MolOptimizer: A Molecular Optimization Toolkit for Fragment-Based Drug Design
Abstract
1. Author Summary
2. Introduction
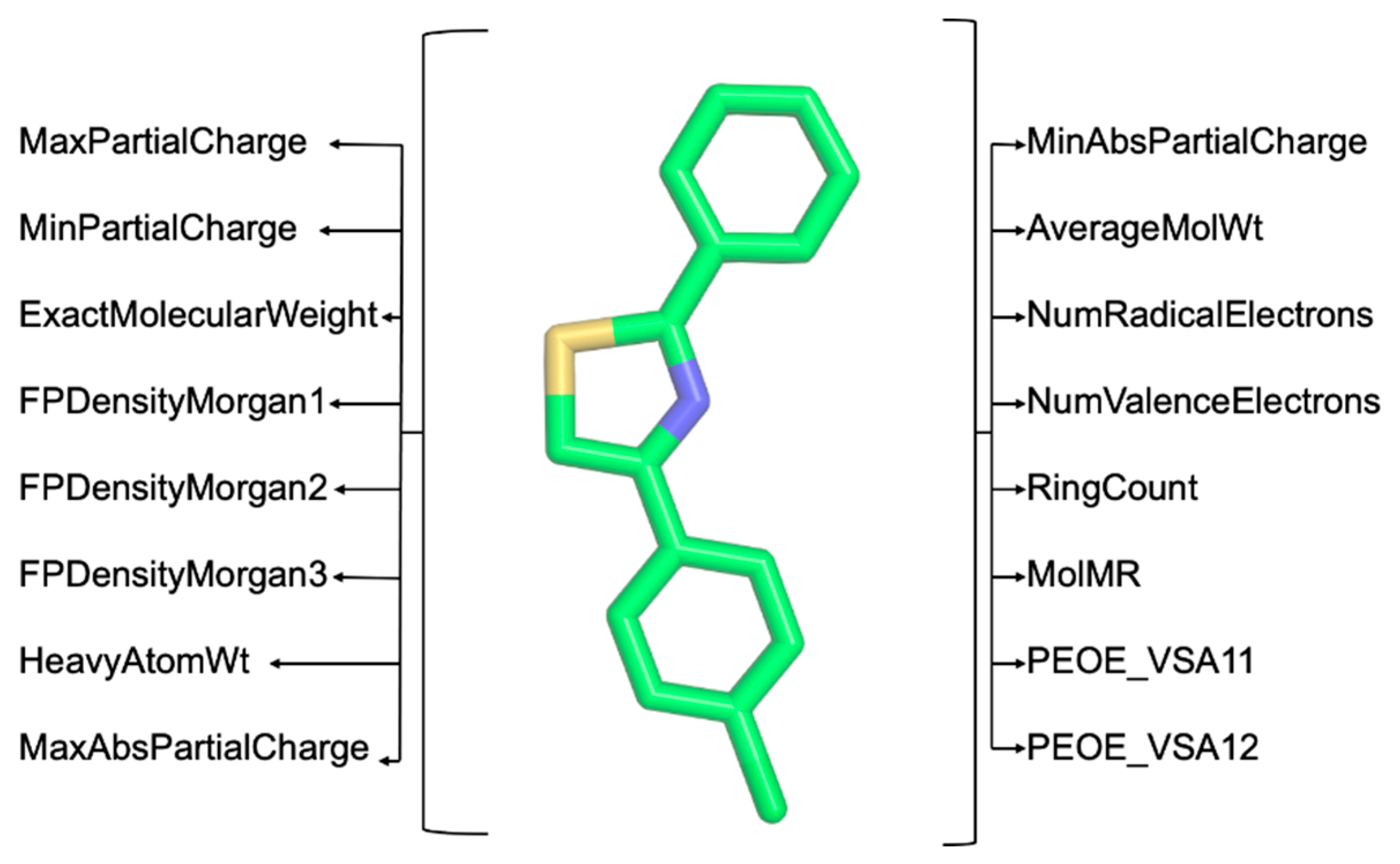
3. Implementation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leach, A.R.; Hann, M.M.; Burrows, J.N.; Griffen, E.J. Fragment screening: An introduction. Mol. Biosyst. 2006, 2, 429. [Google Scholar] [CrossRef] [PubMed]
- Siegal, G.; Ab, E.; Schultz, J. Integration of fragment screening and library design. Drug Discov. Today 2007, 12, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C. NMR methods in fragment screening: Theory and a comparison with other biophysical techniques. Drug Discov. Today 2009, 14, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Tam, B.; Akabayov, B. NMR-Fragment Based Virtual Screening: A Brief Overview. Molecules 2018, 23, 233. [Google Scholar] [CrossRef] [PubMed]
- Tam, B.; Sherf, D.; Cohen, S.; Eisdorfer, S.A.; Perez, M.; Soffer, A.; Vilenchik, D.; Akabayov, S.R.; Wagner, G.; Akabayov, B. Discovery of small-molecule inhibitors targeting the ribosomal peptidyl transferase center (PTC) of M. tuberculosis. Chem. Sci. 2019, 10, 8764–8767. [Google Scholar] [CrossRef] [PubMed]
- Grimberg, H.; Tiwari, V.S.; Tam, B.; Gur-Arie, L.; Gingold, D.; Polachek, L.; Akabayov, B. Machine learning approaches to optimize small-molecule inhibitors for RNA targeting. J. Cheminform. 2022, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Eyal, Z.; Matzov, D.; Krupkin, M.; Wekselman, I.; Paukner, S.; Zimmerman, E.; Rozenberg, H.; Bashan, A.; Yonath, A. Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2015, 112, E5805–E5814. [Google Scholar] [CrossRef] [PubMed]
- Goodsell, D.S.; Morris, G.M.; Olson, A.J. Automated docking of flexible ligands: Applications of AutoDock. J. Mol. Recognit. JMR 1996, 9, 1–5. [Google Scholar] [CrossRef]
