Comparison of Antibacterial Activities of Korean Pine (Pinus densiflora) Needle Steam Distillation Extract on Escherichia coli and Staphylococcus aureus Focusing on Membrane Fluidity and Genes Involved in Membrane Lipids and Stress
Abstract
:1. Introduction
2. Results
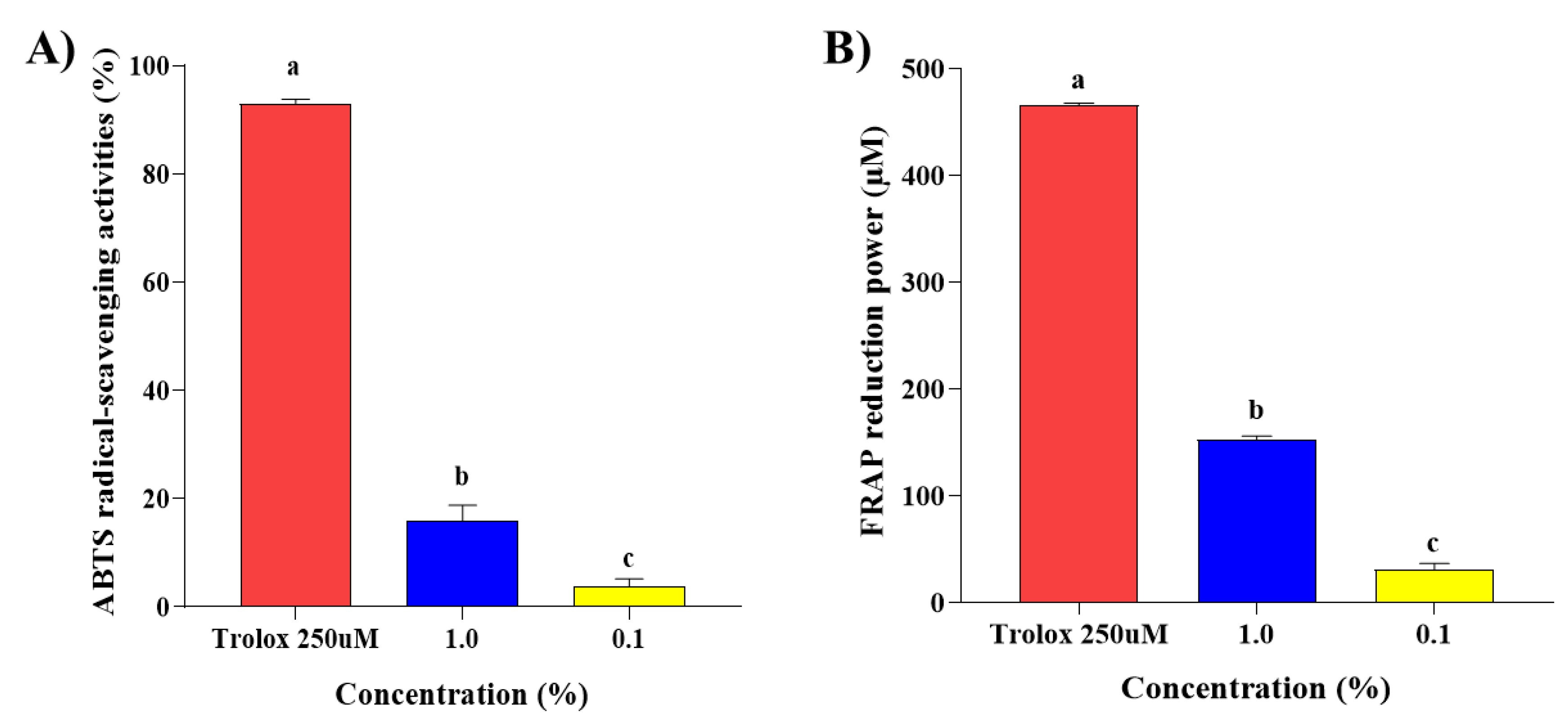
2.1. Antioxidant Effects of Steam Distilled Pine Needle Extracts
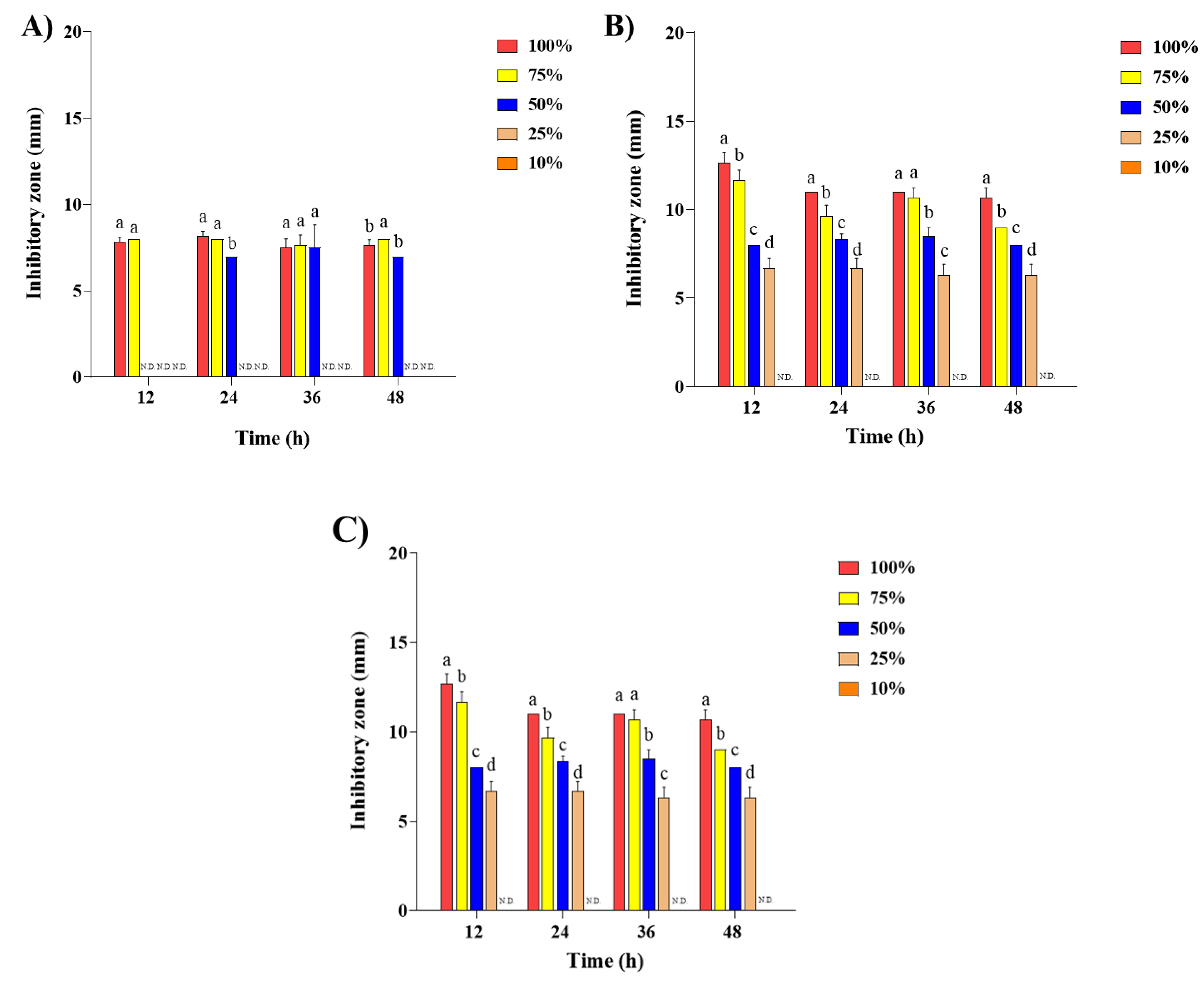
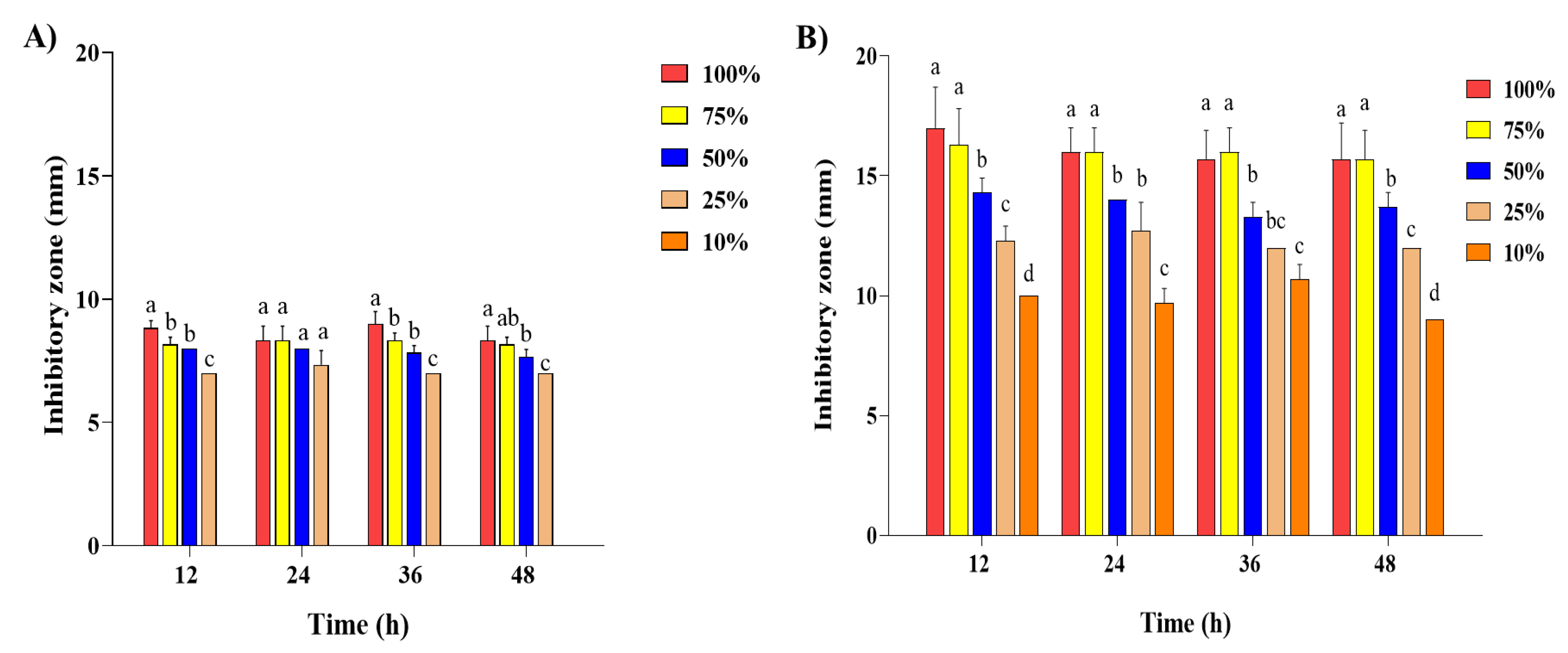
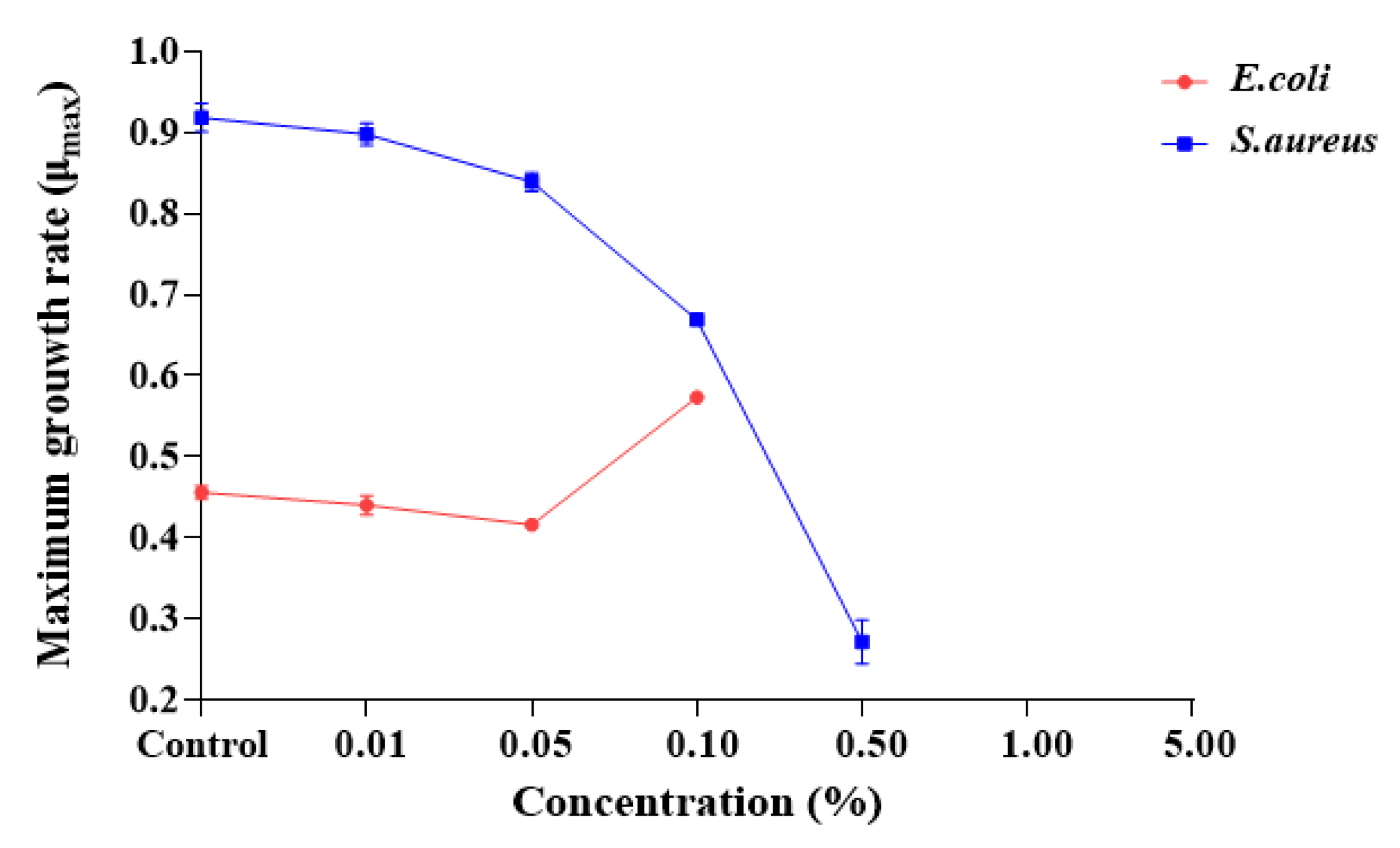
2.2. Antibacterial Activity of Steam Distilled Pine Needle Extract
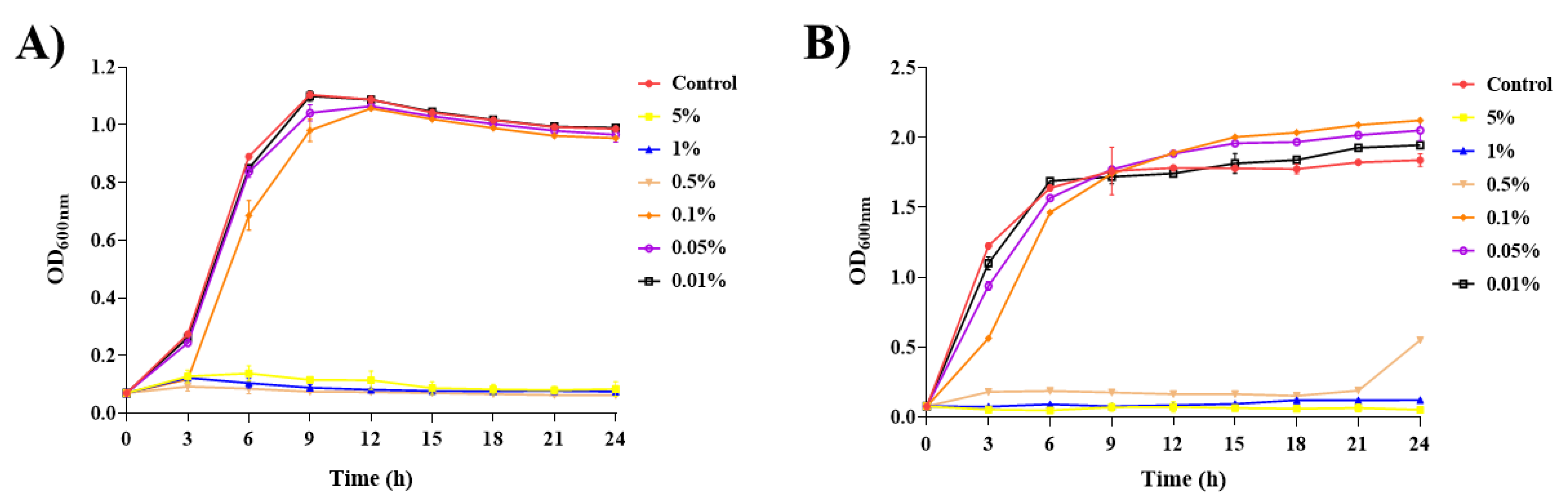
2.3. Membrane Fluidity Changes under SDPNE Treatments
2.4. Membrane Fatty Acid Compositions of Escherichia coli and Staphylococcus aureus with SDPNE
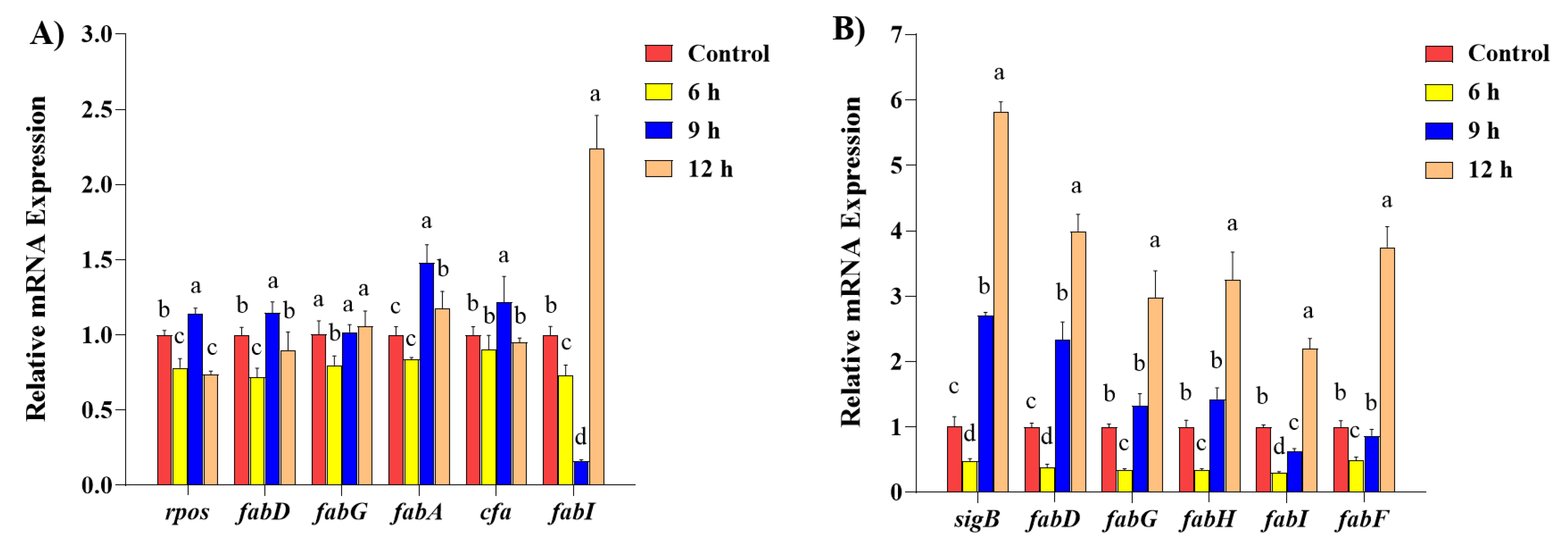
2.5. The Transcription of Fatty Acid Biosynthesis Gene
3. Discussion
4. Materials and Methods
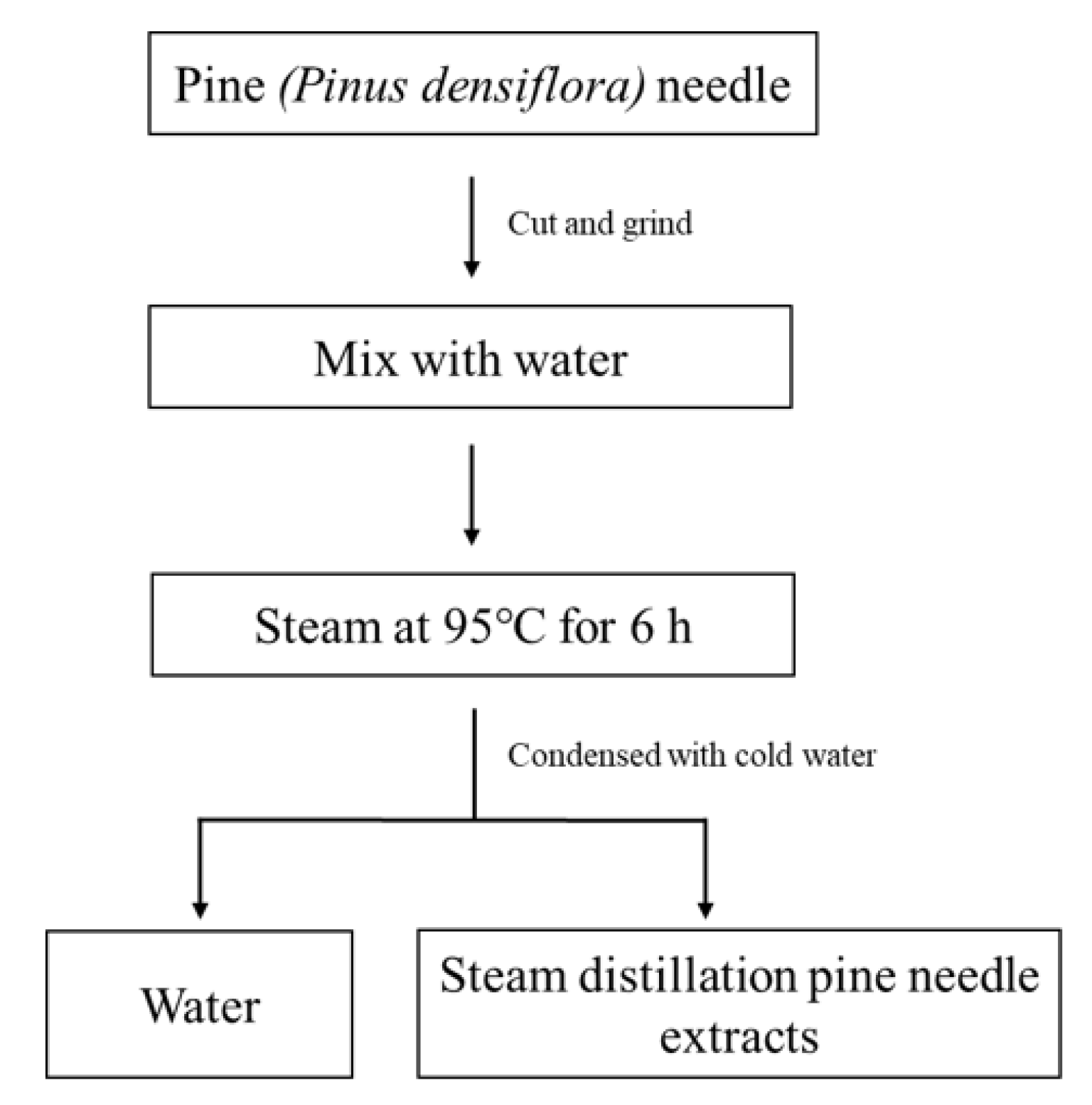
4.1. Sample Preparation
4.2. Total Phenolic Content
4.3. Total Flavonoid Content
4.4. 2,2′-Azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) Radical Scavenging Activity
4.5. Ferric Reducing Antioxidant Power (FRAP) Assay
4.6. Microorganisms and Materials
4.7. Antibacterial Activity
4.8. Measurement of Bacterial Growth
4.9. Membrane Fluidity Analysis
4.10. Membrane Lipid Extraction
4.11. Total RNA and Gene Expression Analysis (Realtime qRT-PCR)
4.12. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duan, X.; Jia, Z.; Li, J.; Wu, S. The influencing factors of leaf functional traits variation of Pinus densiflora Sieb. et Zucc. GECCO 2022, 38, e02177. [Google Scholar] [CrossRef]
- Lim, J.H.; Kim, J.C.; Kim, K.J.; Son, Y.S.; Sunwoo, Y.; Han, J.S. Seasonal variations of monoterpene emissions from Pinus densiflora in East Asia. Chemosphere 2008, 73, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cho, K.S.; Jeon, Y.; Kim, J.B.; Lim, Y.R.; Lee, K.; Lee, I.S. Characteristics and distribution of terpenes in South Korean forests. J. Ecol. Environ. 2017, 41, 19. [Google Scholar] [CrossRef]
