Exploring Antioxidant and α-Glucosidase Inhibitory Activities in Mulberry Leaves (Morus alba L.) across Growth Stages: A Comprehensive Metabolomic Analysis with Chemometrics
Abstract
:1. Introduction
2. Results and Discussion
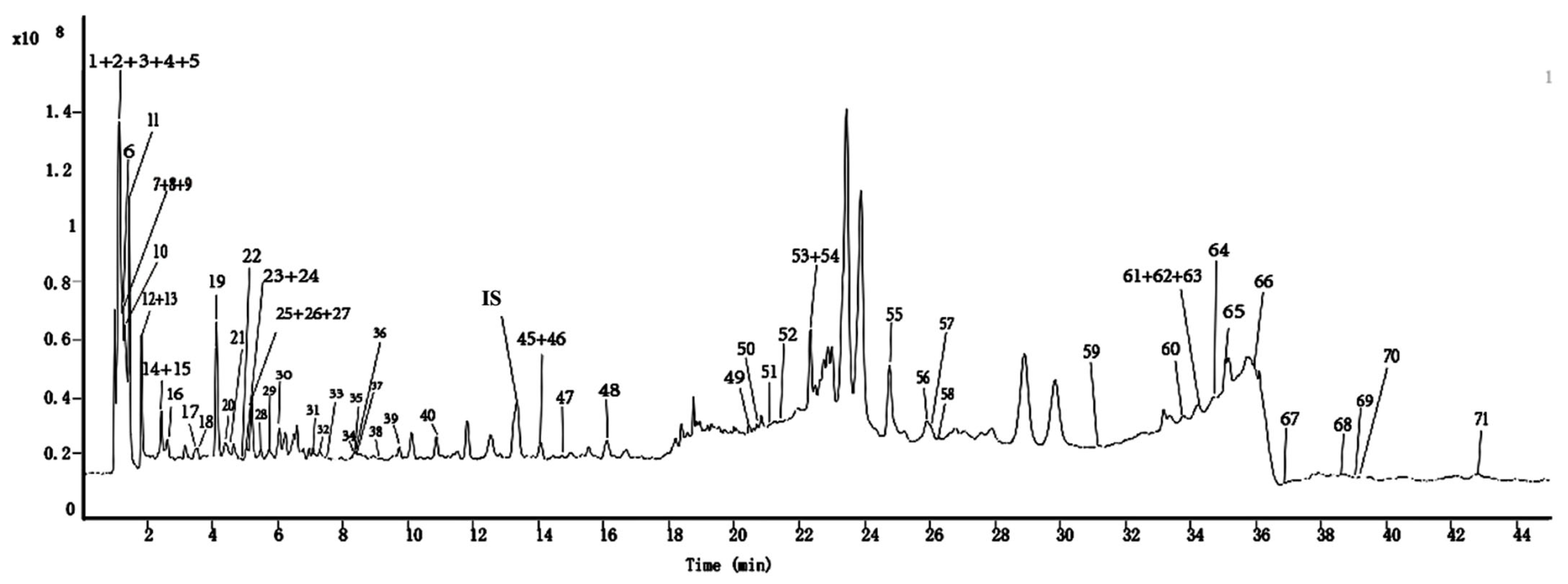
2.1. GC-MS Profiling of Metabolites in ML Extract
2.1.1. GC-MS Profiling of Metabolites
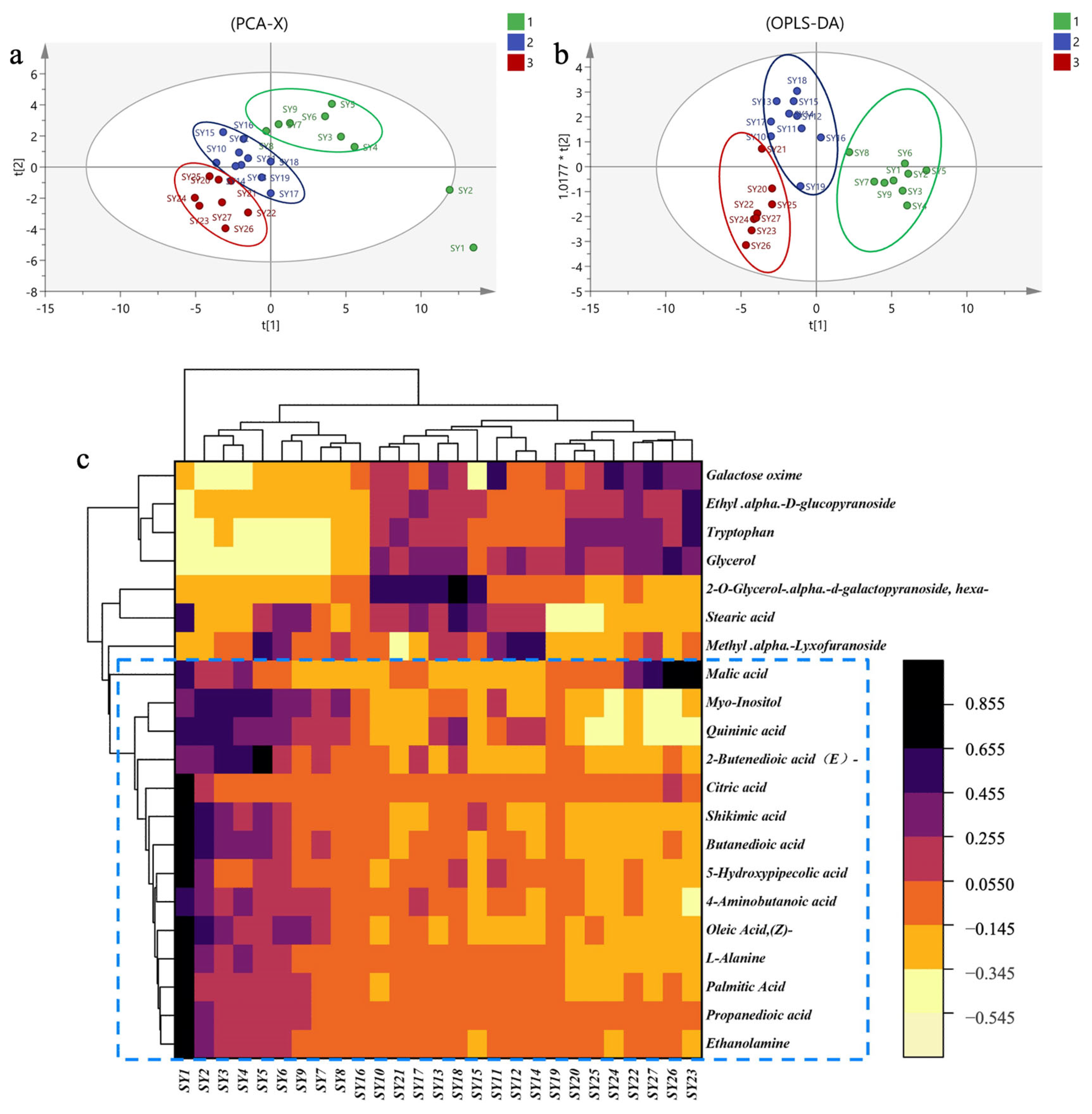
2.1.2. Multivariate Data Analysis of MLs at Different Growth Periods
2.2. Secondary Metabolic Analysis
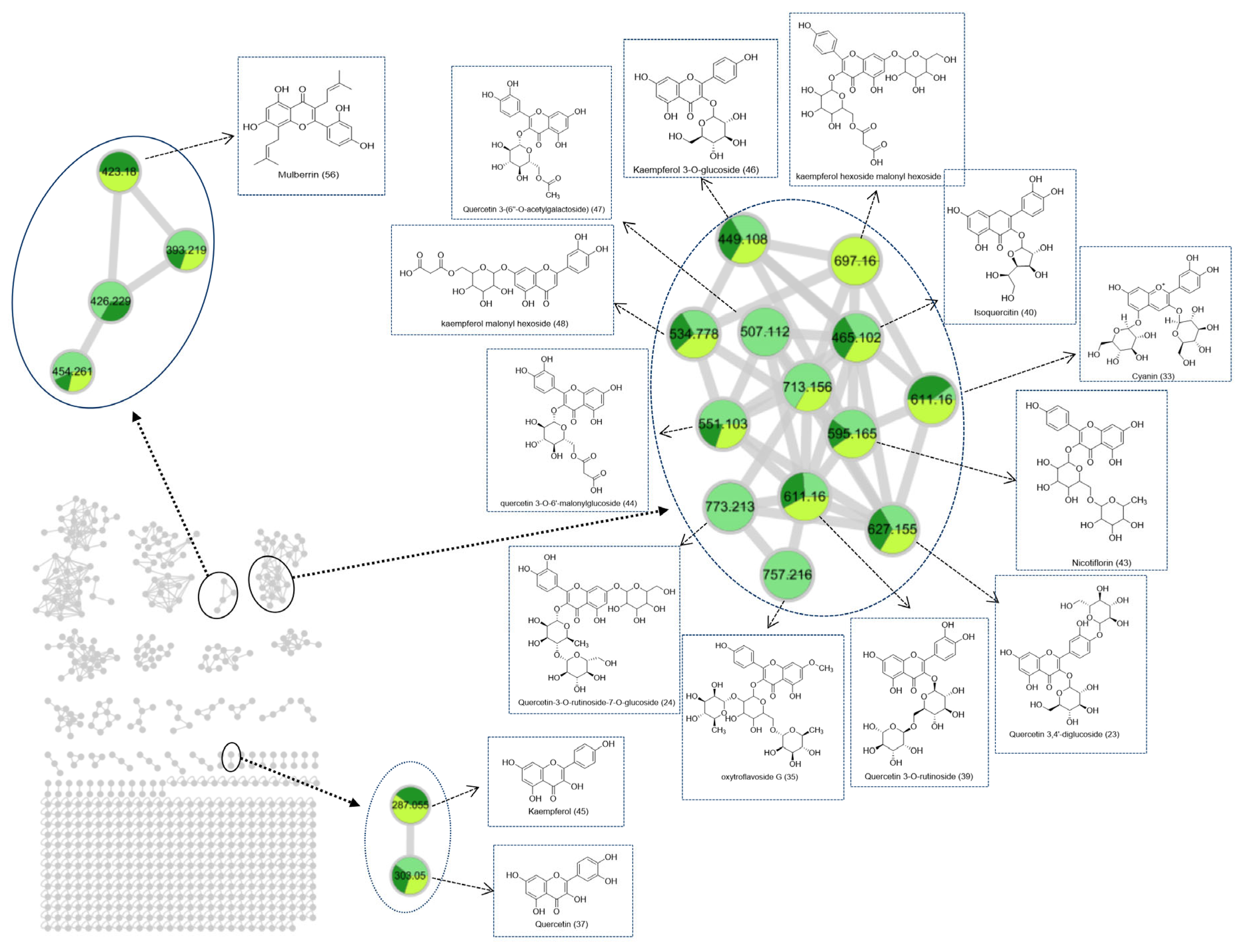
2.2.1. UPLC–Quadrupole Time-of-Flight (QTOF)–MS Data and Molecular Networking (MN) Analysis
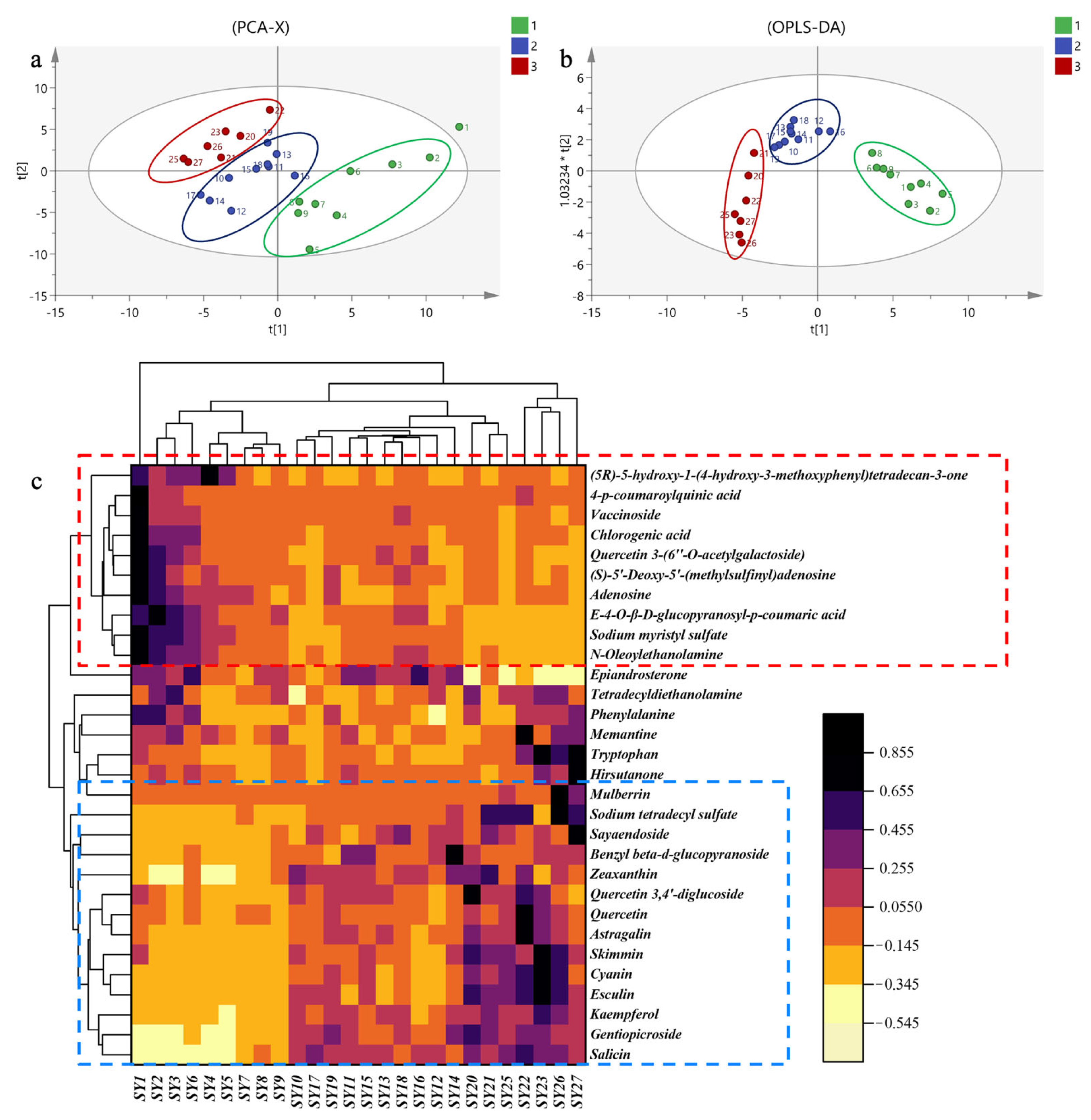
2.2.2. Secondary Metabolites of MLs at Different Growth Periods
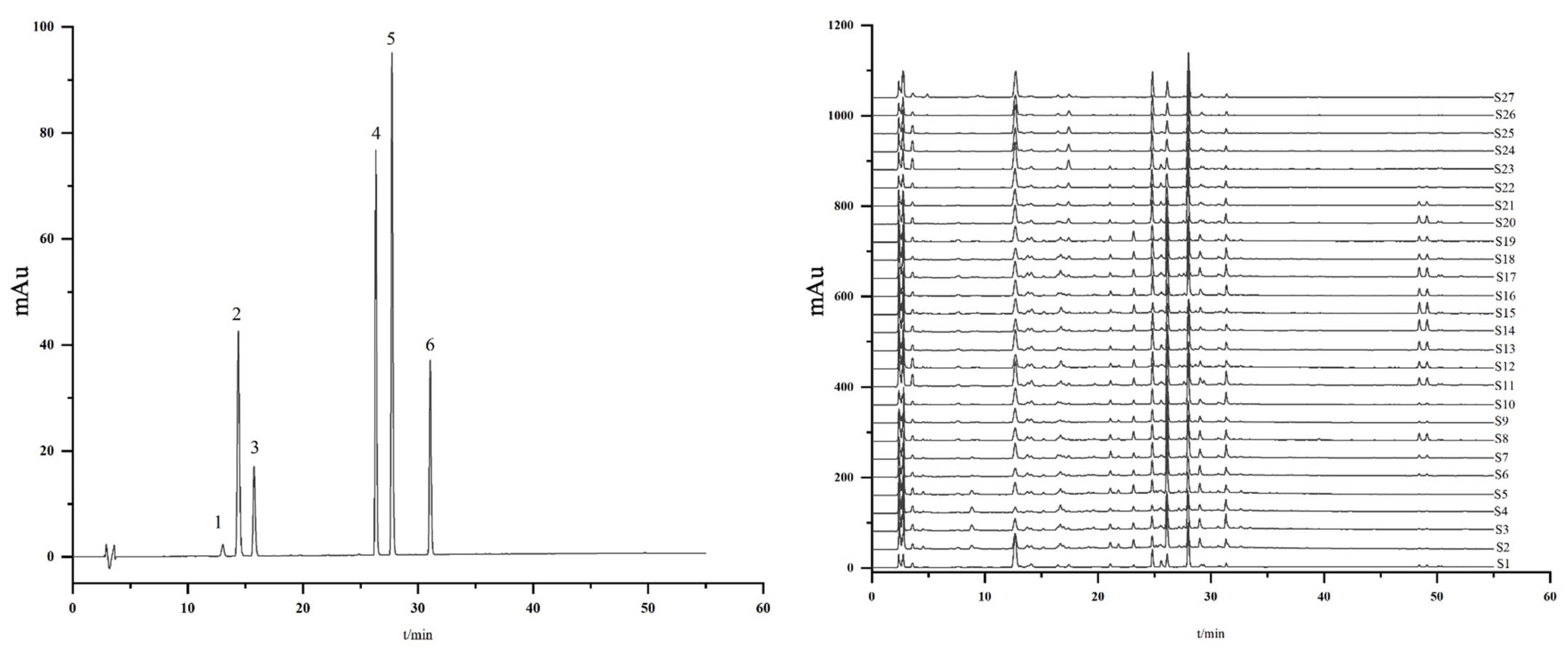
2.3. The Determination of Six Compounds from MLs by HPLC Analysis
2.3.1. Linearity and Method Validation
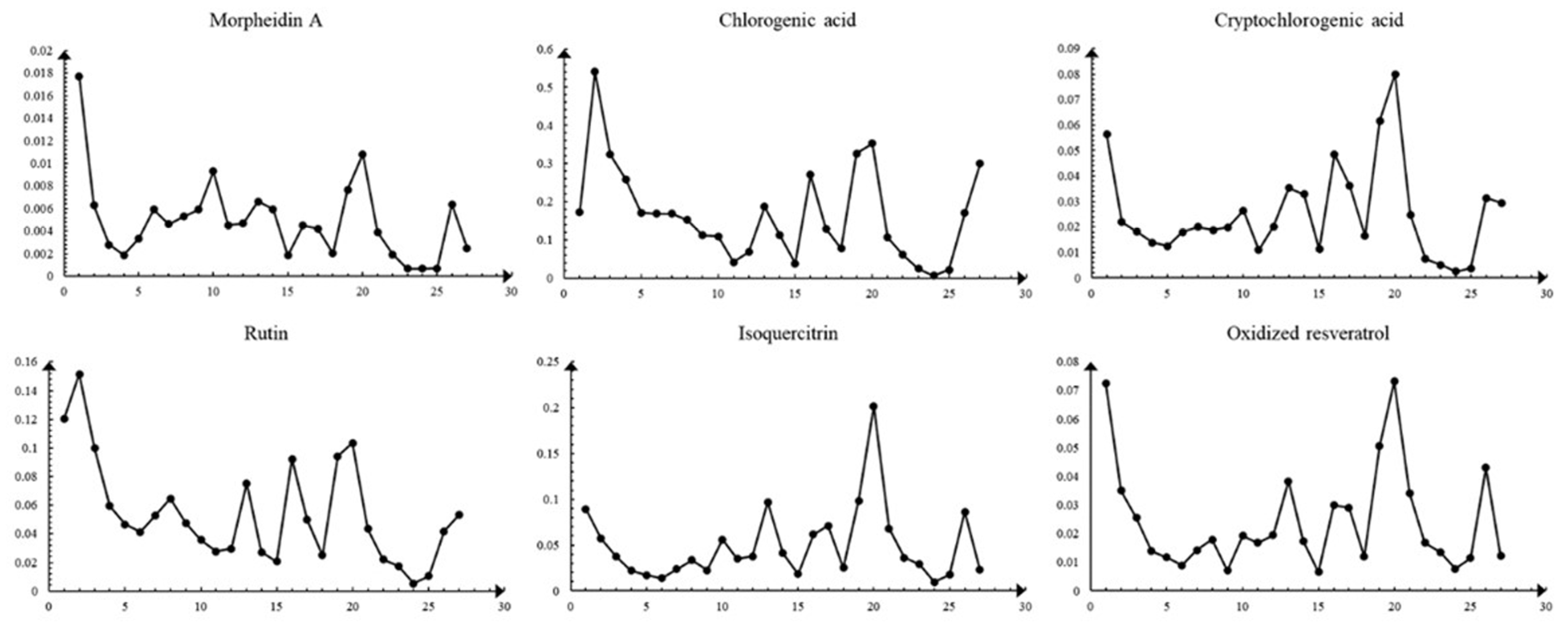
2.3.2. Simultaneous Quantitative Analysis of Six Constituents of MLs
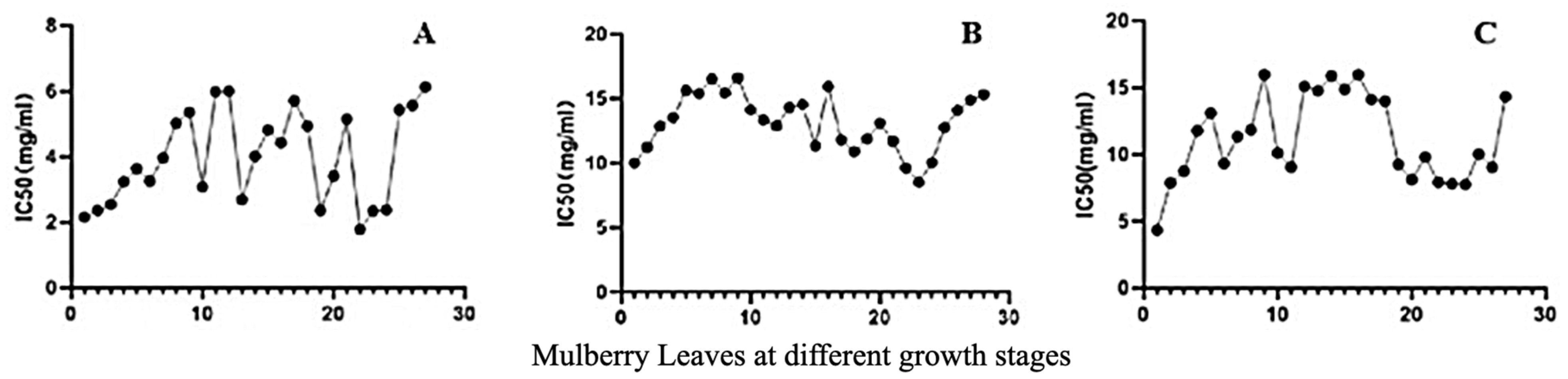
2.4. Antioxidant Activities of MLs at Different Growth Stages
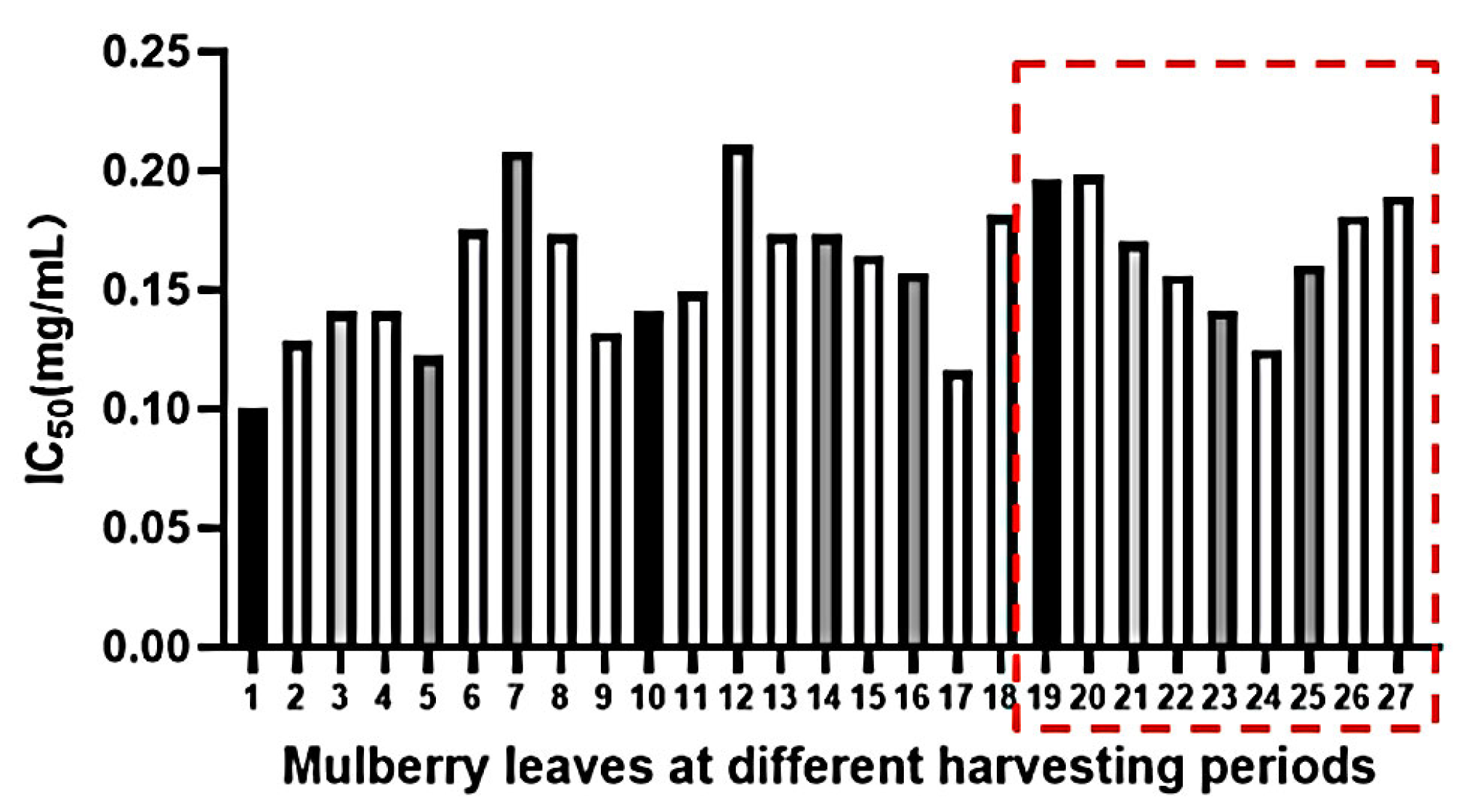
2.5. The Inhibitory Effects of MLs at Different Growth Stages on α-Glucosidase Activities
3. Materials and Methods
3.1. Plant Materials and Chemicals
3.1.1. Plant Materials
3.1.2. Chemicals
3.2. Sample Preparation for MS and HPLC Analyses
3.2.1. GC-MS Analysis
3.2.2. UPLC-MS Analysis
3.2.3. HPLC Analysis
3.3. GC-MS Profiling and Modeling of Silylated Primary Metabolites
3.4. Analysis, Modeling, and Quantification of the UPLC-QTOF-MS Dataset
3.4.1. GNPS Molecular MS/MS Network
3.4.2. Data Processing and Statistical Analysis
3.5. Standard Substance Solution and Sample Solution Preparation
3.6. Determination of Antioxidant Activities at Different Growth Stages of MLs
3.6.1. Inhibition of 1,1 diphenyl 2-picryl hydrazine (DPPH)
3.6.2. Inhibition of Hydroxyl Radical
3.6.3. ABTS Radical Scavenging Activity
3.7. Determination of α-Glucosidase Inhibitory Activity at Different Growth Stages of MLs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sugiyama, M.; Katsube, T.; Koyama, A.; Itamura, H. Effect of solar radiation on the functional components of mulberry (Morus alba L.) leaves. J. Sci. Food Agric. 2016, 96, 3915–3921. [Google Scholar] [CrossRef] [PubMed]
- Singab, A.N.; El-Beshbishy, H.A.; Yonekawa, M.; Nomura, T.; Fukai, T. Hypoglycemic effect of Egyptian Morus alba root bark extract: Effect on diabetes and lipid peroxidation of streptozotocin-induced diabetic rats. J. Ethnopharmacol. 2005, 100, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Song, C.; Feng, X.; Wang, Y.; Meng, T.; Li, S.; Bai, Y.; Du, B.; Sun, Q. Isolation and hypoglycemic effects of water extracts from mulberry leaves in Northeast China. Food Funct. 2020, 11, 3112–3125. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, T.; Xiao, L.; Chen, R.Y. Two novel flavanes from the leaves of Morus alba L. J. Asian Nat. Prod. Res. 2010, 12, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Chen, G.; Ma, B.; Zhong, C.; He, N. Metabolic Profiling and Transcriptome Analysis of Mulberry Leaves Provide Insights into Flavonoid Biosynthesis. J. Agric. Food Chem. 2020, 68, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.Y.; Chen, Y.C. A review of the dietary flavonoid, kaempferol on human health and cancer chemoprevention. Food Chem. 2013, 138, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Chai, X.; Hou, G.; Zhao, F.; Meng, Q. Phytochemistry, bioactivities and future prospects of mulberry leaves: A review. Food Chem. 2022, 372, 131335. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.Q.; Cheng, S.Y.; Zhang, J.Q.; Lin, H.F.; Chen, Y.Y.; Yue, S.J.; Tian, M.; Tang, Y.P.; Zhao, Y.C. Morus alba L. Leaves—Integration of Their Transcriptome and Metabolomics Dataset: Investigating Potential Genes Involved in Flavonoid Biosynthesis at Different Harvest Times. Front. Plant Sci. 2021, 12, 736332. [Google Scholar] [CrossRef]
- Yu, X.; Zhu, Y.; Fan, J.; Wang, D.; Gong, X.; Ouyang, Z. Accumulation of Flavonoid Glycosides and UFGT Gene Expression in Mulberry Leaves (Morus alba L.) before and after Frost. Chem. Biodivers. 2017, 14, e8760. [Google Scholar] [CrossRef]
- Padda, M.S.; Picha, D.H. Effect of low temperature storage on phenolic composition and antioxidant activity of sweetpotatoes. Postharvest Biol. Technol. 2008, 47, 176–180. [Google Scholar] [CrossRef]
