Computational 3D Modeling-Based Identification of Inhibitors Targeting Cysteine Covalent Bond Catalysts for JAK3 and CYP3A4 Enzymes in the Treatment of Rheumatoid Arthritis
Abstract
1. Introduction
2. Results and Discussion
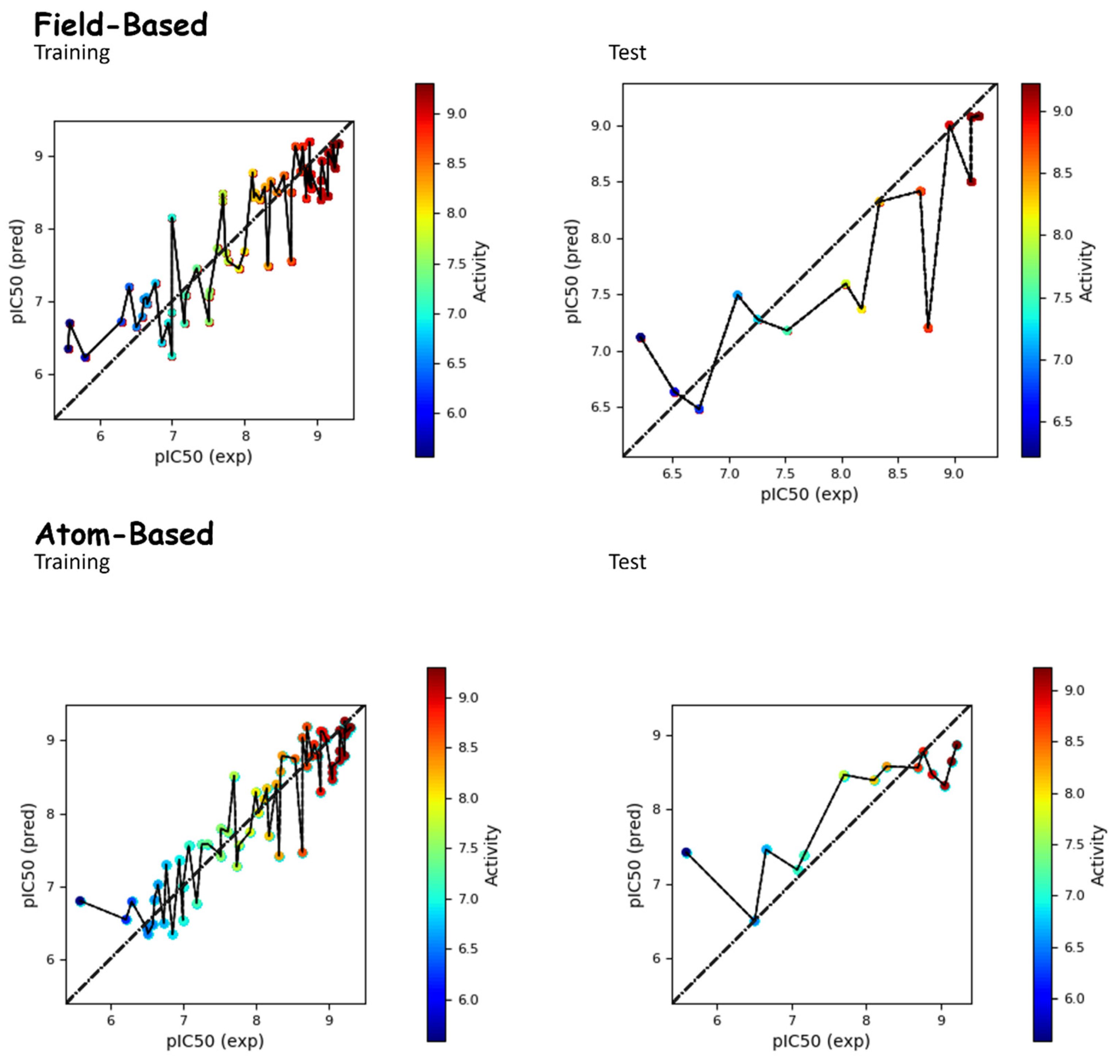
2.1. Static Results of QSAR Models
Static Analysis of Field-Based and Atom-Based Models
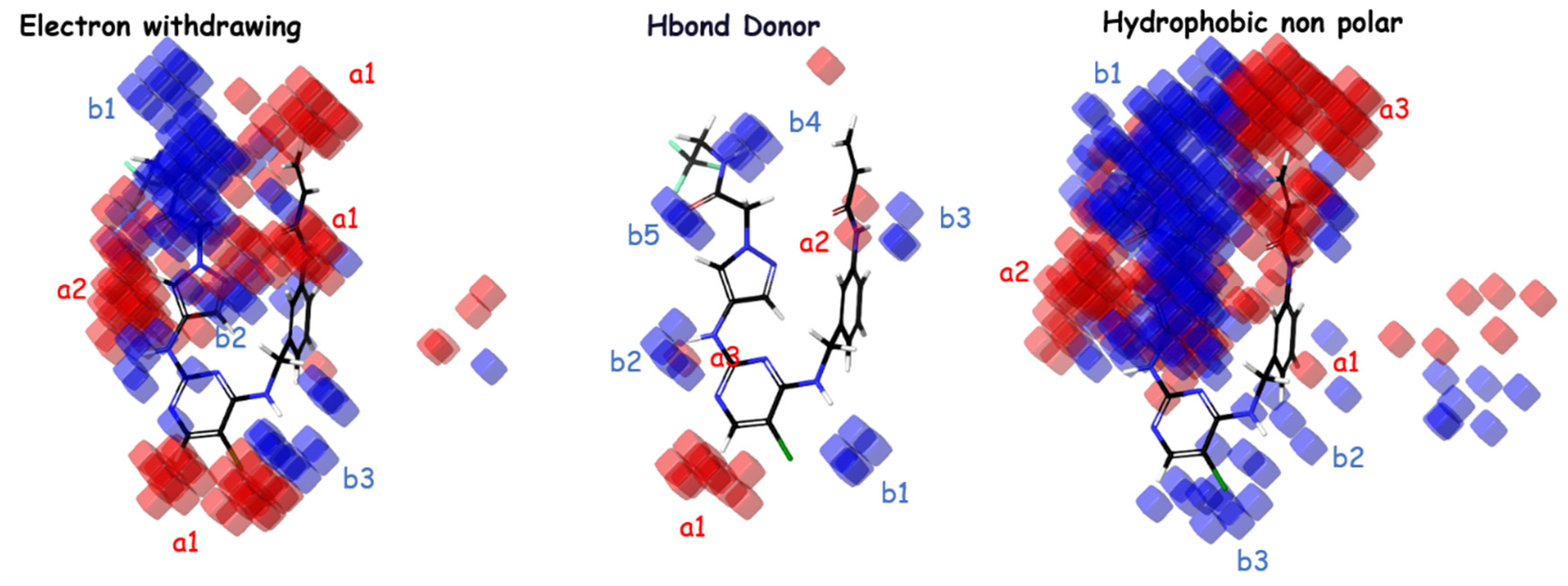
2.2. Contour Maps
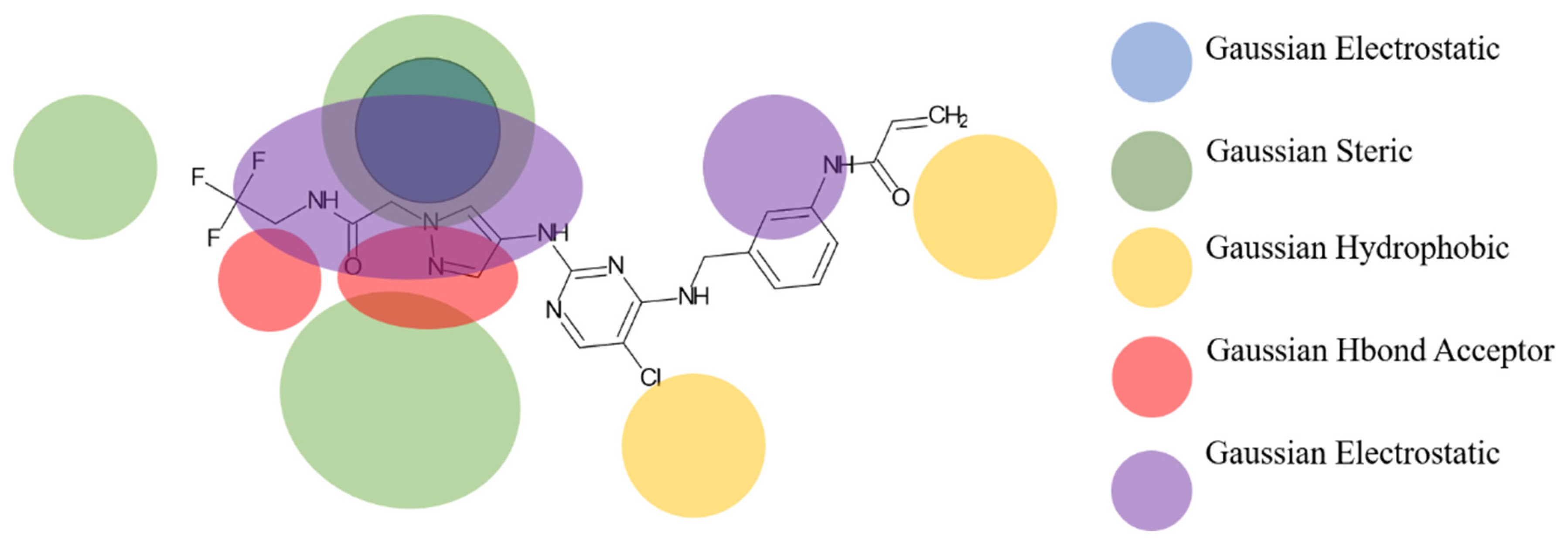
2.3. Design of New Molecules Based on QSAR Models
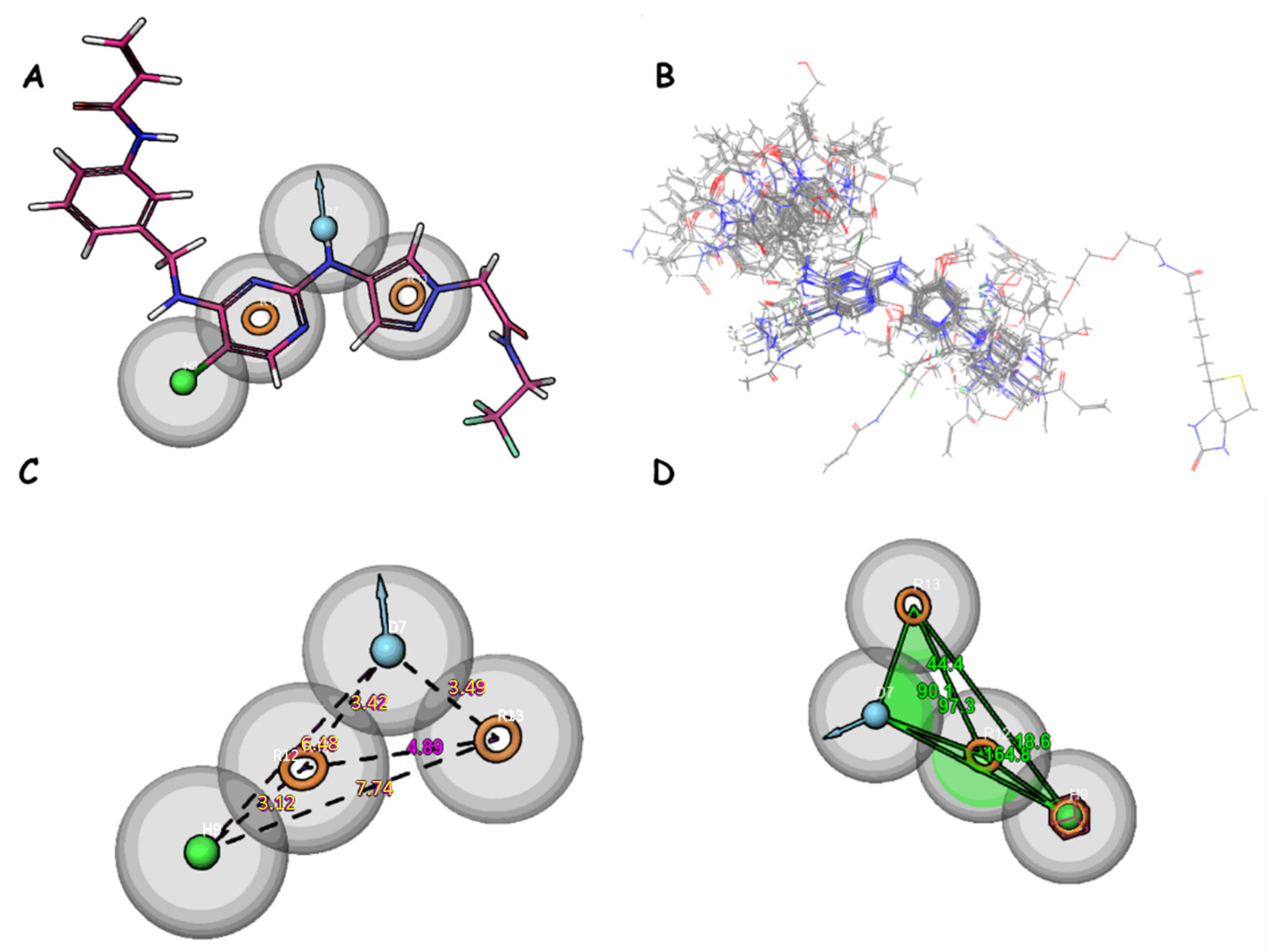
2.4. Pharmacophore Model Analysis
Validation of Pharmacophore Models
2.5. Identification of Compounds Using Pharmacophore Model DHRRR_1
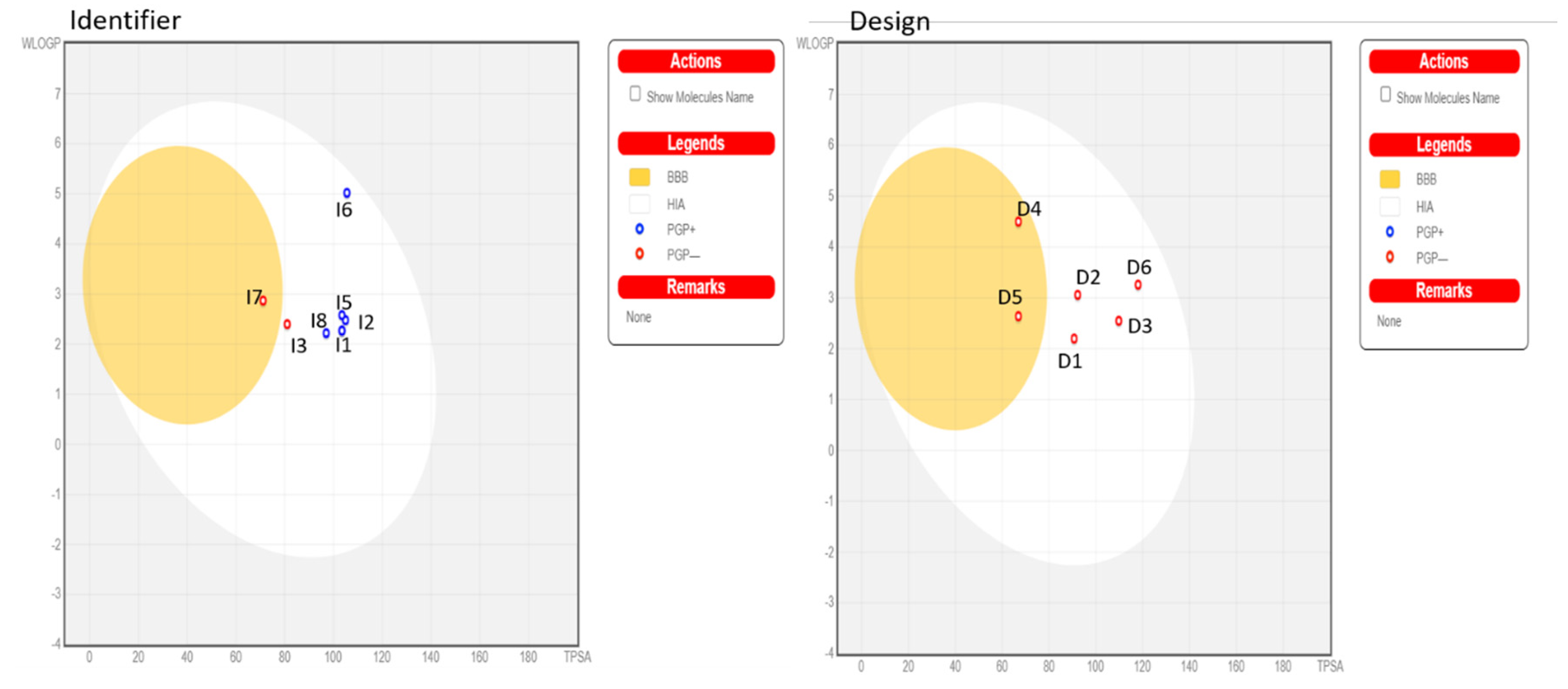
2.6. ADMET Analysis
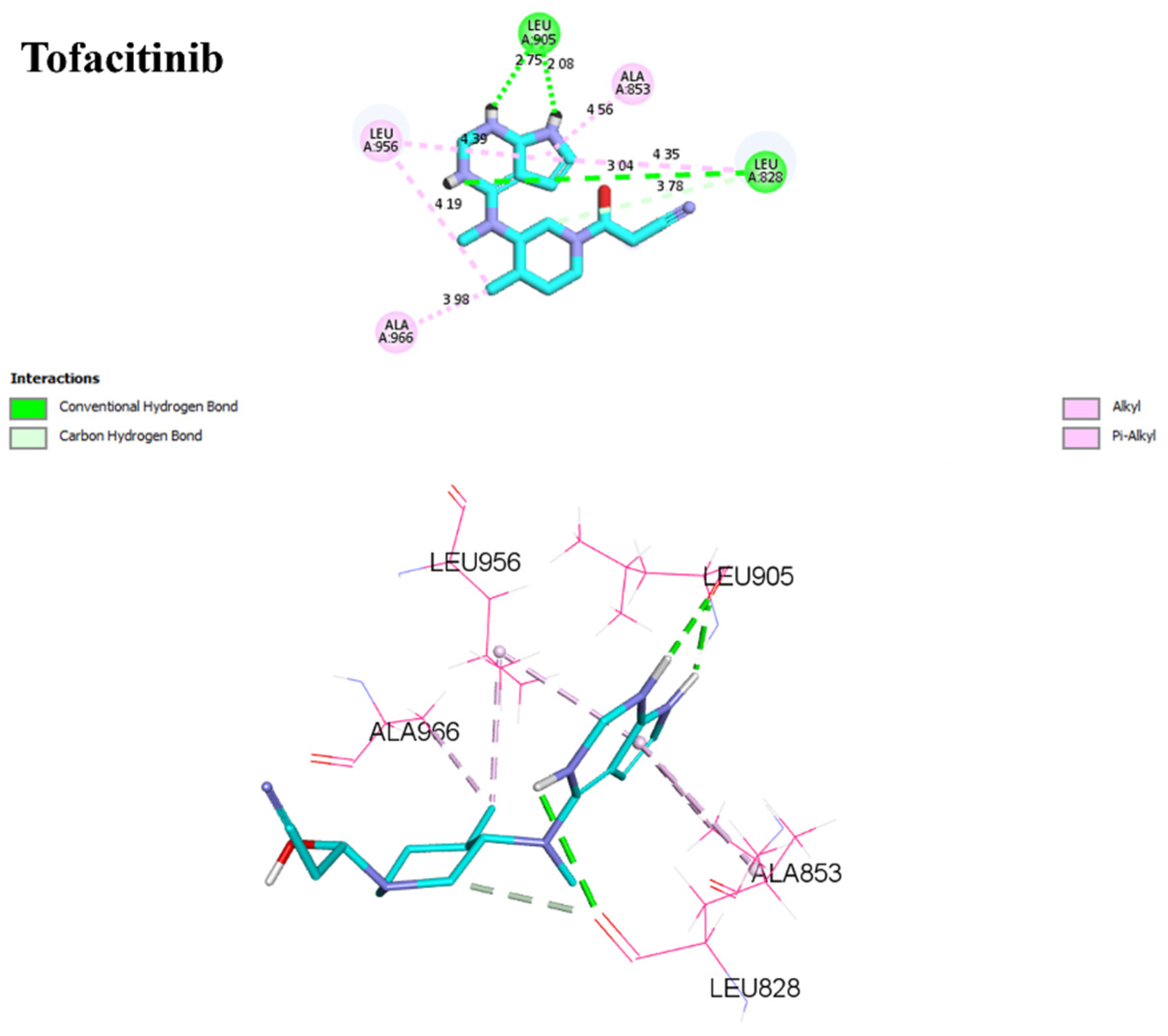
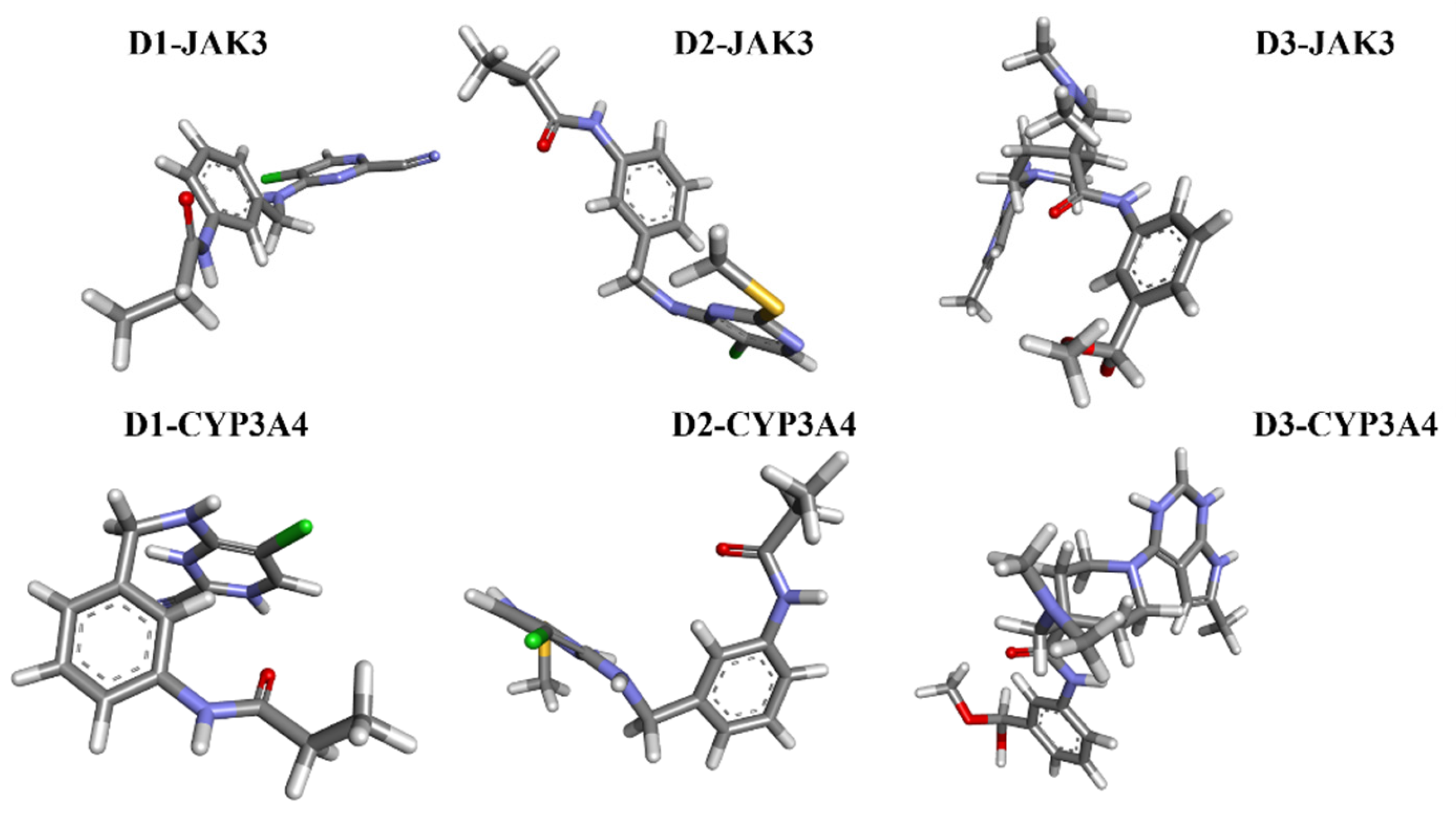
2.7. Analysis Docking of Selected Molecules (Covalent Docking between Identified and Designed Molecules)
2.8. Molecular Docking with CYP3A4
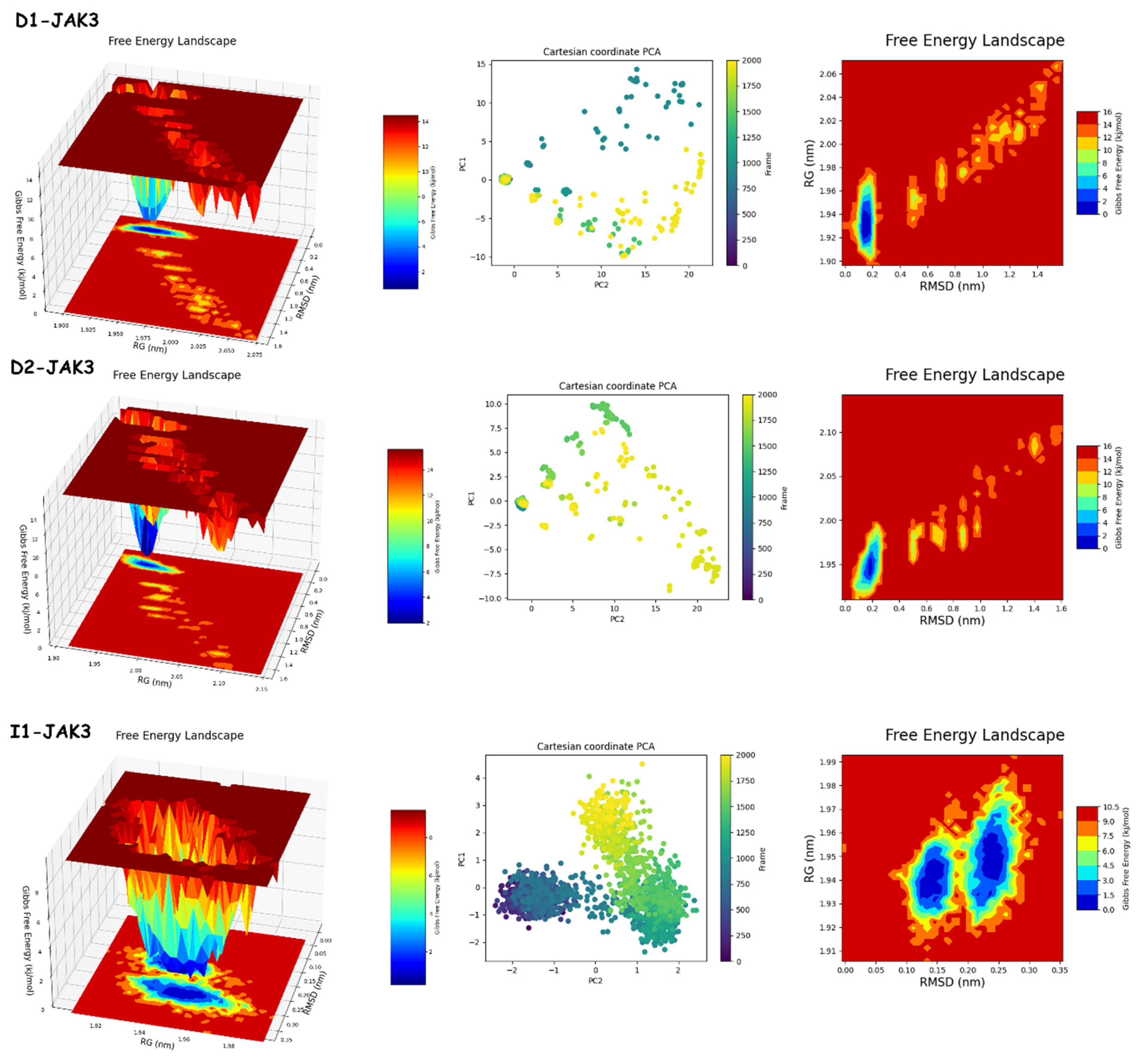
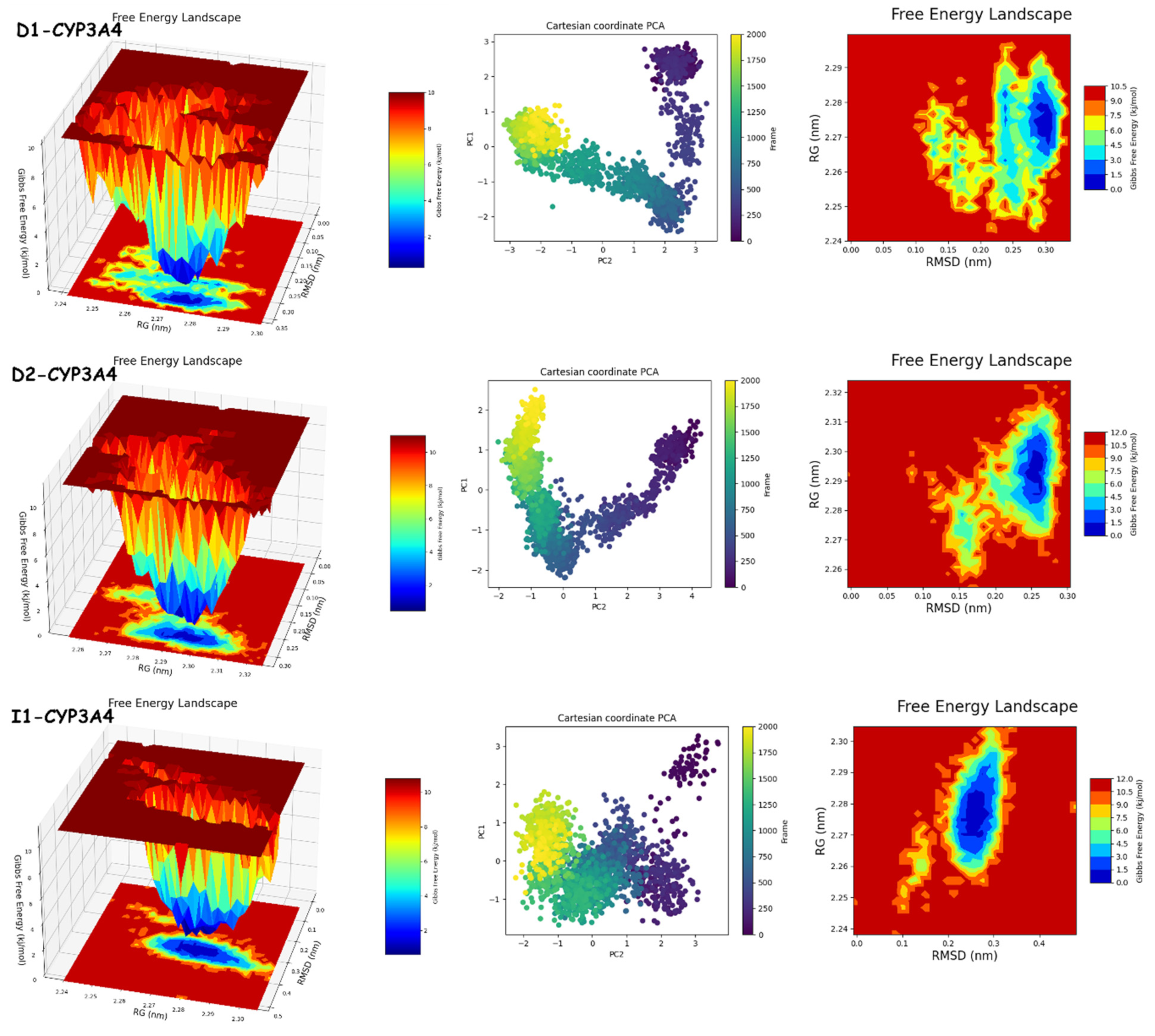
2.9. Molecular Dynamics Simulation Analysis
2.9.1. RMSD, RMSF, RoG, and SASA Analyses
2.9.2. PCA and FEL Analyses
2.10. MM/GBSA Analysis
3. Methods and Materials
3.1. Dataset
3.2. Building Robust Models: Exploring Three-Dimensional QSAR in Development
3.2.1. Preparation of Ligands
3.2.2. Field-Based and Atom-Based 3D-QSAR
3.2.3. Evaluating the Predictive Power of 3D-QSAR Models: A Comparative Analysis
3.3. Pharmacophore Hypothesis Generation
3.4. Molecular Docking
Irreversible (Covalent Docking)
3.5. Protein Structure Preparation
3.6. Schrödinger Covalent Docking
Reversible (Non-Covalent)
3.7. Predictive Toxicity Analysis and Bioactivity Assessment
3.8. Molecular Dynamics Simulation
3.9. Evaluating Binding Free Energy with Molecular Mechanics/Generalized Born Surface Area (MM/GBSA)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Liu, Z.; Pang, Y.; Zhou, H. The interaction between nanoparticles and immune system: Application in the treatment of inflammatory diseases. J. Nanobiotechnol. 2022, 20, 127. [Google Scholar] [CrossRef] [PubMed]