- Lee, J.; Kang, H.K.; Cheong, H.; Park, Y. A novel antimicrobial peptides from pine needles of Pinus densiflora Sieb. et Zucc. against foodborne bacteria. Front. Microbiol. 2021, 12, 662462. [Google Scholar]
- Feng, S.; Zeng, W.; Luo, F.; Zhao, J.; Yang, Z.; Sun, Q. Antibacterial activity of organic acids in aqueous extracts from pine needles (Pinus massoniana Lamb.). Food Sci. Biotechnol. 2010, 19, 35–41. [Google Scholar] [CrossRef]
- Rana, A.K.; Guleria, S.; Gupta, V.K.; Thakur, V.K. Cellulosic pine needles-based biorefinery for a circular bioeconomy. Bioresour. Technol. 2022, 367, 128255. [Google Scholar] [CrossRef] [PubMed]
- Metsämuuronen, S.; Sirén, H. Bioactive phenolic compounds, metabolism and properties: A review on valuable chemical compounds in Scots pine and Norway spruce. Phytochem. Rev. 2019, 18, 623–664. [Google Scholar] [CrossRef]
- Jeon, Y.H.; Seo, J.E.; Kim, J.H.; Lee, Y.J.; Choi, S.W. Quantitative changes of flavonol glycosides from pine needles by cultivar, harvest season, and thermal process. Prev. Nutr. Food Sci. 2021, 26, 100. [Google Scholar] [CrossRef]
- Cushnie, T.T.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents. 2005, 26, 343–356. [Google Scholar] [CrossRef]
- Tarahovsky, Y.S.; Kim, Y.A.; Yagolnik, E.A.; Muzafarov, E.N. Flavonoid–membrane interactions: Involvement of flavonoid–metal complexes in raft signaling. Biochim. Biophys. Acta Biomembr. BBA-Biomembr. 2014, 1838, 1235–1246. [Google Scholar] [CrossRef]
- Kopeć, W.; Telenius, J.; Khandelia, H. Molecular dynamics simulations of the interactions of medicinal plant extracts and drugs with lipid bilayer membranes. FEBS J. 2013, 280, 2785–2805. [Google Scholar] [CrossRef] [PubMed]