- Farag, M.A.; Baky, M.H.; Morgan, I.; Khalifa, M.R.; Rennert, R.; Mohamed, O.G.; El-Sayed, M.M.; Porzel, A.; Wessjohann, L.A.; Ramadan, N.S. Comparison of Balanites aegyptiaca parts: Metabolome providing insights into plant health benefits and valorization purposes as analyzed using multiplex GC-MS, LC-MS, NMR-based metabolomics, and molecular networking. RSC Adv. 2023, 13, 21471–21493. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhong, F.; Zhu, J. Bridging Targeted and Untargeted Mass Spectrometry-Based Metabolomics via Hybrid Approaches. Metabolites 2020, 10, 348. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Shaari, K. LC-MS metabolomics analysis of Stevia rebaudiana Bertoni leaves cultivated in Malaysia in relation to different developmental stages. Phytochem. Anal. 2022, 33, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.A.; Perez de Souza, L.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the Context of Plant Natural Products Research: From Sample Preparation to Metabolite Analysis. Metabolites 2020, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zeng, Z.; Ye, G.; Zhao, C.; Lu, X.; Xu, G. Non-targeted metabolomics study for the analysis of chemical compositions in three types of tea by using gas chromatograph-mass spectrometry and liquid chromatography-mass spectrometry. Se Pu Chin. J. Chromatogr. 2014, 32, 804–816. [Google Scholar] [CrossRef] [PubMed]
- Carazzone, C.; Rodríguez, J.P.; Gonzalez, M.; López, G.-D. Volatilomics of natural products: Whispers from nature. In Metabolomics-Methodology and Applications in Medical Sciences and Life Sciences; IntechOpen: London, UK, 2021. [Google Scholar]
- Duan, P.B.; Xiao, P.T.; Yang, X.; Hao, J.H.; Li, K.; Liu, E.H. Screening of hypoglycemic components in Platycladi Cacumen by phytochemical investigation, spectrum-effect relationship, and chemometric methods. J. Sep. Sci. 2022, 45, 2591–2602. [Google Scholar] [CrossRef] [PubMed]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- An, Q.; Wang, L.; Ding, X.-Y.; Shen, Y.-J.; Hao, S.-H.; Li, W.-J.; Li, H.-Y.; Wang, T.; Zhan, Z.-L.; Zheng, Y.-G. Validation of Sennae Folium specification grade classification based on UPLC-Q-TOF/MS spectrum-effect relationship. Arab. J. Chem. 2022, 15, 104223. [Google Scholar] [CrossRef]
- Zou, X.-Y.; He, Y.-J.; Yang, Y.-H.; Yan, X.-P.; Li, Z.-B.; Yang, H. Systematic Identification of Bioactive Compositions in Leaves of Morus Cultivars Using UHPLC-ESI-QTOF-MS/MS and Comprehensive Screening of High-Quality Resources. Separations 2022, 9, 76. [Google Scholar] [CrossRef]
- Wang, Z.; Tang, C.; Xiao, G.; Dai, F.; Lin, S.; Li, Z.; Luo, G. Comparison of free and bound phenolic compositions and antioxidant activities of leaves from different mulberry varieties. BMC Chem. 2021, 15, 21. [Google Scholar] [CrossRef] [PubMed]
- D’urso, G.; Mes, J.J.; Montoro, P.; Hall, R.D.; de Vos, R.C. Identification of bioactive phytochemicals in mulberries. Metabolites 2020, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L.; Bai, Y.L.; Shu, S.L.; Qian, D.W.; Ou-yang, Z.; Liu, L.; Duan, J.A. Simultaneous quantitation of nucleosides, nucleobases, amino acids, and alkaloids in mulberry leaf by ultra high performance liquid chromatography with triple quadrupole tandem mass spectrometry. J. Sep. Sci. 2014, 37, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wei, M.; Guo, G.; Li, Y.; Pan, X.; Song, X.; Rong, L.; Chen, Y.; Chen, B. Analysis of Main Components in Jujube and Mulberry Extracts by High-Sensitive HPLC-ESI-Q-TOF-MS/MS. J. Chromatogr. Sci. 2021, 59, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ding, M.; Cui, C.; An, Q.; Wu, J.; Zhou, G.; Wan, Y.; Bao, W. Overexpression of a Gene Encoding Trigonelline Synthase from Areca catechu L. Promotes Drought Resilience in Transgenic Arabidopsis. Plants 2022, 11, 487. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, M.; Roy, S.; Ravichandran, K.; Mishra, B.; Gupta, D.K.; Nagarajan, S.; Arul Selvaraj, R.C.; Provaznik, I. In silico screening and molecular dynamics of phytochemicals from Indian cuisine against SARS-CoV-2 M(Pro). J. Biomol. Struct. Dyn. 2022, 40, 3155–3169. [Google Scholar] [CrossRef] [PubMed]