- Gautam, S.; Kumar, U.; Dada, R. Yoga and its impact on chronic inflammatory autoimmune arthritis. Front. Biosci. Elite 2020, 13, 77–116. [Google Scholar] [CrossRef]
- Sangha, P.S.; Thakur, M.; Akhtar, Z.; Ramani, S.; Gyamfi, R.S. The Link between Rheumatoid Arthritis and Dementia: A Review. Cureus 2020, 12, e7855. [Google Scholar] [CrossRef] [PubMed]
- de Jong, T.A.; Semmelink, J.F.; Denis, S.W.; van de Sande, M.G.H.; Houtkooper, R.H.L.; van Baarsen, L.G.M. Altered lipid metabolism in synovial fibroblasts of individuals at risk of developing rheumatoid arthritis. J. Autoimmun. 2023, 134, 102974. [Google Scholar] [CrossRef] [PubMed]
- Haville, S.; Deane, K.D. Pre-RA: Can early diagnosis lead to prevention? Best Pract. Res. Clin. Rheumatol. 2022, 36, 101737. [Google Scholar] [CrossRef] [PubMed]
- Padyukov, L. Genetics of rheumatoid arthritis. Semin. Immunopathol. 2022, 44, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Franklin, R.J.M.; Simons, M. CNS remyelination and inflammation: From basic mechanisms to therapeutic opportunities. Neuron 2022, 110, 3549–3565. [Google Scholar] [CrossRef]
- Godoi, M.A.; Camilli, A.C.; Gonzales, K.G.A.; Costa, V.B.; Papathanasiou, E.; Leite, F.R.M.; Guimarães-Stabili, M.R. JAK/STAT as a Potential Therapeutic Target for Osteolytic Diseases. Int. J. Mol. Sci. 2023, 24, 10290. [Google Scholar] [CrossRef]
- Winthrop, K.L. The emerging safety profile of JAK inhibitors in rheumatic disease. Nat. Rev. Rheumatol. 2017, 13, 234–243. [Google Scholar] [CrossRef]
- Nezamololama, N.; Fieldhouse, K.; Metzger, K.; Gooderham, M. Emerging systemic JAK inhibitors in the treatment of atopic dermatitis: A review of abrocitinib, baricitinib, and upadacitinib. Drugs Context 2020, 9, 2020-8-5. [Google Scholar] [CrossRef]
- Liu, C.; Kieltyka, J.; Fleischmann, R.; Gadina, M.; O’Shea, J.J. A Decade of JAK Inhibitors: What Have We Learned and What May Be the Future? Arthritis Rheumatol. 2021, 73, 2166–2178. [Google Scholar] [CrossRef] [PubMed]
- Faris, A.; Ibrahim, I.M.; Al Kamaly, O.; Saleh, A.; Elhallaoui, M. Computer-Aided Drug Design of Novel Derivatives of 2-Amino-7,9-dihydro-8H-purin-8-one as Potent Pan-Janus JAK3 Inhibitors. Molecules 2023, 28, 5914. [Google Scholar] [CrossRef] [PubMed]