- Hendrich, A.B. Flavonoid-membrane interactions: Possible consequences for biological effects of some polyphenolic com—Pounds 1. Acta Pharmacol. Sin. 2006, 27, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Sun, Z.; Chen, W. Antimicrobial susceptibility and antibacterial mechanism of limonene against Listeria monocytogenes. Molecules 2019, 25, 33. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M.; Walker, C.; Epand, R.F.; Magarvey, N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Biophys. Acta Biomembr. BBA-Biomembr. 2016, 1858, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Mingeot-Leclercq, M.P.; Décout, J.L. Bacterial lipid membranes as promising targets to fight antimicrobial resistance, molecular foundations and illustration through the renewal of aminoglycoside antibiotics and emergence of amphiphilic aminoglycosides. MedChemComm 2016, 7, 586–611. [Google Scholar] [CrossRef]
- Siliakus, M.F.; van der Oost, J.; Kengen, S.W. Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 2017, 21, 651–670. [Google Scholar] [CrossRef] [PubMed]
- Murínová, S.; Dercová, K. Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int. J. Microbiol. 2014, 2014, 873081. [Google Scholar] [CrossRef]
- Hassan, N.; Anesio, A.M.; Rafiq, M.; Holtvoeth, J.; Bull, I.; Haleem, A.; Hasan, F. Temperature driven membrane lipid adaptation in glacial psychrophilic bacteria. Front. Microbiol. 2020, 11, 824. [Google Scholar] [CrossRef]
- Belete, T.M. Novel targets to develop new antibacterial agents and novel alternatives to antibacterial agents. Hum. Microbiome J. 2019, 11, 100052. [Google Scholar] [CrossRef]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482. [Google Scholar] [CrossRef]