- Kırmızıbekmez, H.; Tatar, D.; Erdoğan, M.; Kúsz, N.; Hohmann, J. A new depside and a new secoiridoid from the aerial parts of Gentiana olivieri from flora of Turkey. Nat. Prod. Res. 2022, 36, 2208–2214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Z.; Luo, Z.; Chen, Y.; Huang, X. Insights into the conversion of dissolved organic phosphorus favors algal bloom, arsenate biotransformation and microcystins release of Microcystis aeruginosa. J. Environ. Sci. 2023, 125, 205–214. [Google Scholar] [CrossRef]
- Bati, B.; Celik, I.; Vuran, N.E.; Turan, A.; Alkan, E.E.; Zirek, A.K. Effects of Gundelia tournefortii L. on biochemical parameters, antioxidant activities and DNA damage in a rat model of experimental obesity. Braz. J. Biol. 2021, 83, e251198. [Google Scholar] [CrossRef]
- Adamiak, K.; Lewandowska, K.; Sionkowska, A. The Infuence of Salicin on Rheological and Film-Forming Properties of Collagen. Molecules 2021, 26, 1661. [Google Scholar] [CrossRef]
- Ilic-Tomic, T.; Sokovic, M.; Vojnovic, S.; Ciric, A.; Veljic, M.; Nikodinovic-Runic, J.; Novakovic, M. Diarylheptanoids from Alnus viridis ssp. viridis and Alnus glutinosa: Modulation of quorum sensing activity in Pseudomonas aeruginosa. Planta Med. 2017, 83, 117–125. [Google Scholar] [CrossRef]
- Luo, L.; Liu, X.; Jin, X.; Liu, Y.; Ma, J.; Zhang, S.; Zhang, D.; Chen, X.; Sheng, L.; Li, Y. Simultaneous determination of skimmin, apiosylskimmin, 7-hydroxycoumarin and 7-hydroxycoumarin glucuronide in rat plasma by liquid chromatography-Orbitrap mass spectrometry and its application to pharmacokinetics. Biomed. Chromatogr. 2022, 36, e5223. [Google Scholar] [CrossRef] [PubMed]
- Abe, C.; Soma, A.; Tun, T.N.N.; Zhang, Y.; Nishitani, Y.; Kayaki, H.; Kawakami, H.; Matsui, T. Pharmacokinetic profiles of 3-(4-hydroxy-3-methoxyphenyl) propionic acid and its conjugates in Sprague-Dawley rats. Biosci. Biotechnol. Biochem. 2023, 87, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Peng, L.; Fu, Y.; Wang, W.; Wang, P.; Zhou, F. Ginnalin A Binds to the Subpockets of Keap1 Kelch Domain To Activate the Nrf2-Regulated Antioxidant Defense System in SH-SY5Y Cells. ACS Chem. Neurosci. 2021, 12, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Bao, T.; Xu, Y.; Gowd, V.; Zhao, J.C.; Xie, J.H.; Liang, W.K.; Chen, W. Systematic study on phytochemicals and antioxidant activity of some new and common mulberry cultivars in China. J. Funct. Foods 2016, 25, 537–547. [Google Scholar] [CrossRef]
- Basit, A.; Ahmad, S.; Naeem, A.; Usman, M.; Ahmed, I.; Shahzad, M.N. Chemical profiling of Justicia vahlii Roth.(Acanthaceae) using UPLC-QTOF-MS and GC-MS analysis and evaluation of acute oral toxicity, antineuropathic and antioxidant activities. J. Ethnopharmacol. 2022, 287, 114942. [Google Scholar] [CrossRef] [PubMed]
- Mieres-Castro, D.; Theoduloz, C.; Sus, N.; Burgos-Edwards, A.; Schmeda-Hirschmann, G.; Frank, J.; Jiménez-Aspee, F. Iridoids and polyphenols from chilean Gaultheria spp. berries decrease the glucose uptake in Caco-2 cells after simulated gastrointestinal digestion. Food Chem. 2022, 369, 130940. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yin, X.; Sun, Y.-p.; Liu, Y.; Zhou, Y.-Y.; Pan, J.; Guan, W.; Yang, B.-Y.; Kuang, H.-X. Chemical constituent from the roots of Solanum melongena L. and their potential anti-inflammatory activity. Nat. Product. Res. 2022, 36, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dong, Y.; Song, R.; Yu, A.; Wei, J.; Fan, Q.; Yao, J.; Shan, D.; Zhong, X.; Lv, F.; et al. Intestinal metabolism and absorption mechanism of multi-components in Gaultheria leucocarpa var. yunnanensis—An assessment using in situ and in vitro models, comparing gut segments in pathological with physiological conditions. J. Ethnopharmacol. 2022, 286, 114844. [Google Scholar] [CrossRef]
- Viet Thanh, N.T.; Thu Hien, D.T.; Minh, T.T.; Nhiem, N.X.; Yen, P.H.; Van Kiem, P. Flavonol glycosides from Phoebe poilanei Kosterm. Vietnam. J. Chem. 2018, 56, 711–716. [Google Scholar] [CrossRef]
- Miyase, T.; Ueno, A.; Takizawa, N.; Kobayashi, H.; Oguchi, H. Ionone and lignan glycosides from Epimedium diphyllum. Phytochemistry 1989, 28, 3483–3485. [Google Scholar] [CrossRef]