- Faris, A.; Ibrahim, I.M.; Hadni, H.; Elhallaoui, M. High-throughput virtual screening of phenylpyrimidine derivatives as selective JAK3 antagonists using computational methods. J. Biomol. Struct. Dyn. 2023, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Yang, L.; Addison, G. Current Status in the Discovery of Covalent Janus Kinase 3 (JAK3) Inhibitors. Mini Rev. Med. Chem. 2019, 19, 1531–1543. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, Q.; Li, W.; Ge, G.; Peng, J.; Xu, Y.; Yang, H.; Bai, J.; Geng, D. Comprehensive overview of microRNA function in rheumatoid arthritis. Bone Res. 2023, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yin, Y.; Shi, G.; Zhou, Y.; Shao, S.; Wei, Y.; Wu, L.; Zhang, D.; Sun, L.; Zhang, T. A highly selective JAK3 inhibitor is developed for treating rheumatoid arthritis by suppressing γc cytokine–related JAK-STAT signal. Sci. Adv. 2022, 8, eabo4363. [Google Scholar] [CrossRef]
- Baselga, J. Targeting tyrosine kinases in cancer: The second wave. Science 2006, 312, 1175–1178. [Google Scholar] [CrossRef]
- Wang, X.; Rao, J.; Tan, Z.; Xun, T.; Zhao, J.; Yang, X. Inflammatory signaling on cytochrome P450-mediated drug metabolism in hepatocytes. Front. Pharmacol. 2022, 13, 1043836. [Google Scholar] [CrossRef]
- Veeravalli, V.; Dash, R.P.; Thomas, J.A.; Babu, R.J.; Madgula, L.M.V.; Srinivas, N.R. Critical Assessment of Pharmacokinetic Drug–Drug Interaction Potential of Tofacitinib, Baricitinib and Upadacitinib, the Three Approved Janus Kinase Inhibitors for Rheumatoid Arthritis Treatment. Drug Saf. 2020, 43, 711–725. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, X.; Zhang, P.; Li, Y.; Fu, M.; Lin, H.; Feng, S.; Shen, K.; Yu, G.; Li, X. Effect of CYP3A4 induction and inhibition on the pharmacokinetics of SHR0302 in healthy subjects. Br. J. Clin. Pharmacol. 2023, 89, 2561–2568. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, C.; Liu, G.; Liu, R.; Chen, Y.; Li, W.; Cao, Z.; Zhao, B.; Lu, C.; Liu, Y. Drug-Metabolizing Cytochrome P450 Enzymes Have Multifarious Influences on Treatment Outcomes. Clin. Pharmacokinet. 