- Rodríguez Ayala, F.; Bartolini, M.; Grau, R. The stress-responsive alternative sigma factor SigB of Bacillus subtilis and its relatives: An old friend with new functions. Front. Microbiol. 2020, 11, 1761. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S. Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. JBC 2019, 294, 11685–11700. [Google Scholar] [CrossRef] [PubMed]
- Ait-Ouazzou, A.; Espina, L.; Gelaw, T.K.; de Lamo-Castellví, S.; Pagán, R.; García-Gonzalo, D. New insights in mechanisms of bacterial inactivation by carvacrol. J. Appl. Microbiol. 2013, 114, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; Jang, M.K.; Jeon, M.J.; Lee, D.G.; Jang, H.J.; Lee, S.W.; Lee, S.H. Verification of antimicrobial activities of various pine needle extracts against antibiotic resistant strains of Staphylococcus aureus. J. Life. Sci. 2010, 20, 589–596. [Google Scholar] [CrossRef]
- Wu, Y.; Bai, J.; Zhong, K.; Huang, Y.; Qi, H.; Jiang, Y.; Gao, H. Antibacterial activity and membrane-disruptive mechanism of 3-p-trans-coumaroyl-2-hydroxyquinic acid, a novel phenolic compound from pine needles of Cedrus deodara, against Staphylococcus aureus. Molecules 2016, 21, 1084. [Google Scholar] [CrossRef] [PubMed]
- Aung, T.; Bibat, M.A.D.; Zhao, C.C.; Eun, J.B. Bioactive compounds and antioxidant activities of Quercus salicina Blume extract. Food Sci. Biotechnol. 2020, 29, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Ju, H.K.; Cho, E.J.; Jang, M.H.; Lee, Y.Y.; Hong, S.S.; Park, J.H.; Kwon, S.W. Characterization of increased phenolic compounds from fermented Bokbunja (Rubus coreanus Miq.) and related antioxidant activity. J. Pharm. Biomed. Anal. 2019, 49, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Chen, Q.; Chen, W.; Yang, J.; Zhou, X.; Zhang, C.; Wang, L. A comprehensive review of Rubia cordifolia L.: Traditional uses, phytochemistry, pharmacological activities, and clinical applications. Front. Pharmacol. 2022, 13, 965390. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef]