- Zúñiga Santamaría, T.; Yescas Gómez, P.; Fricke Galindo, I.; González González, M.; Ortega Vázquez, A.; López López, M. Pharmacogenetic studies in Alzheimer disease. Neurologia 2022, 37, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Ju, W.T.; Kwon, O.C.; Kim, H.B.; Sung, G.B.; Kim, H.W.; Kim, Y.S. Qualitative and quantitative analysis of flavonoids from 12 species of Korean mulberry leaves. J. Food Sci. Technol. 2018, 55, 1789–1796. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ma, Q.-Y.; Liu, C.; Yang, J.; Lv, Q.-T.; Tian, Z.-H.; Jiang, H.-Q.; Rong, R. Three new C21 steroidal glycosides isolated from Metaplexis japonica and their potential inhibitory effects on tyrosine protein kinases. Nat. Product. Res. 2022, 36, 1988–1995. [Google Scholar] [CrossRef] [PubMed]
- Memon, A.A.; Memon, N.; Bhanger, M.I.; Luthria, D.L. Assay of phenolic compounds from four species of ber (Ziziphus mauritiana L.) fruits: Comparison of three base hydrolysis procedure for quantification of total phenolic acids. Food Chem. 2013, 139, 496–502. [Google Scholar] [CrossRef]
- Ma, X.; Ding, Q.; Hou, X.; You, X. Analysis of flavonoid metabolites in watercress (Nasturtium officinale R. Br.) and the non-heading Chinese cabbage (Brassica rapa ssp. chinensis cv. Aijiaohuang) using UHPLC-ESI-MS/MS. Molecules 2021, 26, 5825. [Google Scholar] [CrossRef] [PubMed]
- Khera, P.S.; Garg, P.K.; Tiwari, S.; Bhargava, N.; Yadav, T.; Sureka, B.; Ghosh, T.; Babu, S.; Dadhich, S.; Singh, S. Retrograde Transvenous Obliteration of Gastric Varices using Sodium Tetradecyl Sulphate: Technical Considerations and Results from a Single Institution Retrospective Study. J. Clin. Interv. Radiol. ISVIR 2021, 6, 10–17. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, X.; Hou, W.; Chen, E.; Ye, C.; Chen, M.; Lu, Q.; Yu, X.; Li, W. Reversing the imbalance in bone homeostasis via sustained release of SIRT-1 agonist to promote bone healing under osteoporotic condition. Bioact. Mater. 2023, 19, 429–443. [Google Scholar] [CrossRef]
- Jiang, X.; Yuan, Y.; Shi, M.; Zhang, S.; Sui, M.; Zhou, H. Bu-shen-zhu-yun decoction inhibits granulosa cell apoptosis in rat polycystic ovary syndrome through estrogen receptor α-mediated PI3K/AKT/mTOR pathway. J. Ethnopharmacol. 2022, 288, 114862. [Google Scholar] [CrossRef]
- Dunaway, S.; Fleischer, A.B., Jr. Assessing adherence to evidence-based guidelines of care for acne vulgaris. J. Dermatol. Treat. 2022, 33, 138–142. [Google Scholar] [CrossRef]
- Ramesh, S.; Khan, S.; Park, Y.; Ford, E.; Menegatti, S.; Genzer, J. Self-healing and repair of fabrics: A comprehensive review of the application toolkit. Mater. Today 2022, 54, 90–109. [Google Scholar] [CrossRef]
- Duan, H.; Wang, W.; Li, Y.; Jilany Khan, G.; Chen, Y.; Shen, T.; Bao, N.; Hua, J.; Xue, Z.; Zhai, K.; et al. Identification of phytochemicals and antioxidant activity of Premna microphylla Turcz. stem through UPLC-LTQ-Orbitrap-MS. Food Chem. 2022, 373, 131482. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Wang, Y.; Lu, A.; Zhang, Q.; Li, X.; Zhang, N.; Zhang, J.; Wang, S. From metabolomic analysis to quality assessment and biosynthetic insight in traditional Chinese medicine: Mulberry tree as a case study. Phytochem. Anal. 2022, 33, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Patil, T.V.; Patel, D.K.; Dutta, S.D.; Ganguly, K.; Santra, T.S.; Lim, K.T. Nanocellulose, a versatile platform: From the delivery of active molecules to tissue engineering applications. Bioact. Mater. 2022, 9, 566–589. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Hu, X. ZIF-derived CoFe2O4/Fe2O3 combined with g-C3N4 as high-efficient photocatalysts for enhanced degradation of levofloxacin in the presence of peroxymonosulfate. J. Alloys Compd. 2022, 914, 165338. [Google Scholar] [CrossRef]
- Liu, X.; Wu, M.; Wang, M.; Duan, Y.; Phan, C.U.; Chen, H.; Tang, G.; Liu, B. AIEgen-Lipid Conjugate for Rapid Labeling of Neutrophils and Monitoring of Their Behavior. Angew. Chem. 2021, 133, 3212–3218. [Google Scholar] [CrossRef]
- El-Hawwary, S.S.; Saber, F.R.; Abd Almaksoud, H.M.; Elimam, H.; Sayed, A.M.; Abdelmohsen, U.R. Cytotoxic potential of three Sabal species grown in Egypt: A metabolomic and docking-based study. Nat. Product. Res. 2022, 36, 1109–1114. [Google Scholar] [CrossRef]
- Ortiz-Alvarez, L.; Xu, H.; Di, X.; Kohler, I.; Osuna-Prieto, F.J.; Acosta, F.M.; Vilchez-Vargas, R.; Link, A.; Plaza-Díaz, J.; van der Stelt, M.; et al. Plasma Levels of Endocannabinoids and Their Analogues Are Related to Specific Fecal Bacterial Genera in Young Adults: Role in Gut Barrier Integrity. Nutrients 2022, 14, 2143. [Google Scholar] [CrossRef]