2021, 60, 585–601. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, W.; Li, Q.; Chen, Y.; Zhao, G.; Peng, Y.; Zheng, J. Tofacitinib Is a Mechanism-Based Inactivator of Cytochrome P450 3A4. Chem. Res. Toxicol. 2019, 32, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Radu, A.-F.; Bungau, S.G.; Negru, A.P.; Uivaraseanu, B.; Bogdan, M.A. Novel Potential Janus Kinase Inhibitors with Therapeutic Prospects in Rheumatoid Arthritis Addressed by In Silico Studies. Molecules 2023, 28, 4699. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Chen, M.; Hu, Y.; Lan, Y.; Gan, L.; You, G.; Yue, M.; Wang, H.; Xia, B.; Zhao, J.; et al. CYP3A4/5 mediates the metabolic detoxification of humantenmine, a highly toxic alkaloid from Gelsemium elegans Benth. J. Appl. Toxicol. 2019, 39, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Wall, M.; Corboy, G.P.; Taubenheim, N.; Gregory, G.P.; Opat, S.; Shortt, J. Failure of tofacitinib to achieve an objective response in a DDX3X-MLLT10 T-lymphoblastic leukemia with activating JAK3 mutations. Mol. Case Stud. 2020, 6, a004994. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Yu, C.; Denman, R.J.; Zhang, W. Recent advances in dynamic covalent chemistry. Chem. Soc. Rev. 2013, 42, 6634–6654. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.S.; Jacobsen, E.N. Asymmetric Catalysis by Chiral Hydrogen-Bond Donors. Angew. Chem. Int. Ed. 2006, 45, 1520–1543. [Google Scholar] [CrossRef]
- Climent, M.J.; Corma, A.; Iborra, S. Heterogeneous Catalysts for the One-Pot Synthesis of Chemicals and Fine Chemicals. Chem. Rev. 2011, 111, 1072–1133. [Google Scholar] [CrossRef]
- Kapetanovic, I.M. Computer-aided drug discovery and development (CADDD): In silico-chemico-biological approach. Chem. Biol. Interact. 2008, 171, 165–176. [Google Scholar] [CrossRef]
- Lu, F.; Wang, D.; Li, R.-L.; He, L.-Y.; Ai, L.; Wu, C.-J. Current strategies and technologies for finding drug targets of active components from traditional Chinese medicine. Front. Biosci. Landmark 2021, 26, 572–589. [Google Scholar] [CrossRef]
