. Microb. Biotechnol. 2020, 13, 1314–1335. [Google Scholar] [CrossRef] [PubMed]
- Haque, N.; Peralta-Videa, J.R.; Jones, G.L.; Gill, T.E.; Gardea-Torresdey, J.L. Screening the Phytoremediation Potential of Desert Broom (Baccharis sarothroides Gray) Growing on Mine Tailings in Arizona, USA. Environ. Pollut. 2008, 153, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Mendel, R.R.; Bittner, F. Cell Biology of Molybdenum. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 621–635. [Google Scholar] [CrossRef]
- Schwarz, G.; Mendel, R.R.; Ribbe, M.W. Molybdenum Cofactors, Enzymes and Pathways. Nature 2009, 460, 839–847. [Google Scholar] [CrossRef]
- Wagner, U.G.; Stupperich, E.; Kratky, C. Structure of the Molybdate/Tungstate Binding Protein Mop from Sporomusa Ovata. Structure 2000, 8, 1127–1136. [Google Scholar] [CrossRef]
- Makdessi, K.; Fritsche, K.; Pich, A.; Andreesen, J.R. Identification and Characterization of the Cytoplasmic Tungstate/Molybdate- Binding Protein (Mop) from Eubacterium acidaminophilum. Arch. Microbiol. 2004, 181, 45–51. [Google Scholar] [CrossRef]
- Demtröder, L.; Narberhaus, F.; Masepohl, B. Coordinated Regulation of Nitrogen Fixation and Molybdate Transport by Molybdenum. Mol. Microbiol. 2019, 111, 17–30. [Google Scholar] [CrossRef]
- Kraepiel, A.M.L.; Bellenger, J.P.; Wichard, T.; Morel, F.M.M. Multiple Roles of Siderophores in Free-Living Nitrogen-Fixing Bacteria. BioMetals 2009, 22, 573–581. [Google Scholar] [CrossRef]
- Thomas, W.; Bellenger, J.P.; Morel, F.M.M.; Kraepiel, A.M.L. Role of the Siderophore Azotobactin in the Bacterial Acquisition of Nitrogenase Metal Cofactors. Environ. Sci. Technol. 2009, 43, 7218–7224. [Google Scholar] [CrossRef]
- Duhme-Klair, A.K. From Siderophores and Self-Assembly to Luminescent Sensors: The Binding of Molybdenum by Catecholamides. Eur. J. Inorg. Chem. 2009, 2009, 3689–3701. [Google Scholar] [CrossRef]
- Schalk, I.J.; Hannauer, M.; Braud, A. New Roles for Bacterial Siderophores in Metal Transport and Tolerance. Environ. Microbiol. 2011, 13, 2844–2854. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, T.C.; Nolan, E.M. Beyond Iron: Non-Classical Biological Functions of Bacterial Siderophores. Dalton Trans. 2015, 44, 6320–6339. [Google Scholar] [CrossRef] [PubMed]
- Vatansever, R.; Filiz, E.; Ozyigit, I.I. In Silico Identification and Comparative Analysis of Molybdenum (Mo) Transporter Genes in Plants. Rev. Bras. Bot. 2016, 39, 87–99. [Google Scholar] [CrossRef]
- Gil-Díez, P.; Tejada-Jiménez, M.; León-Mediavilla, J.; Wen, J.; Mysore, K.S.; Imperial, J.; González-Guerrero, M. MtMOT1.2 Is Responsible for Molybdate Supply to Medicago Truncatula Nodules. Plant Cell Environ. 2019, 42, 310–320. [Google Scholar] [CrossRef]
- Tejada-Jiménez, M.; Gil-Díez, P.; León-Mediavilla, J.; Wen, J.; Mysore, K.S.; Imperial, J.; González-Guerrero, M. Medicago Truncatula Molybdate Transporter Type 1 (MtMOT1.3) Is a Plasma Membrane Molybdenum Transporter Required for Nitrogenase Activity in Root Nodules under Molybdenum Deficiency. New Phytol. 2017, 216, 1223–1235. [Google Scholar] [CrossRef]
- Gao, J.S.; Wu, F.F.; Shen, Z.L.; Meng, Y.; Cai, Y.P.; Lin, Y. A Putative Molybdate Transporter LjMOT1 Is Required for Molybdenum Transport in Lotus Japonicus. Physiol. Plant. 2016, 158, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Minner-Meinen, R.; Weber, J.N.; Kistner, S.; Meyfarth, P.; Saudhof, M.; van den Hout, L.; Schulze, J.; Mendel, R.R.; Hänsch, R.; Kaufholdt, D. Physiological Importance of Molybdate Transporter Family 1 in Feeding the Molybdenum Cofactor Biosynthesis Pathway in Arabidopsis thaliana. Molecules 2022, 27, 3158. [Google Scholar] [CrossRef] [PubMed]
- Tejada-Jiménez, M.; Galván, A.; Fernández, E. Algae and Humans Share a Molybdate Transporter. Proc. Natl. Acad. Sci. USA 2011, 108, 6420–6425. [Google Scholar] [CrossRef] [PubMed]
- Leves, F.P.; Tierney, M.L.; Howitt, S.M. Polar Residues in a Conserved Motif Spanning Helices 1 and 2 Are Functionally Important in the SulP Transporter Family. Int. J. Biochem. Cell Biol. 2008, 40, 2596–2605. [Google Scholar] [CrossRef] [PubMed]
- Shibagaki, N.; Grossman, A.R. The Role of the STAS Domain in the Function and Biogenesis of a Sulfate Transporter as Probed by Random Mutagenesis. J. Biol. Chem. 2006, 281, 22964–22973. [Google Scholar] [CrossRef]