- Choi, H.J.; Naznin, M.; Alam, M.B.; Javed, A.; Alshammari, F.H.; Kim, S.; Lee, S.H. Optimization of the extraction conditions of Nypa fruticans Wurmb. using response surface methodology and artificial neural network. Food Chem. 2022, 381, 132086. [Google Scholar] [CrossRef]
- Kumar, J.B.S.; Sharma, B. A review on neuropharmacological role of erucic acid: An omega-9 fatty acid from edible oils. Nutr. Neurosci. 2022, 25, 1041–1055. [Google Scholar] [CrossRef]
- Ahmed, A.S.; Diab, H.M.; Alkahtani, M.A.; Alshehri, M.A.; Saber, H.; Badr, H.; Dandrawy, M.K.; El-Mansi, A.A.; Shati, A.A.; Ahmed, A.E. Molecular epidemiology of virulent E. coli among rural small scale dairy herds and shops: Efficacy of selected marine algal extracts and disinfectants. Int. J. Environ. Health Res. 2022, 32, 72–94. [Google Scholar] [CrossRef] [PubMed]
- Eldeigdhye, S.M.; Abdelaleem, M.A.; Mater, S.N.A.; Kamal, A.M. The bioprotective role of irradiated hibiscus against harmful effects of using repeatedly frying edible oil. Braz. J. Biol. 2021, 84, e253084. [Google Scholar] [CrossRef] [PubMed]
- Chebbi, M.; Beltifa, A.; Alibi, S.; Di Bella, G.; Loturco, V.; Sire, O.; Ben Mansour, H.; Le Tilly, V. Estrogenic hazards of short chain phthalates and bisphenols found in cosmetic products. Int. J. Environ. Health Res. 2022, 32, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Moazezi, M.R.; Bayat, H.; Tavakoli, O.; Hallajisani, A. Hydrothermal liquefaction of Chlorella vulgaris and catalytic upgrading of product: Effect of process parameter on bio-oil yield and thermodynamics modeling. Fuel 2022, 318, 123595. [Google Scholar] [CrossRef]
- De Lima, R.M.T.; Dos Reis, A.C.; de Oliveira Santos, J.V.; de Oliveira Ferreira, J.R.; de Oliveira Filho, J.W.G.; Dias, A.C.S.; de Menezes, A.-A.P.M.; da Mata, A.M.O.F.; de Alencar, M.V.O.B.; Dos Santos, T.d.J.A. Antitumoral effects of [6]-gingerol [(S)-5-hydroxy-1-(4-hydroxy-3-methoxyphenyl)-3-decanone] in sarcoma 180 cells through cytogenetic mechanisms. Biomed. Pharmacother. 2020, 126, 110004. [Google Scholar] [CrossRef] [PubMed]
- Yilmazer-Musa, M.; Griffith, A.M.; Michels, A.J.; Schneider, E.; Frei, B. Grape seed and tea extracts and catechin 3-gallates are potent inhibitors of α-amylase and α-glucosidase activity. J. Agric. Food Chem. 2012, 60, 8924–8929. [Google Scholar] [CrossRef] [PubMed]
- Weckwerth, W.; Wenzel, K.; Fiehn, O. Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 2004, 4, 78–83. [Google Scholar] [CrossRef]
- Guo, M.; Wu, Z.; An, Q.; Li, H.; Wang, L.; Zheng, Y.; Guo, L.; Zhang, D. Comparison of Volatile Oils and Primary Metabolites of Raw and Honey-Processed Ephedrae Herba by GC–MS and Chemometrics. J. AOAC Int. 2022, 105, 576–586. [Google Scholar] [CrossRef]
- Biswas, A.; Dey, S.; Xiao, A.; Huang, S.; Birhanie, Z.M.; Deng, Y.; Liu, L.; Li, D. Phytochemical content and antioxidant activity of different anatomical parts of Corchorus olitorius and C. capsularis during different phenological stages. Heliyon 2023, 9, e16494. [Google Scholar] [CrossRef]
- Guo, W.; Xiao, Y.; Fu, X.; Long, Z.; Wu, Y.; Lin, Q.; Ren, K.; Jiang, L. Identification of novel α-glucosidase and ACE inhibitory peptides from Douchi using peptidomics approach and molecular docking. Food Chem. X 2023, 19, 100779. [Google Scholar] [CrossRef]

| No. | tR/min | Metabolite Name | m/z | Molecular Formula |
|---|---|---|---|---|
| 1 | 3.976 | Liactic Acid, 2TMS derivative | 219.1 | C9H22O3Si2 |
| 2 | 4.096 | Glycolic acid, 2TMS derivative | 218.1 | C8H20O3Si2 |
| 3 | 4.215 | L-Valine, TMS derivative | 217 | C8H19NO2Si |
| 4 | 4.355 | L-Alanine, 2TMS derivative | 232.1 | C9H23NO2Si2 |
| 5 | 4.744 | Hydracrylic acid, 2TMS derivative | 235.1 | C9H22O3Si2 |
| 6 | 5.445 | Propanedioic acid, 2TMS derivative | 248.1 | C9H20O4Si2 |
| 7 | 5.626 | Valine, 2TMS derivative | 246.1 | C11H27NO2Si2 |
| 8 | 5.823 | Urea, 2TMS derivative | 233.1 | C7H20N2OSi2 |
| 9 | 6.316 | Ethanolamine, 3TMS derivative | 262.1 | C11H31NOSi3 |
| 10 | 6.415 | Glycerol, 3TMS derivative | 314.11 | C12H32O3Si3 |
| 11 | 6.669 | Niacin, TMS derivative | 301 | C9H13NO2Si |
| 12 | 6.778 | L-Proline, 2TMS derivative | 314.1 | C11H25NO2Si2 |
| 13 | 6.944 | Butanedioic acid, 2TMS derivative | 299 | C10H22O4Si2 |
| 14 | 7.313 | Glyceric acid, 3TMS derivative | 322.1 | C12H30O4Si3 |