- Nascimento, I.J.d.S.; de Aquino, T.M.; da Silva-Júnior, E.F. The New Era of Drug Discovery: The Power of Computer-aided Drug Design (CADD). Lett. Drug Des. Discov. 2022, 19, 951–955. [Google Scholar] [CrossRef]
- da Silva-Júnior, E.F. “You’ve got the Body I’ve got the Brains”—Could the current AI-based tools replace the human ingenuity for designing new drug candidates? Bioorg. Med. Chem. 2023, 94, 117475. [Google Scholar] [CrossRef] [PubMed]
- Azam, M.A.; Thathan, J.; Jupudi, S. Pharmacophore modeling, atom based 3D-QSAR, molecular docking and molecular dynamics studies on Escherichia coli ParE inhibitors. Comput. Biol. Chem. 2020, 84, 107197. [Google Scholar] [CrossRef] [PubMed]
- Rondla, R.; PadmaRao, L.S.; Ramatenki, V.; Haredi-Abdel-Monsef, A.; Potlapally, S.R.; Vuruputuri, U. Selective ATP competitive leads of CDK4: Discovery by 3D-QSAR pharmacophore mapping and molecular docking approach. Comput. Biol. Chem. 2017, 71, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef]
- Shu, L.; Chen, C.; Huan, X.; Huang, H.; Wang, M.; Zhang, J.; Yan, Y.; Liu, J.; Zhang, T.; Zhang, D. Design, synthesis, and pharmacological evaluation of 4- or 6-phenyl-pyrimidine derivatives as novel and selective Janus kinase 3 inhibitors. Eur. J. Med. Chem. 2020, 191, 112148. [Google Scholar] [CrossRef]
- Tan, L.; Akahane, K.; McNally, R.; Reyskens, K.M.; Ficarro, S.B.; Liu, S.; Herter-Sprie, G.S.; Koyama, S.; Pattison, M.J.; Labella, K. Development of selective covalent Janus kinase 3 inhibitors. J. Med. Chem. 2015, 58, 6589–6606. [Google Scholar] [CrossRef]
- Yadav, D.K.; Saloni; Sharma, P.; Misra, S.; Singh, H.; Mancera, R.L.; Kim, K.; Jang, C.; Kim, M.; Pérez-Sánchez, H.; et al. Studies of the benzopyran class of selective COX-2 inhibitors using 3D-QSAR and molecular docking. Arch. Pharm. Res. 2018, 41, 1178–1189. [Google Scholar] [CrossRef]
- Ozgencil, F.; Eren, G.; Ozkan, Y.; Guntekin-Ergun, S.; Cetin-Atalay, R. Identification of small-molecule urea derivatives as novel NAMPT inhibitors via pharmacophore-based virtual screening. Bioorg. Med. Chem. 2020, 28, 115217. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2021-1, Maestro—Schrödinger, LLC.: New York, NY, USA, 2021.