- Baxter, I.; Muthukumar, B.; Hyeong, C.P.; Buchner, P.; Lahner, B.; Danku, J.; Zhao, K.; Lee, J.; Hawkesford, M.J.; Guerinot, M.L.; et al. Variation in Molybdenum Content across Broadly Distributed Populations of Arabidopsis thaliana is Controlled by a Mitochondrial Molybdenum Transporter (MOT1). PLoS Genet. 2008, 4, e1000004. [Google Scholar] [CrossRef]
- Tejada-Jiménez, M.; Llamas, Á.; Sanz-Luque, E.; Galván, A.; Fernández, E. A High-Affinity Molybdate Transporter in Eukaryotes. Proc. Natl. Acad. Sci. USA 2007, 104, 20126–20130. [Google Scholar] [CrossRef]
- Gasber, A.; Klaumann, S.; Trentmann, O.; Trampczynska, A.; Clemens, S.; Schneider, S.; Sauer, N.; Feifer, I.; Bittner, F.; Mendel, R.R.; et al. Identification of an Arabidopsis Solute Carrier Critical for Intracellular Transport and Inter-Organ Allocation of Molybdate. Plant Biol. 2011, 13, 710–718. [Google Scholar] [CrossRef]
- Temple, H.; Phyo, P.; Yang, W.; Lyczakowski, J.J.; Echevarría-Poza, A.; Yakunin, I.; Parra-Rojas, J.P.; Terrett, O.M.; Saez-Aguayo, S.; Dupree, R.; et al. Discovery of Putative Golgi S-Adenosyl Methionine Transporters Reveals the Importance of Plant Cell Wall Polysaccharide Methylation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Yadav, S.K. Heavy Metals Toxicity in Plants: An Overview on the Role of Glutathione and Phytochelatins in Heavy Metal Stress Tolerance of Plants. S. Afr. J. Bot. 2010, 76, 167–179. [Google Scholar] [CrossRef]
- Edwards, R.; Dixon, D.P.; Walbot, V. Plant Glutathione S-Transferases: Enzymes with Multiple Functions in Sickness and in Health. Trends Plant Sci. 2000, 5, 193–198. [Google Scholar] [CrossRef]
- Klein, M.; Burla, B.; Martinoia, E. The Multidrug Resistance-Associated Protein (MRP/ABCC) Subfamily of ATP-Binding Cassette Transporters in Plants. FEBS Lett. 2006, 580, 1112–1122. [Google Scholar] [CrossRef]
- Kosakivska, I.V.; Babenko, L.M.; Romanenko, K.O.; Korotka, I.Y.; Potters, G. Molecular Mechanisms of Plant Adaptive Responses to Heavy Metals Stress. Cell Biol. Int. 2021, 45, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Kutrowska, A.; Szelag, M. Low-Molecular Weight Organic Acids and Peptides Involved in the Long-Distance Transport of Trace Metals. Acta Physiol. Plant. 2014, 36, 1957–1968. [Google Scholar] [CrossRef]
- Kuang, R.; Chan, K.H.; Yeung, E.; Lim, B.L. Molecular and Biochemical Characterization of AtPAP15, a Purple Acid Phosphatase with Phytase Activity, in Arabidopsis. Plant Physiol. 2009, 151, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Frascaroli, E.; Tuberosa, R. Effect of Abscisic Acid on Pollen Germination and Tube Growth of Maize Genotypes. Plant Breed. 1993, 110, 250–254. [Google Scholar] [CrossRef]
- Zhao, S.; Wu, Y.; He, Y.; Wang, Y.; Xiao, J.; Li, L.; Wang, Y.; Chen, X.; Xiong, W.; Wu, Y. RopGEF2 Is Involved in ABA-Suppression of Seed Germination and Post-Germination Growth of Arabidopsis. Plant J. 2015, 84, 886–899. [Google Scholar] [CrossRef]
- Frey, A.; Godin, B.; Bonnet, M.; Sotta, B.; Marion-Poll, A. Maternal Synthesis of Abscisic Acid Controls Seed Development and Yield in Nicotiana Plumbaginifolia. Planta 2004, 218, 958–964. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weber, J.-N.; Minner-Meinen, R.; Kaufholdt, D. The Mechanisms of Molybdate Distribution and Homeostasis with Special Focus on the Model Plant Arabidopsis thaliana. Molecules 2024, 29, 40. https://doi.org/10.3390/molecules29010040
Weber J-N, Minner-Meinen R, Kaufholdt D. The Mechanisms of Molybdate Distribution and Homeostasis with Special Focus on the Model Plant Arabidopsis thaliana. Molecules. 2024; 29(1):40. https://doi.org/10.3390/molecules29010040
Chicago/Turabian StyleWeber, Jan-Niklas, Rieke Minner-Meinen, and David Kaufholdt. 2024. "The Mechanisms of Molybdate Distribution and Homeostasis with Special Focus on the Model Plant Arabidopsis thaliana" Molecules 29, no. 1: 40. https://doi.org/10.3390/molecules29010040
APA StyleWeber, J.-N., Minner-Meinen, R., & Kaufholdt, D. (2024). The Mechanisms of Molybdate Distribution and Homeostasis with Special Focus on the Model Plant Arabidopsis thaliana. Molecules, 29(1), 40. https://doi.org/10.3390/molecules29010040