- Park, Y.S.; Jeon, M.H.; Hwang, H.J.; Park, M.R.; Lee, S.H.; Kim, S.G.; Kim, M. Antioxidant activity and analysis of proanthocyanidins from pine (Pinus densiflora) needles. Nutr. Res. Pract. 2011, 5, 281–287. [Google Scholar] [CrossRef]
- Marielle, B.; Sarrah, G. Assessment of bacterial membrane fluidity by flow cytometry. J. Microbiol. Methods 2017, 143, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Gohrbandt, M.; Lipski, A.; Grimshaw, J.W.; Buttress, J.A.; Baig, Z.; Herkenhoff, B.; Strahl, H. Low membrane fluidity triggers lipid phase separation and protein segregation in living bacteria. EMBO J. 2022, 41, e109800. [Google Scholar] [CrossRef]
- Khameneh, B.; Eskin, N.M.; Iranshahy, M.; Fazly Bazzaz, B.S. Phytochemicals: A promising weapon in the arsenal against antibiotic-resistant bacteria. Antibiotics 2021, 10, 1044. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Sheng, W.; Sun, G.Y.; Lee, J.C.M. Effects of fatty acid unsaturation numbers on membrane fluidity and α-secretase-dependent amyloid precursor protein processing. Neurochem. Int. 2011, 58, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, E.L.; Zhang, R.; Ross, D.H.; Ashford, N.K.; Chen, X.; Alonzo, F., III; Xu, L. Elucidating the Impact of Bacterial Lipases, Human Serum Albumin, and FASII Inhibition on the Utilization of Exogenous Fatty Acids by Staphylococcus aureus. bioRxiv. 2023. [Google Scholar] [CrossRef]
- Mauger, M.; Ferreri, C.; Chatgilialoglu, C.; Seemann, M. The bacterial protective armor against stress: The cis-trans isomerase of unsaturated fatty acids, a cytochrome-c type enzyme. J. Inorg. Biochem. 2021, 224, 111564. [Google Scholar] [CrossRef]
- Zhu, Y.; Ouyang, Z.; Du, H.; Wang, M.; Wang, J.; Sun, H.; Sun, Y. New opportunities and challenges of natural products research: When target identification meets single-cell multiomics. Acta Pharm. Sin. B 2022, 12, 4011–4039. [Google Scholar] [CrossRef]