- Janet, J.P.; Kulik, H.J. Resolving Transition Metal Chemical Space: Feature Selection for Machine Learning and Structure–Property Relationships. J. Phys. Chem. A 2017, 121, 8939–8954. [Google Scholar] [CrossRef] [PubMed]
- Jawarkar, R.D.; Zaki, M.E.A.; Al-Hussain, S.A.; Abdullah Alzahrani, A.Y.; Ming, L.C.; Samad, A.; Rashid, S.; Mali, S.; Elossaily, G.M. Mechanistic QSAR analysis to predict the binding affinity of diverse heterocycles as selective cannabinoid 2 receptor inhibitor. J. Taibah Univ. Sci. 2023, 17, 2265104. [Google Scholar] [CrossRef]
- Clark, M.; Cramer, R.D.; Opdenbosch, N.V. Validation of the general purpose tripos 5.2 force field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar] [CrossRef]
- Shinde, M.G.; Modi, S.J.; Kulkarni, V.M. QSAR and Molecular Docking of Phthalazine Derivatives as Epidermal Growth Factor Receptor (EGFR) Inhibitors. J. Appl. Pharm. Sci. 2017, 7, 181–191. [Google Scholar] [CrossRef][Green Version]
- Giordano, D.; Biancaniello, C.; Argenio, M.A.; Facchiano, A. Drug Design by Pharmacophore and Virtual Screening Approach. Pharmaceuticals 2022, 15, 646. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-Y. Pharmacophore modeling and applications in drug discovery: Challenges and recent advances. Drug Discov. Today 2010, 15, 444–450. [Google Scholar] [CrossRef]
- Aljoundi, A.; Bjij, I.; El Rashedy, A.; Soliman, M.E.S. Covalent Versus Non-covalent Enzyme Inhibition: Which Route Should We Take? A Justification of the Good and Bad from Molecular Modelling Perspective. Protein J. 2020, 39, 97–105. [Google Scholar] [CrossRef]
- Tivon, B.; Gabizon, R.; Somsen, B.A.; Cossar, P.J.; Ottmann, C.; London, N. Covalent flexible peptide docking in Rosetta. Chem. Sci. 2021, 12, 10836–10847. [Google Scholar] [CrossRef]
- Faris, A.; Ibrahim, I.M.; Alnajjar, R.; Hadni, H.; Bhat, M.A.; Yaseen, M.; Chakraborty, S.; Alsakhen, N.; Shamkh, I.M.; Mabood, F.; et al. QSAR-driven screening uncovers and designs novel pyrimidine-4,6-diamine derivatives as potent JAK3 inhibitors. J. Biomol. Struct. Dyn. 2023, 1–30. [Google Scholar] [CrossRef]
- Stortz, C.A.; Johnson, G.P.; French, A.D.; Csonka, G.I. Comparison of different force fields for the study of disaccharides. Carbohydr. Res. 2009, 344, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Olson, A.J. Using AutoDock for Ligand-Receptor Docking. Curr. Protoc. Bioinform. 2008, 24, 8.14.1–8.14.40. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Faris, A.; Ibrahim, I.M.; Chakraborty, S.; Kamaly, O.A.; Alshawwa, S.Z.; Hallaoui, M.E. Identification of Selective JAK3/STAT1 and CYP34A from Pyrazolopyrimidine Derivatives: A Search for Potential Drug Targets for Rheumatoid Arthritis using In-silico Drug Discovery Techniques. Lett. Drug Des. Discov. 2023, 20, 1–26. [Google Scholar] [CrossRef]
- Faris, A.; Hadni, H.; Saleh, B.A.; Khelfaoui, H.; Harkati, D.; Ait Ahsaine, H.; Elhallaoui, M.; El-Hiti, G.A. In silico screening of a series of 1,6-disubstituted 1H-pyrazolo[3,4-d]pyrimidines as potential selective inhibitors of the Janus kinase 3. J. Biomol. Struct. Dyn. 2023, 1–19. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef]
- Ziada, S.; Diharce, J.; Raimbaud, E.; Aci-Sèche, S.; Ducrot, P.; Bonnet, P. Estimation of Drug-Target Residence Time by Targeted Molecular Dynamics Simulations. J. Chem. Inf. Model. 2022, 62, 5536–5549. [Google Scholar] [CrossRef]
- Zhang, X.; Perez-Sanchez, H.; Lightstone, F.C. A Comprehensive Docking and MM/GBSA Rescoring Study of Ligand Recognition upon Binding Antithrombin. Curr. Top. Med. Chem. 2017, 17, 1631–1639. [Google Scholar] [CrossRef]




| Formation of a Covalent Bond with α,β-Unsaturated Ketone: | |
|---|---|
| Advantages | Disadvantages |
| 1. Wide applicability | 1. Limited reactivity of some α,β-unsaturated ketones |
| 2. Generally efficient reaction | 2. Incompatibility with certain functional groups |
| 3. Structural diversity | 3. Formation of undesired by-products |
| Michael 1,4 Reaction: | |
| Advantages | Disadvantages |
| 1. Formation of C-C bonds | 1. Limited reactivity of some nucleophiles |
| 2. Simple reaction steps | 2. Competitive side reactions |
| 3. Wide range of acceptor enones | 3. Sensitivity to steric effects |
| Factors | SD | R2 | R2CV | R2 Scramble | Stability | F | P | RMSE | Q2 | Pearson-r |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.7667 | 0.4738 | 0.3882 | 0.1235 | 0.99 | 44.1 | 2.38 × 10−8 | 0.59 | 0.5667 | 0.8013 |
| 2 | 0.6364 | 0.6449 | 0.4337 | 0.2443 | 0.936 | 43.6 | 1.62 × 10−11 | 0.52 | 0.6729 | 0.8383 |