| 15 | 7.452 | 2-Butenedioic acid (E)-, 2TMS derivative | 299 | C10H20O4Si2 |
| 16 | 7.805 | Serine, 3TMS derivative | 306.1 | C12H31NO3Si3 |
| 17 | 8.288 | L-Threonine, 3TMS derivative | 320.1 | C13H33NO3Si3 |
| 18 | 8.407 | Ethanolamine, 3TMS derivative | 299 | C11H31NOSi3 |
| 19 | 8.952 | Beta-Alanine, 3TMS derivative | 299.1 | C12H31NO2Si3 |
| 20 | 10.265 | Malic acid, 3TMS derivative | 350.1 | C13H30O5Si3 |
| 21 | 10.924 | Pipecolic acid, 2TMS derivative | 258.1 | C12H27NO2Si2 |
| 22 | 11.095 | 4-Aminobutanoic acid, 3TMS de rivative | 332.1 | C13H33NO2Si3 |
| 23 | 12.849 | 5-Hydroxypipecolic acid, 3TMS derivative | 360.2 | C15H35NO3Si3 |
| 24 | 13.352 | L-Glutamic acid, 3TMS derivative | 363.11 | C14H33NO4Si3 |
| 25 | 13.430 | L-Phenylalanine, 2TMS derivative | 331.1 | C15H27NO2Si2 |
| 26 | 14.748 | Asparagine, 3TMS derivative | 348.2 | C13H32N2O3Si3 |
| 27 | 15.791 | D-Lyxose, 4TMS derivative | 333.2 | C17H42O5Si4 |
| 28 | 17.088 | Glycerol, 3TMS derivative | 394.2 | C12H32O3Si3 |
| 29 | 18.899 | Shikimic acid, 4TMS derivative | 462.2 | C19H42O5Si4 |
| 30 | 19.200 | Citric acid, 4TMS derivative | 465.2 | C18H40O7Si4 |
| 31 | 20.606 | Quininic acid, 5TMS derivative | 462.2 | C22H52O6Si5 |
| 32 | 21.087 | Alloxanic acid, 4TMS derivative | 446.2 | C16H36N2O5Si4 |
| 33 | 22.303 | Methyl-α-Lyxofuranoside, 3TMS derivative | 446.3 | C15H36O5Si3 |
| 34 | 25.032 | Palmitic Acid, TMS derivative | 456.1 | C19H40O2Si |
| 35 | 27.237 | Myo-Inositol, 6TMS derivative | 432.2 | C24H60O6Si6 |
| 36 | 28.545 | Galactose oxime, 6TMS derivative | 319.2 | C24H61NO6Si6 |
| 37 | 29.676 | Tryptophan, 4TMS derivative | 337.2 | C23H44N2O2Si4 |
| 38 | 29.873 | Oleic Acid (Z)-, TMS derivative | 354.3 | C21H42O2Si |
| 39 | 30.340 | L-Rhamnose, 4TMS derivative | 361.2 | C18H44O5Si4 |
| 40 | 30.605 | Stearic acid, TMS derivative | 356.3 | C21H44O2Si |
| 41 | 34.081 | 2-O-Glycerol-α-d-galactopyranoside, hexa-, TMS | 361.2 | C27H66O8Si6 |
| 42 | 38.689 | Ethyl-α-d-glucopyranoside, 4TMS derivative | 439.2 | C20H48O6Si4 |
| 43 | 39.275 | 1-Monopalmitin, 2TMS derivative | 459.3 | C25H54O4Si2 |
| 44 | 41.268 | Sucrose, 8TMS derivative | 481.3 | C36H86O11Si8 |
| 45 | 41.729 | Lactose, 8TMS derivative | 450.2 | C36H86O11Si8 |
| 46 | 43.275 | Glycerol monostearate, 2TMS derivative | 487.3 | C27H58O4Si2 |
| No. | tR/min | Precursor Ion Type | Precursor Ion (m/z) | Formula | Fragment Ions (m/z) | Identification | Ref. |
|---|---|---|---|---|---|---|---|
| 1 | 1.06 | [M + H]+ | 175.1189 | C6H14N4O2 | 130.0973, 116.0701, 70.0651, 60.0556 | Arginine | [21,24] |
| 2 | 1.079 | [M + H]+ | 164.0916 | C6H13NO4 | 146.0803, 128.0703, 110.0596, 82.0651, 80.0495, 69.0335 | 1-Deoxynojirimycin (1-DNJ) | [21,24,23] |
| 3 | 1.089 | [M + H]+ | 156.0420 | C6H9N3O2 | 110.0715, 93.0440, 82.0261 | Histidine | [21,24] |
| 4 | 1.172 | [M + H]+ | 146.0810 | C6H11NO3 | 128.0701, 100.0755 | N-isobutyrylglycine | [21] |
| 5 | 1.179 | [M + NH4]+ | 360.1504 | C12H22O11 | 101.0232, 71.0491 | Sucrose | [25] |
| 6 | 1.202 | [M + H]+ | 116.0707 | C5H9NO2 | 116.0706, 70.0652 | Proline | [21,24] |
| 7 | 1.22 | [M + H]+ | 137.9750 | C7H7NO2 | 138.0546, 94.0650, 92.0492, 78.0336 | Trigonelline | [26] |
| 8 | 1.31 | [M + H]+ | 314.0920 | C11H15N5O4S | 164.0565, 136.0618, 97.0248 | (S)-5′-Deoxy-5′-(methylsulfinyl)adenosine | [27] |
| 9 | 1.308 | [M + H]+ | 358.1498 | C16H20O9 | 196.0965, 178.0859, 150.0906 | Gentiopicroside | [28] |
| 10 | 1.375 | [M + H]+ | 268.1043 | C10H13N5O4 | 137.0646, 136.0617 | Adenosine | [29] |
| 11 | 1.463 | [M + H]+ | 132.1018 | C6H13NO2 | 86.0964, 69.0697 | Isoleucine | [21,24] |
| 12 | 1.82 | [M + H]+ | 284.0989 | C10H13N5O5 | 286.1451, 154.0846, 153.0584, 152.514 | Guanosine | [30] |
| 13 | 1.887 | [M + H]+ | 287.1102 | C13H18O7 | 107.0296 | Salicin | [31] |
| 14 | 2.413 | [M + H]+ | 166.0862 | C9H11NO2 | 103.0539, 121.0842, 120.0805, 107.0494 | Phenylalanine | [24] |
| 15 | 2.477 | [M + H]+ | 120.0809 | C4H9NO3 | 119.0705, 102.0116, 84.9590, 75.9350 | l-Threonine | [21] |
| 16 | 2.536 | [M + H]+ | 220.1177 | C9H17NO5 | 184.0949, 116.0323, 90.0548 | Pantothenic acid | [21] |
| 17 | 3.431 | [M + K]+ | 367.1500 | C19H20O5 | 131.0680, 103.0388 | Hirsutanone | [32] |
| 18 | 3.452 | [M + H]+ | 298.0964 | C11H15N5O3S | 163.0414, 145.0314, 136.0615 | Vitamin L2 | [21] |