- Folin, O.; Dennis, W. On phosphotungstic-phosphomolybdic compounds as color reagents. JBC 1992, 12, 239–243. [Google Scholar] [CrossRef]
- Moreno, M.I.N.; Isla, M.I.; Sampietro, A.R.; Vattuone, M.A. Comparison of the free radical-scavenging activity of propolis from several regions of Argentina. J. Ethnopharmacol. 2000, 71, 109–114. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Benzie, I.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Qadrie, Z.L.; Jacob, B.; Anandan, R.; Rajkapoor, B.; Ulla, M.R. Anti-bacterial activity of ethanolic extract of Indoneesiella echioides (L) nees. evaluated by the filter paper disc method. Pak. J. Pharm. Sci. 2009, 22. [Google Scholar]



| Components | Steam Distillation |
|---|---|
| Total phenolic content (mg GAE/g (1)) | 3.610 ± 0.002 |
| Total flavonoid content (mg QE/100 g (2)) | 24.970 ± 1.550 |
| Bacteria | Fatty Acids | Fatty Acid Composition (%) | |||||
|---|---|---|---|---|---|---|---|
| 6 h | 9 h | 12 h | |||||
| CG (1) | 0.1% SDPNE | CG (1) | 0.1% SDPNE | CG (1) | 0.1% SDPNE | ||
| E. coli | SFAs (2) | 55.94 | 57.37 | 57.43 | 58.14 | 60.71 | 61.19 |
| UFAs (2) | 44.06 | 42.63 | 42.57 | 41.86 | 39.29 | 38.81 | |
| Ratio (3) | 1.27:1 | 1.35:1 | 1.35:1 | 1.39:1 | 1.55:1 | 1.58:1 | |
| S. aureus | SFAs (2) | 59.54 | 69.77 | 53.96 | 61.31 | 54.02 | 59.21 |
| UFAs (2) | 40.46 | 30.23 | 46.04 | 38.69 | 45.98 | 40.79 | |
| Ratio (3) | 1.47:1 | 2.31:1 | 1.17:1 | 1.58:1 | 1.18:1 | 1.45:1 | |
| Bacteria | Fatty Acids | Fatty Acid Composition (%) | |||||
|---|---|---|---|---|---|---|---|
| 6 h | 9 h | 12 h | |||||
| CG (1) | 0.1% SDPNE | CG (1) | 0.1% SDPNE | CG (1) | 0.1% SDPNE | ||
| E. coli | C12:0 | 2.82 | 3.23 | 2.94 | 3.10 | 3.10 | 3.25 |
| C14:0 | 7.85 | 8.53 | 7.89 | 7.49 | 7.86 | 8.03 | |
| C15:0 | 1.01 | 1.38 | 1.28 | - | 0.95 | 0.76 | |