- Moriwaki, H.; Tian, Y.S.; Kawashita, N.; Takagi, T. Mordred: A molecular descriptor calculator. J. Cheminform. 2018, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Basak, S.C.; Grunwald, G.D.; Niemi, G.J. Use of Graph-Theoretic and Geometrical Molecular Descriptors in Structure-Activity Relationships. In Chemical Topology to Three-Dimensional Geometry; Balaban, A.T., Ed.; Plenum Press: New York, NY, USA, 1997. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Muthukrishnan, R.; Rohini, R. LASSO: A feature selection technique in predictive modeling for machine learning. In Proceedings of the 2016 IEEE International Conference on Advances in Computer Applications (ICACA), Coimbatore, India, 24 October 2016; pp. 18–20. [Google Scholar]
- Loh, W.-Y. Classification and regression trees. WIREs Data Min. Knowl. Discov. 2011, 1, 14–23. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, O.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Lundberg, S.M.; Nair, B.; Vavilala, M.S.; Horibe, M.; Eisses, M.J.; Adams, T.; Liston, D.E.; Low, D.K.; Newman, S.F.; Kim, J.; et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2018, 2, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Farris, F.A. The Gini Index and Measures of Inequality. Am. Math. Mon. 2010, 117, 851–864. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soffer, A.; Viswas, S.J.; Alon, S.; Rozenberg, N.; Peled, A.; Piro, D.; Vilenchik, D.; Akabayov, B. MolOptimizer: A Molecular Optimization Toolkit for Fragment-Based Drug Design. Molecules 2024, 29, 276. https://doi.org/10.3390/molecules29010276
Soffer A, Viswas SJ, Alon S, Rozenberg N, Peled A, Piro D, Vilenchik D, Akabayov B. MolOptimizer: A Molecular Optimization Toolkit for Fragment-Based Drug Design. Molecules. 2024; 29(1):276. https://doi.org/10.3390/molecules29010276
Chicago/Turabian StyleSoffer, Adam, Samuel Joshua Viswas, Shahar Alon, Nofar Rozenberg, Amit Peled, Daniel Piro, Dan Vilenchik, and Barak Akabayov. 2024. "MolOptimizer: A Molecular Optimization Toolkit for Fragment-Based Drug Design" Molecules 29, no. 1: 276. https://doi.org/10.3390/molecules29010276
APA StyleSoffer, A., Viswas, S. J., Alon, S., Rozenberg, N., Peled, A., Piro, D., Vilenchik, D., & Akabayov, B. (2024). MolOptimizer: A Molecular Optimization Toolkit for Fragment-Based Drug Design. Molecules, 29(1), 276. https://doi.org/10.3390/molecules29010276