| 19 | 4.08 | [M + H]+ | 205.0973 | C11H12N2O2 | 188.07606, 159.0917, 146.0599, 144.0808 | Tryptophan | [24] |
| 20 | 4.25 | [M + H]+ | 341.0867 | C15H16O9 | 180.0373, 179.0336, 133.0874 | Esculin | [21] |
| 21 | 4.446 | [M + H]+ | 325.0920 | C15H16O8 | 164.0425, 163.0387, 107.0494 | Skimmin | [33] |
| 22 | 4.816 | [M − H]+ | 194.1150 | C10H12O4 | 195.1212, 136.0472, 135.0435, 59.0714 | 3-(4-Hydroxy-3-methoxyphenyl) propionic acid | [34] |
| 23 | 4.999 | [M + H]+ | 627.1554 | C27H30O17 | 466.1064, 465.1033, 304.0536, 303.0498, 85.0281 | Quercetin 3,4′-diglucoside | [23] |
| 24 | 5.08 | [M + H]+ | 773.2125 | C33H40O21 | 465.1025, 304.0517, 303.0489, 85.0275 | Quercetin-3-O-rutinoside-7-O-glucoside | [23] |
| 25 | 5.11 | [M + H]+ | 467.1174 | C20H20O13 | 154.0219, 153.0171 | Ginnalin A | [35] |
| 26 | 5.189 | [M + H]+ | 355.1027 | C16H18O9 | 163.0388, 145.0281, 135.0438, 117.0334 | Chlorogenic acid | [21,36] |
| 27 | 5.276 | [M + NH4]+ | 344.1340 | C15H18O8 | 165.0538, 119.0482, 91.0407 | E-4-O-β-d-glucopyranosyl-p-coumaric acid | [37] |
| 28 | 5.575 | [M + H-H2O]+ | 518.1870 | C25H28O13 | 148.0472, 147.0435, 119.0484, 91.0531 | Vaccinoside | [38] |
| 29 | 5.74 | [M + NH4]+ | 288.1400 | C13H18O6 | 213.0523, 138.9628 | Benzyl beta-d-glucopyranoside | [39] |
| 30 | 5.983 | [M + H]+ | 713.1558 | C30H32O20 | 551.1035, 465.1018, 303.0498 | Quercetin 3-O-(6″-malonyl-glucoside) 7-O-glucoside | [21,23] |
| 31 | 7.27 | [M + H]+ | 611.1604 | C27H31O16+ | 615.1748, 613.1666, 612.1619, 611.1609 | Cyanin | [27] |
| 32 | 7.298 | [M + H]+ | 339.1070 | C16H18O8 | 148.0469, 147.0438, 120.0520, 119.0487, 91.0533 | 4-p-Coumaroylquinic acid | [40] |
| 33 | 7.46 | [M + H]+ | 697.1603 | C30H32O19 | 697.1054, 449.1069, 288.0577, 287.0547 | Kaempferol hexoside malonyl hexoside | [21,23] |
| 34 | 8.349 | [M + H]+ | 757.2164 | C33H40O20 | 611.1635, 465.0997, 303.0492 | Quercetin-3-O-rutinoside-7-Orhamnoside | [21,23] |
| 35 | 8.36 | [M + H]+ | 757.2164 | C34H42O19 | 85.0281, 71.0344 | Oxytroflavoside G | [41] |
| 36 | 8.377 | [M + NH4]+ | 434.2020 | C19H28O10 | 145.0479, 133.0550, 115.0369, 97.0284, 85.0279 | Sayaendoside | [42] |
| 37 | 8.41 | [M + H]+ | 303.0500 | C15H10O7 | 257.0420, 229.0490, 165.0149, 153.0172, 137.0226 | Quercetin | [21,22] |
| 38 | 9.457 | [M + H]+ | 180.1017 | C12H21N | 180.1023, 163.0960, 135.1002, 121.0835 | Memantine | [43] |
| 39 | 9.73 | [M + H]+ | 611.1600 | C27H30O16 | 465.9610, 303.0491 | Quercetin 3-O-rutinoside (rutin) | [21,44] |
| 40 | 11.741 | [M + H]+ | 465.1206 | C21H20O12 | 305.0577, 304.0529, 303.0492 | Isoquercitin | [22] |
| 41 | 13.02 | [M + H]+ | 373.2220 | C19H32O7 | 135.1148, 109.1007 | Byzantionoside B | [45] |
| 42 | 13.048 | [M + H]+ | 595.1604 | C27H30O15 | 287.0547, 129.0562, 127.0406, 85.0299, 71.0488 | Kaempferol-3-O-rutinoside | [23] |
| 43 | 13.084 | [M + H]+ | 595.1649 | C27H30O15 | 287.0535, 129.0521, 127.0404, 85.0274, 71.0489 | Nicotiflorin | [23] |
| 44 | 13.151 | [M + H]+ | 551.1207 | C24H22O15 | 304.0525, 303.0491, 159.0282, 127.0382, 109.0280 | Quercetin 3-O-6′-malonylglucoside | [19,46] |
| 45 | 14.04 | [M + H]+ | 287.0548 | C15H10O6 | 213.0570, 165.0185, 153.01732, 121.0267 | Kaempferol | [21] |
| 46 | 14.045 | [M + H]+ | 449.1079 | C21H20O11 | 451.3095, 289.0580, 288.0580, 287.0545 | Kaempferol 3-O-glucoside (astragalin) | [21,23,44] |
| 47 | 14.809 | [M + H]+ | 507.1124 | C23H22O13 | 507.3026, 303.0493 | Quercetin 3-(6″-O-acetylgalactoside) | [47] |
| 48 | 16.012 | [M + H]+ | 535.1080 | C24H22O14 | 289.0606, 288.0571, 287.0544 | Kaempferol malonyl hexoside | [23] |
| 49 | 20.35 | [M + H]+ | 316.3207 | C14H29NaO4S | 319.1866, 317.3239, 316.3207, 106.0861 | Sodium tetradecyl sulfate | [48] |
| 50 | 20.81 | [M + Na]+ | 264.2320 | C14H8O4 | 203.1431 | Alizarin | [49] |
| 51 | 21.11 | [M + H]+ | 361.1389 | C21H28O5 | 361.1395, 344.1892, 325.1344, 307.1182 | Prednisolone | [23] |
| 52 | 21.66 | [M + H]+ | 291.1953 | C19H30O2 | 273.1829, 161.1278, 147.1172 | Epiandrosterone | [50] |
| 53 | 22.22 | [M + K]+ | 225.5670 | C9H16O4 | 125.0944, 123.1161, 97.1005 | Azelaic acid | [51] |
| 54 | 22.277 | [M + H]+ | 274.2741 | C16H35NO2 | 274.2740, 256.2633 | N-Lauryldiethanolamine | [52] |
| 55 | 24.615 | M + ACN + H | 432.2378 | C22H30O6 | 135.0799, 119.0852, 107.0861 | Prostratin | [53] |
| 56 | 25.95 | [M + H]+ | 423.1803 | C25H26O6 | 423.1795, 311.0543, 241.0496 | Mulberrin | [54] |