| C16:0 | 40.64 | 39.63 | 41.65 | 42.38 | 44.29 | 45.12 | |
| C17:0 | 1.21 | 1.15 | 1.10 | 1.55 | 1.90 | 1.15 | |
| C18:0 | 2.41 | 3.46 | 2.57 | 3.62 | 2.62 | 2.87 | |
| C16:1 | 12.88 | 13.13 | 12.29 | 8.53 | 9.05 | 8.41 | |
| C20:1 | - | - | - | - | 0.71 | 0.57 | |
| Cis-C18:1n-9 | 28.77 | 25.35 | 27.89 | 27.13 | 25.95 | 26.39 | |
| Trans-C18:1n-9 | - | 2.30 | - | 2.07 | - | - | |
| Isomerization (2) | - | 8.33 | - | 7.08 | - | - | |
| Cis-C18:2n-6 | 0.80 | - | 0.73 | 1.55 | 1.19 | 1.34 | |
| Trans-C18:2n-6 | 1.61 | 1.84 | 1.65 | 2.58 | 2.38 | 2.10 | |
| Isomerization (2) | 66.67 | 100.00 | 69.23 | 62.50 | 66.67 | 61.11 | |
| S. aureus | C14:0 | 2.31 | 2.33 | - | 2.38 | - | 1.97 |
| C16:0 | 5.20 | 5.81 | 5.04 | 4.76 | 6.32 | 6.58 | |
| C18:0 | 21.97 | 24.42 | 20.86 | 21.43 | 21.84 | 21.71 | |
| C20:0 | 30.06 | 37.21 | 28.06 | 32.74 | 25.86 | 28.95 | |
| C18:3n-6 | 2.31 | - | - | - | - | - | |
| C20:1 | 2.89 | 2.91 | 2.88 | 2.98 | 4.02 | 2.63 | |
| Cis-C18:1n-9 | 11.56 | 10.47 | 12.95 | 11.90 | 11.49 | 11.84 | |
| Trans-C18:1n-9 | 21.39 | 13.95 | 28.06 | 21.43 | 27.01 | 23.03 | |
| Isomerization (2) | 35.09 | 42.86 | 31.58 | 35.71 | 29.85 | 33.96 | |
| Cis-C18:2n-6 | - | - | - | - | - | - | |
| Trans-C18:2n-6 | 2.31 | 2.91 | 2.16 | 2.38 | 3.45 | 3.29 | |
| Bacteria | Target | Primer (5′→3′) |
|---|---|---|
| E. coli | fabA | Fw—CCAGGAACGTATCGCACAAG Rv—CGCTGAACAAGTCCGATCAGT |
| fabD | Fw—CGTTGGAATGCTGGCTGATA Rv—CTGCTGCCATACGCGATACA | |
| fabG | Fw—CGCTCAGGCGATCAGTGATT Rv—ACCGGCATTATTGACCAGGA | |
| fabI | Fw—CGGTAAGCGCATTCTGGTAA Rv—TGCTGGCATCTTCTGCAACA | |
| cfa | Fw—CTTAGCCGTGCCGGTATAGC Rv—CGAGACCTGCGCGTAAGACT | |
| rpos | Fw—CGCCGGATGATCGAGAGTAA Rv—CCACCAGGTTGCGTATGTTG | |
| gyrB | Fw—TGCGTGGCTTGCTGGAAGAA Rv—CGGGTTCATCTCGCCCAGAC | |
| S. aureus | fabD | Fw—TTGACGCATAGTTCGGCATT Rv—ACTGCAGCCATGCTTCCTACA |
| fabF | Fw—GACGTGTGAGTCCATTTTTC Rv—ACCACCAGTAATCATTGCAT | |