| 3 | 0.5389 | 0.7506 | 0.4161 | 0.3818 | 0.818 | 47.2 | 3.24 × 10−14 | 0.52 | 0.6686 | 0.8181 |
| 4 | 0.5023 | 0.7880 | 0.5334 | 0.4656 | 0.805 | 42.7 | 6.13 × 10−15 | 0.44 | 0.7630 | 0.8775 |
| Factors | SD | R2 | R2CV | R2 Scramble | Stability | F | P | RMSE | Q2 | Pearson-r |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.6463 | 0.5585 | 0.47 | 0.1595 | 0.987 | 62 | 3.00 × 10−10 | 1.04 | 0.2246 | 0.4915 |
| 2 | 0.535 | 0.7037 | 0.5033 | 0.2836 | 0.909 | 57 | 2.10 × 10−13 | 0.91 | 0.4079 | 0.6392 |
| 3 | 0.4215 | 0.8199 | 0.4942 | 0.4679 | 0.810 | 71.3 | 1.61 × 10−17 | 0.77 | 0.5738 | 0.7576 |
| 4 | 0.3533 | 0.8761 | 0.5683 | 0.5665 | 0.655 | 81.3 | 2.91 × 10−20 | 0.69 | 0.6633 | 0.818 |
| Factors | Steric | Electrostatic | Hydrophobic | H-Bond Acceptor | H-Bond Donor |
|---|---|---|---|---|---|
| 1 | 0.41 | 0.115 | 0.149 | 0.261 | 0.066 |
| 2 | 0.287 | 0.095 | 0.223 | 0.29 | 0.105 |
| 3 | 0.319 | 0.122 | 0.219 | 0.19 | 0.149 |
| 4 | 0.333 | 0.125 | 0.209 | 0.184 | 0.149 |
| Compound | 2D | Affinity (Kcal/mol) | pIC50 (Pred) Field-Based | pIC50 (Pred) Atom-Based |
|---|---|---|---|---|
| D1 | 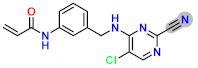 | −8.67 | 8.42353 | 7.75 |
| D2 |  | −8.18 | 7.47945 | 7.00 |
| D3 | 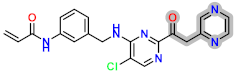 | −7.48 | 8.34337 | 8.30 |
| D4 | 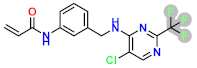 | −8.56 | 8.28205 | 7.94 |
| D5 | 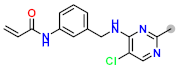 | −7.87 | 8.01863 | 7.48 |
| D6 | 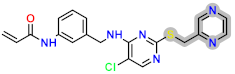 | −7.77 | 8.0394 | 7.54 |
| D7 | 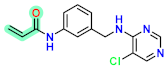 | −7.38 | 8.42353 | 7.75 |
| Model | Survival Score | Site Score | Vector Score | Volume Score | Score | PhaseHypoScore |
|---|---|---|---|---|---|---|
| DHRRR_2 | 6.102 | 0.776 | 0.908 | 0.754 | 0.973 | 1.339 |
| DHRRR_1 | 6.168 | 0.794 | 0.927 | 0.777 | 0.972 | 1.342 |
| DHRR_2 | 5.773 | 0.861 | 0.954 | 0.761 | 0.804 | 1.150 |
| DHRR_1 | 5.813 | 0.897 | 0.973 | 0.765 | 0.961 | 1.310 |
| DDRRR_2 | 5.985 | 0.764 | 0.933 | 0.793 | 0.948 | 1.307 |
| DDRRR_1 | 6.033 | 0.792 | 0.929 | 0.812 | 0.956 | 1.318 |
| DDHRR_2 | 6.225 | 0.825 | 0.963 | 0.737 | 0.945 | 1.318 |
| DDHRR_1 | 6.265 | 0.837 | 0.951 | 0.765 | 0.932 | 1.308 |
| DDHR_1 | 5.772 | 0.895 | 0.948 | 0.760 | 0.765 | 1.111 |
| AHRR_1 | 5.742 | 0.895 | 0.977 | 0.729 | 0.812 | 1.156 |
| ADRR_1 | 5.674 | 0.941 | 0.975 | 0.782 | 0.950 | 1.290 |
| ADHRR_3 | 6.003 | 0.751 | 0.913 | 0.730 | 0.886 | 1.246 |
| ADHRR_2 | 6.152 | 0.844 | 0.979 | 0.729 | 0.906 | 1.276 |
| ADHRR_1 | 6.255 | 0.904 | 0.983 | 0.743 | 0.948 | 1.324 |
| ADHR_3 | 5.665 | 0.885 | 0.951 | 0.722 | 0.920 | 1.260 |
| ADHR_2 | 5.717 | 0.901 | 0.957 | 0.726 | 0.815 | 1.158 |
| ADDHR_1 | 5.980 | 0.845 | 0.956 | 0.777 | 0.928 | 1.286 |
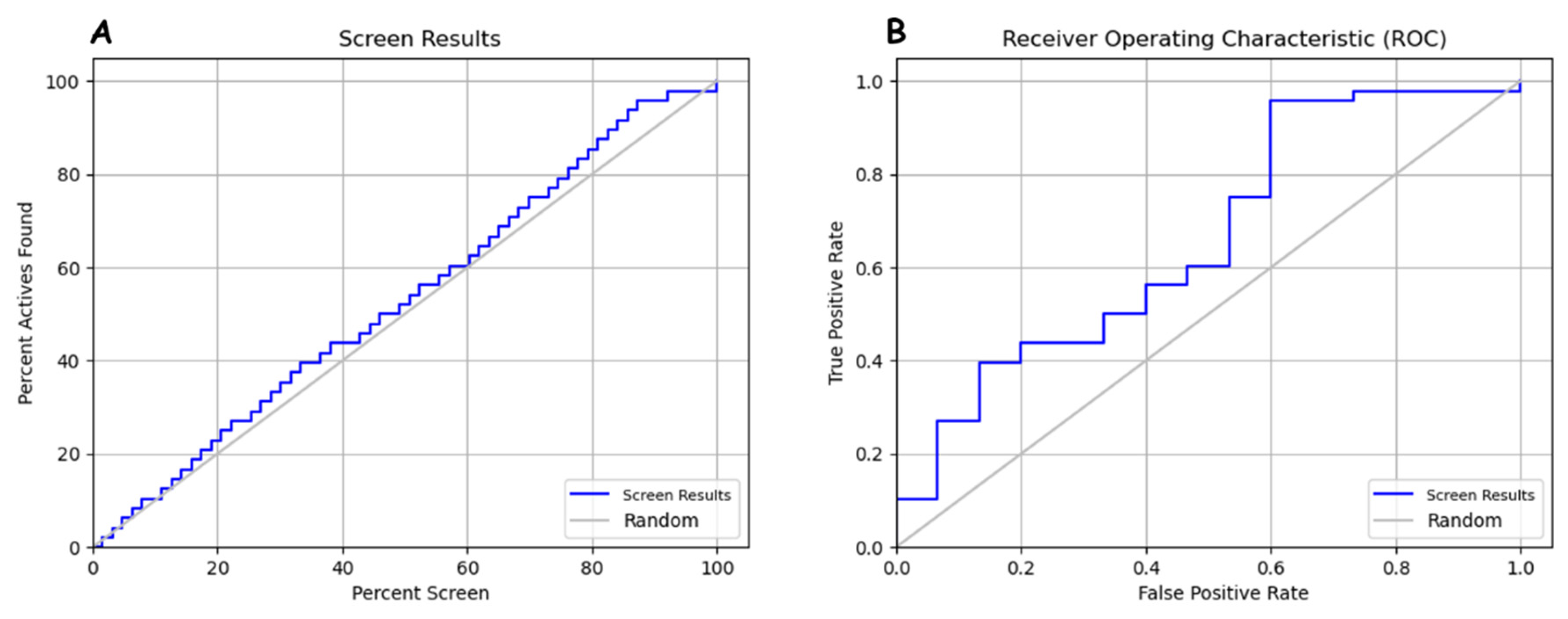
| Hypothesis | PhaseHypoScore | EF1% | BEDROC160.9 | Matches | ||
|---|---|---|---|---|---|---|
| DHRR_1 | 1.3 | 1.31 | 1 | 4 of 4 | ||
| BEDROC:1 | ||||||
| alpha*Ra | alpha | |||||
| 160.9, | 122.5905 | |||||
| BEDROC: 0.940 | ||||||
| alpha*Ra | alpha | |||||
| 15.2381 | 20.0 | |||||
| BEDROC: 0.874 | ||||||
| alpha*Ra | alpha | |||||
| 6.0952 | 8.0 | |||||
| ROC | 0.66 | |||||
| Count and percentage of actives in top N% of decoy results. | ||||||
| % Decoys | 1% | 2% | 5% | 10% | 20% | |
| # Actives | 0 | 0 | 0 | 5 | 19 | |
| % Actives | ||||||
| Count and percentage of actives in top N% of results. | ||||||
| % Results | 1% | 2% | 5% | 10% | 15% | |
| # Actives | 0 | 1 | 3 | 5 | 11 | |
| % Actives | 0 | 2.1 | 6.2 | 10.4 | 11 | |
| Enrichment factors concerning N% sample size. | ||||||
| % Sample | 0.01 | 0.02 | 0.05 | 0.1 | 0.2 | |
| EF | 1.3 | 1.3 | 1.3 | 1.1 | 1.2 | |
| EF* | inf | inf | 1.6 | 2 | 2 | |
| EF’ | inf | inf | 1.6 | 2.5 | 2.5 | |
| DEF | n/a | n/a | n/a | n/a | n/a | |
| DEF* | n/a | n/a | n/a | n/a | n/a | |
| DEF’ | n/a | n/a | n/a | n/a | n/a | |
| Eff | −1 | −1 | −1 | 0.0204 | 0.329 | |
| Enrichment factors concerning N% actives recovered. | ||||||
| % Actives | 40% | 50% | 60% | 70% | 80% | |
| EF | 1.1 | 1.1 | 1.1 | 1.1 | 1.1 | |
| EF* | 3 | 1.5 | 1.3 | 1.3 | 1.3 | |
| EF’ | 2.5 | 2.1 | 1.9 | 1.7 | 1.7 | |
| FOD | 0.07 | 0.1 | 0.2 | 0.2 | 0.3 | |
| Compounds | ID | Affinity (kcal/mol) | Smiles | pIC50 (Pred) Field-Based | pIC50 (Pred) Atom-Based |
|---|---|---|---|---|---|
| I1 | BDBM50117501 | −8.72 |  | 8.51 | 7.98 |
| I2 | SCHEMBL20184389 | −5.63 | 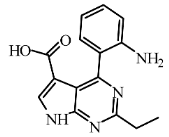 | 7.16 | 7.49 |