| 57 | 26.139 | [M + H]+ | 302.3051 | C18H39NO2 | 302.2048, 284.2936, 106.0851, 102.0911, 88.0755 | Tetradecyldiethanolamine | [55] |
| 58 | 26.216 | [M + H]+ | 302.3053 | C18H39NO2 | 302.3050, 284.2935, 106.0861, 88.0752 | Ethanol, 2,2′-(tetradecylimino)bis- | [56] |
| 59 | 31.35 | [M + H]+ | 496.3389 | C24H50NO7P | 478.3274, 258.1112, 184.0726 | 1-Palmitoyl-sn-glycero-3-phosphocholine | [57] |
| 60 | 33.871 | [M + H]+ | 317.1800 | C14H29NaO4S | 317.3232, 106.0857 | Sodium myristyl sulfate | [58] |
| 61 | 34.09 | [M + H]+ | 322.2739 | C20H35NO2 | 322.2739, 261.2212, 243.2112 | α-Linolenoyl ethanolamide | [59] |
| 62 | 34.23 | [M + H]+ | 421.1640 | C25H24O6 | 365.1019, 323.0605 | Kuwanon A | [54,60] |
| 63 | 34.27 | [M + H]+ | 326.2687 | C20H39NO2 | 326.3762, 310.0664 | N-oleoylethanolamine | [61] |
| 64 | 34.814 | [M + K]+ | 470.4199 | C21H20O10 | 283.1673 | Isovitexin | [58] |
| 65 | 34.99 | [M + H]+ | 407.1852 | C25H26O5 | 283.0596, 255.0648 | Rubraflavone A | [55] |
| 66 | 36.10 | [M + H]+ | 403.2328 | C20H34O8 | 259.1531, 157.0129, 185.0806, 139.0022, 129.0180 | Tributyl acetylcitrate | [62] |
| 67 | 36.95 | [M +H]+ | 282.2790 | C18H34O2 | 69.0698, 57.0307, 55.0540 | cis-Octadecenoic acid | [63] |
| 68 | 38.90 | [M + H]+ | 391.2838 | C24H38O4 | 167.0331, 149.0230, 71.0854, 57.0698 | Bis(2-ethylhexyl) phthalate | [64] |
| 69 | 39.363 | [M + H]+ | 284.2942 | C18H37NO | 285.2801, 284.2927 | N,N-dimethylpalmitamide | [65] |
| 70 | 39.47 | [M + H]+ | 352.3200 | C21H34O4 | 177.1252, 137.0933 | 5-hydroxy-1-(4-hydroxy-3-methoxyphenyl) tetradecan-3-one | [66] |
| Compound | Linearity Range (mg/mL) | Calibration Equation | Correlation Factor (r) |
|---|---|---|---|
| Mulberroside A | 0.0003~0.006 | y = 5,921,740.33 x + 467.35 | 0.9995 |
| Chlorogenic acid | 0.00295~0.0944 | y = 9,556,420.73 x + 3459.11 | 0.9998 |
| Cryptochlorogenic acid | 0.001~0.032 | y = 10,673,897.94 x + 641.53 | 0.9999 |
| Rutin | 0.00215~0.086 | y = 16,281,522.08 x − 332.238 | 0.9999 |
| Isoquercitrin | 0.0016~0.064 | y = 27,860,160.12 x − 666.51 | 0.9999 |
| Oxidized resveratrol | 0.0013~0.052 | y = 15,236,962.42 x − 989.82 | 0.9999 |
| No. | Collecting Time | No. | Collecting Time |
|---|---|---|---|
| Y1 | 13 April 2018 | Y15 | 23 August 2018 |
| Y2 | 20 April 2018 | Y16 | 1 September 2018 |
| Y3 | 27 April 2018 | Y17 | 8 September 2018 |
| Y4 | 4 May 2018 | Y18 | 15 September 2018 |
| Y5 | 11 May 2018 | Y19 | 21 September 2018 |
| Y6 | 25 May 2018 | Y20 | 28 September 2018 |
| Y7 | 1 June 2018 | Y21 | 5 October 2018 |
| Y8 | 8 June 2018 | Y22 | 11 October 2018 |
| Y9 | 22 June 2018 | Y23 | 18 October 2018 |
| Y10 | 6 July 2018 | Y24 | 29 October 2018 |
| Y11 | 13 July 2018 | Y25 | 3 November 2018 |
| Y12 | 23 July 2018 | Y26 | 9 November 2018 |
| Y13 | 1 August 2018 | Y27 | 15 November 2018 |
| Y14 | 8 August 2018 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Hao, S.; Li, H.; An, Q.; Yang, L.; Guo, B.; Xue, Z.; Liu, Y.; Guo, L.; Zheng, Y.; et al. Exploring Antioxidant and α-Glucosidase Inhibitory Activities in Mulberry Leaves (Morus alba L.) across Growth Stages: A Comprehensive Metabolomic Analysis with Chemometrics. Molecules 2024, 29, 171. https://doi.org/10.3390/molecules29010171
Li W, Hao S, Li H, An Q, Yang L, Guo B, Xue Z, Liu Y, Guo L, Zheng Y, et al. Exploring Antioxidant and α-Glucosidase Inhibitory Activities in Mulberry Leaves (Morus alba L.) across Growth Stages: A Comprehensive Metabolomic Analysis with Chemometrics. Molecules. 2024; 29(1):171. https://doi.org/10.3390/molecules29010171
Chicago/Turabian StyleLi, Wenjie, Shenghui Hao, Hengyang Li, Qi An, Lina Yang, Bing Guo, Zijing Xue, Yongli Liu, Long Guo, Yuguang Zheng, and et al. 2024. "Exploring Antioxidant and α-Glucosidase Inhibitory Activities in Mulberry Leaves (Morus alba L.) across Growth Stages: A Comprehensive Metabolomic Analysis with Chemometrics" Molecules 29, no. 1: 171. https://doi.org/10.3390/molecules29010171
APA StyleLi, W., Hao, S., Li, H., An, Q., Yang, L., Guo, B., Xue, Z., Liu, Y., Guo, L., Zheng, Y., & Zhang, D. (2024). Exploring Antioxidant and α-Glucosidase Inhibitory Activities in Mulberry Leaves (Morus alba L.) across Growth Stages: A Comprehensive Metabolomic Analysis with Chemometrics. Molecules, 29(1), 171. https://doi.org/10.3390/molecules29010171