| fabG | Fw—GTTGCCGATGCTGATGAAGT Rv—TCATCCCACTCTTGTTCTTTCA | |
| fabH | Fw—GATAACCGCACCTGCACCAT Rv—TGGATCAACTTGCAGCATGTT | |
| fabI | Fw—GAAGACTTACGCGGACGCTT Rv—TGCTACCACCTTCTGGCATTA | |
| sigB | Fw—TGAAGATGCCAAGATTGCAGT Rv—CTAGGCCACCTTCGCGTAA | |
| 16s RNA | Fw—CGGTGAATACGTTCYCGG Rv—GGWTACCTTGTTACGACTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Chung, W.-K.; Moon, S.-H.; Lee, J.-G.; Om, A.-S. Comparison of Antibacterial Activities of Korean Pine (Pinus densiflora) Needle Steam Distillation Extract on Escherichia coli and Staphylococcus aureus Focusing on Membrane Fluidity and Genes Involved in Membrane Lipids and Stress. Molecules 2024, 29, 165. https://doi.org/10.3390/molecules29010165
Zhang Y, Chung W-K, Moon S-H, Lee J-G, Om A-S. Comparison of Antibacterial Activities of Korean Pine (Pinus densiflora) Needle Steam Distillation Extract on Escherichia coli and Staphylococcus aureus Focusing on Membrane Fluidity and Genes Involved in Membrane Lipids and Stress. Molecules. 2024; 29(1):165. https://doi.org/10.3390/molecules29010165
Chicago/Turabian StyleZhang, Ya, Woo-Kyung Chung, Su-Hyun Moon, Jeoung-Gyu Lee, and Ae-Son Om. 2024. "Comparison of Antibacterial Activities of Korean Pine (Pinus densiflora) Needle Steam Distillation Extract on Escherichia coli and Staphylococcus aureus Focusing on Membrane Fluidity and Genes Involved in Membrane Lipids and Stress" Molecules 29, no. 1: 165. https://doi.org/10.3390/molecules29010165
APA StyleZhang, Y., Chung, W.-K., Moon, S.-H., Lee, J.-G., & Om, A.-S. (2024). Comparison of Antibacterial Activities of Korean Pine (Pinus densiflora) Needle Steam Distillation Extract on Escherichia coli and Staphylococcus aureus Focusing on Membrane Fluidity and Genes Involved in Membrane Lipids and Stress. Molecules, 29(1), 165. https://doi.org/10.3390/molecules29010165