| I3 | SCHEMBL645138 | −8.62 |  | 7.19 | 6.90 |
| I4 | BDBM50117502 | −8.52 | 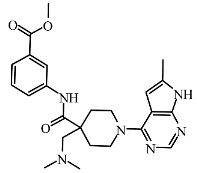 | 8.46 | 7.88 |
| I6 | SCHEMBL5253185 | −7.80 | 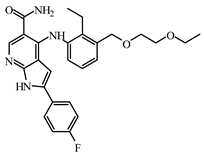 | 8.60 | 7.99 |
| I7 | ZGEWVZMORUOOMV | −6.85 | 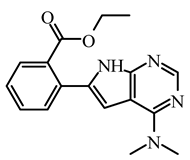 | 8.03 | 7.60 |
| I8 | 60118026 | −7.93 |  | 7.38 | 7.23 |
| Compound | Adsorption | Distribution | Metabolism | Excretion | Toxicity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water Solubility (Logs) | Intestinal Absorption (Human) | Permeability | Substrate | Inhibitor | ||||||||
| BBB | 2D6 | 3A4 | 1A2 | 2C19 | 2C9 | 2D6 | 3A4 | Total Clearance | AMES Toxicity | |||
| D1 | −3.376 | 76.324 | −0.726 | No | Yes | No | No | No | No | Yes | 0.078 | No |
| D2 | −3.931 | 88.657 | −0.796 | No | Yes | No | Yes | Yes | No | Yes | 0.02 | No |
| D3 | −4.456 | 89.197 | −0.024 | No | Yes | Yes | Yes | Yes | No | No | −0.057 | No |
| D4 | −3.825 | 92.308 | −0.007 | No | Yes | Yes | Yes | No | No | No | −0.059 | No |
| D5 | −3.748 | 85.635 | −0.764 | No | Yes | No | Yes | Yes | No | Yes | 0.19 | No |
| D6 | −3.599 | 92.597 | 0.146 | No | Yes | Yes | Yes | No | No | No | −0.004 | No |
| Compound | Adsorption | Distribution | Metabolism | Excretion | Toxicity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water Solubility (Logs) | Intestinal Absorption (Human) | Permeability | Substrate | Inhibitor | ||||||||
| BBB | 2D6 | 3A4 | 1A2 | 2C19 | 2C9 | 2D6 | 3A4 | Total Clearance | AMES Toxicity | |||
| I1 | −3.055 | 81.075 | −1.028 | No | Yes | No | No | No | No | Yes | 0.589 | No |
| I2 | −2.889 | 60.562 | −1.433 | No | No | No | No | No | No | No | 0.709 | No |
| I3 | −2.919 | 95.844 | −1.132 | No | No | Yes | No | No | No | No | 0.62 | Yes |
| I4 | −3.055 | 81.075 | −1.028 | No | Yes | No | No | No | No | Yes | 0.589 | No |
| I5 | −3.084 | 81.619 | −1.022 | No | Yes | No | No | No | No | Yes | 0.598 | No |
| I6 | −4.614 | 89.092 | −1.017 | No | Yes | No | Yes | Yes | No | Yes | 0.477 | Yes |
| I7 | −2.899 | 97.033 | −0.104 | No | No | Yes | Yes | No | No | No | 0.931 | No |
| I8 | −2.995 | 92.516 | −0.859 | No | Yes | Yes | No | No | No | Yes | 0.834 | No |
| Delta Energy Component (Kcal/mol) | D1-JAK3 | D2-JAK3 | I1-JAK3 | D1-CYP3A4 | D2-CYP3A4 | I1-CYP3A4 | Tofacitinib-JAK3 |
|---|---|---|---|---|---|---|---|
| ΔTOTAL | −21.60 | −25.96 | −36.51 | 6.53 | −15.83 | −29.68 | −3.20 |
| ΔGSOLV | 33.97 | 26.49 | 110.50 | −63.87 | 56.84 | −14.72 | 52.55 |
| ΔGGAS | −55.57 | −52.45 | −147.00 | 70.40 | −72.67 | −14.96 | −55.75 |
| ΔESURF | −4.94 | −5.18 | −5.88 | −4.63 | −4.23 | −5.77 | −3.34 |
| ΔEGB | 38.91 | 31.67 | 116.38 | −59.24 | 61.07 | −8.95 | 55.89 |
| ΔEEL | −20.91 | −14.37 | −109.57 | 103.24 | −46.97 | 22.66 | −32.93 |
| ΔVDWAALS | −34.66 | −38.08 | −37.44 | −32.84 | 56.84 | −37.62 | −22.82 |
| Factors | H-Bond Donor | Hydrophobic/Nonpolar | Electron-Withdrawing |
|---|---|---|---|
| 1 | 0.031 | 0.63 | 0.338 |
| 2 | 0.035 | 0.598 | 0.366 |
| 3 | 0.043 | 0.575 | 0.383 |
| 4 | 0.053 | 0.555 | 0.392 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faris, A.; Alnajjar, R.; Guo, J.; AL Mughram, M.H.; Aouidate, A.; Asmari, M.; Elhallaoui, M. Computational 3D Modeling-Based Identification of Inhibitors Targeting Cysteine Covalent Bond Catalysts for JAK3 and CYP3A4 Enzymes in the Treatment of Rheumatoid Arthritis. Molecules 2024, 29, 23. https://doi.org/10.3390/molecules29010023
Faris A, Alnajjar R, Guo J, AL Mughram MH, Aouidate A, Asmari M, Elhallaoui M. Computational 3D Modeling-Based Identification of Inhibitors Targeting Cysteine Covalent Bond Catalysts for JAK3 and CYP3A4 Enzymes in the Treatment of Rheumatoid Arthritis. Molecules. 2024; 29(1):23. https://doi.org/10.3390/molecules29010023
Chicago/Turabian StyleFaris, Abdelmoujoud, Radwan Alnajjar, Jingjing Guo, Mohammed H. AL Mughram, Adnane Aouidate, Mufarreh Asmari, and Menana Elhallaoui. 2024. "Computational 3D Modeling-Based Identification of Inhibitors Targeting Cysteine Covalent Bond Catalysts for JAK3 and CYP3A4 Enzymes in the Treatment of Rheumatoid Arthritis" Molecules 29, no. 1: 23. https://doi.org/10.3390/molecules29010023
APA StyleFaris, A., Alnajjar, R., Guo, J., AL Mughram, M. H., Aouidate, A., Asmari, M., & Elhallaoui, M. (2024). Computational 3D Modeling-Based Identification of Inhibitors Targeting Cysteine Covalent Bond Catalysts for JAK3 and CYP3A4 Enzymes in the Treatment of Rheumatoid Arthritis. Molecules, 29(1), 23. https://doi.org/10.3390/molecules29010023









