Visualization and Comparison of the Level of Apurinic/Apyrimidinic Endonuclease 1 in Live Normal/Cancerous and Neuron Cells with a Fluorescent Nanoprobe
Abstract
1. Introduction
2. Results and Discussion
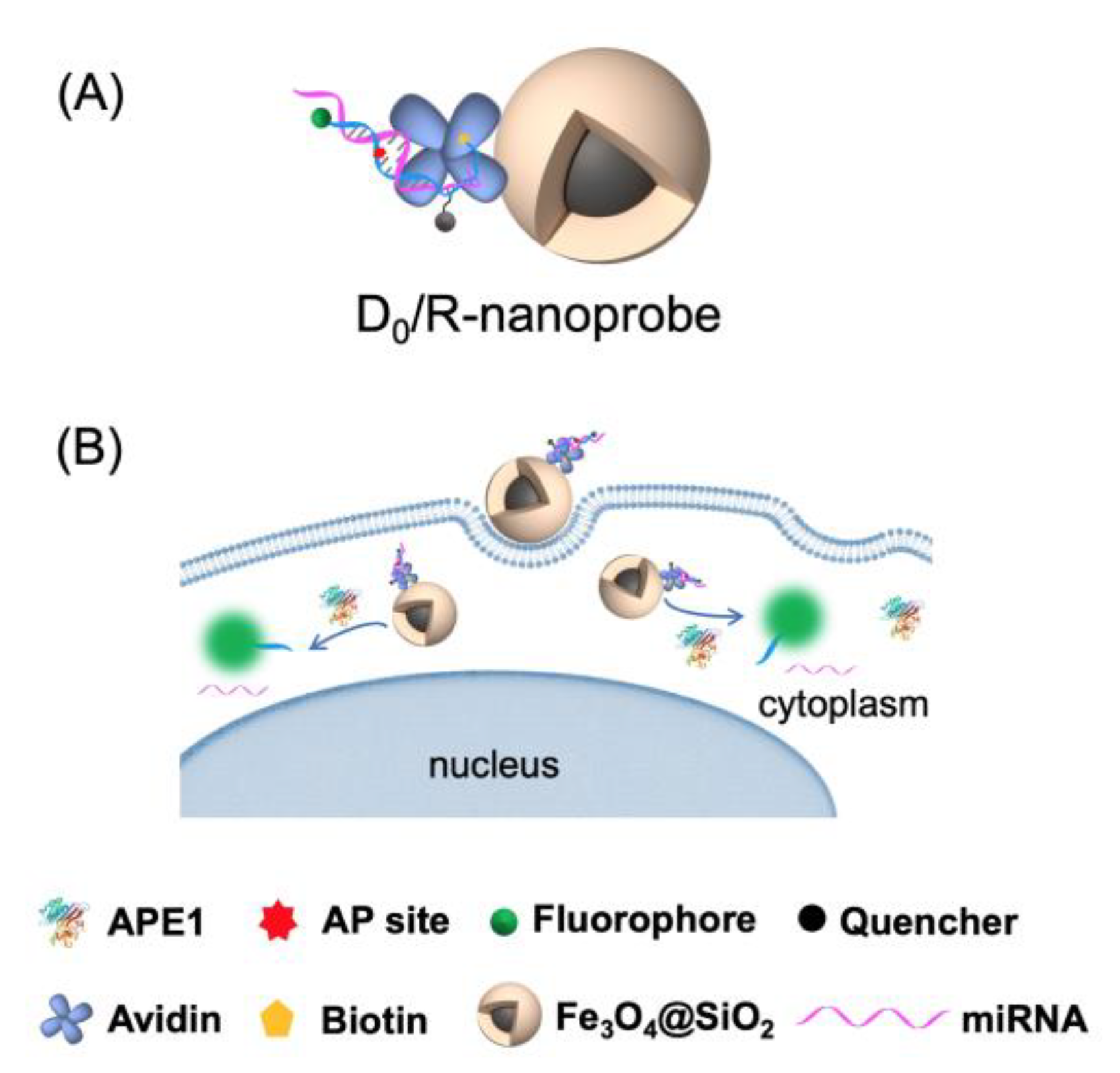
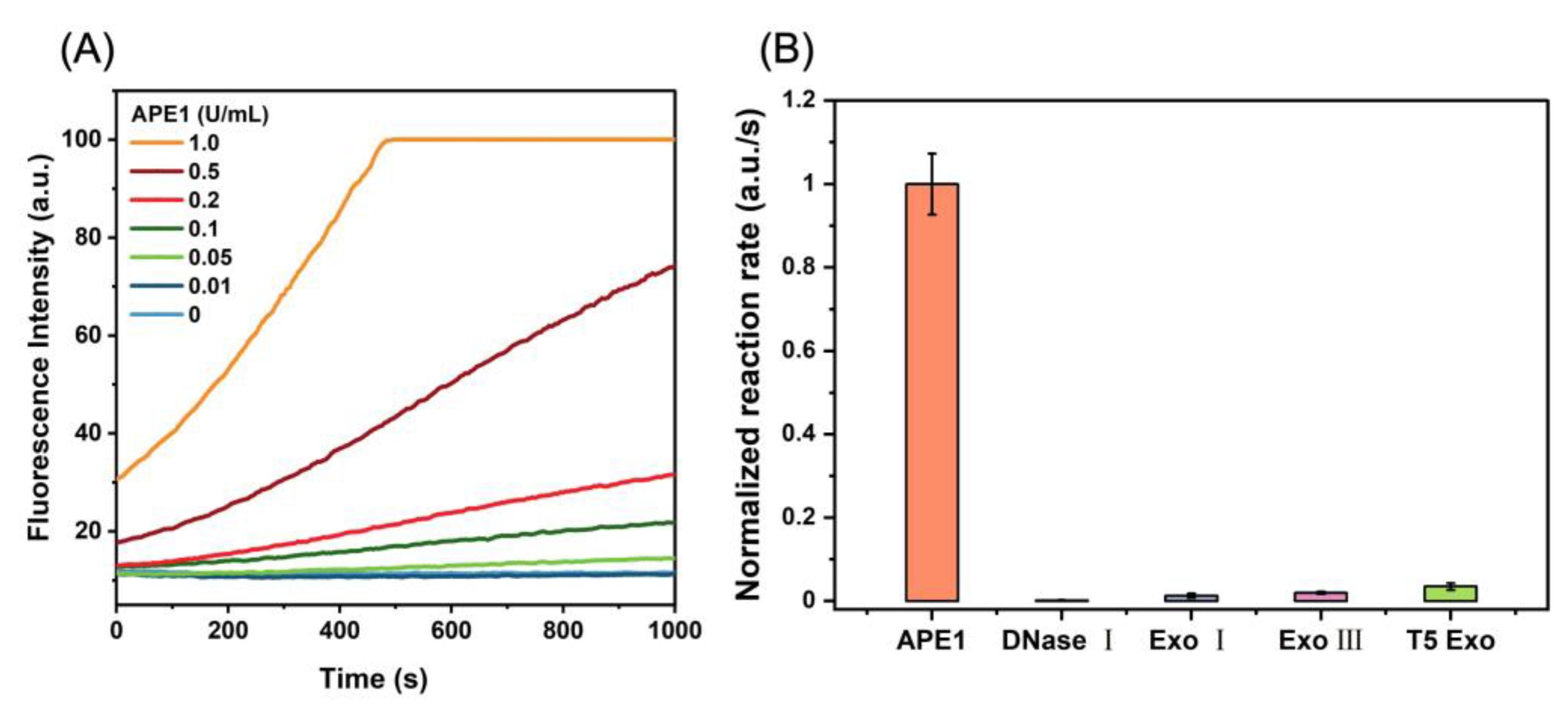
2.1. Construction and Characterization of the DNA/RNA Hybrid Fluorescent Nanoprobe (D0/R-Nanoprobe)
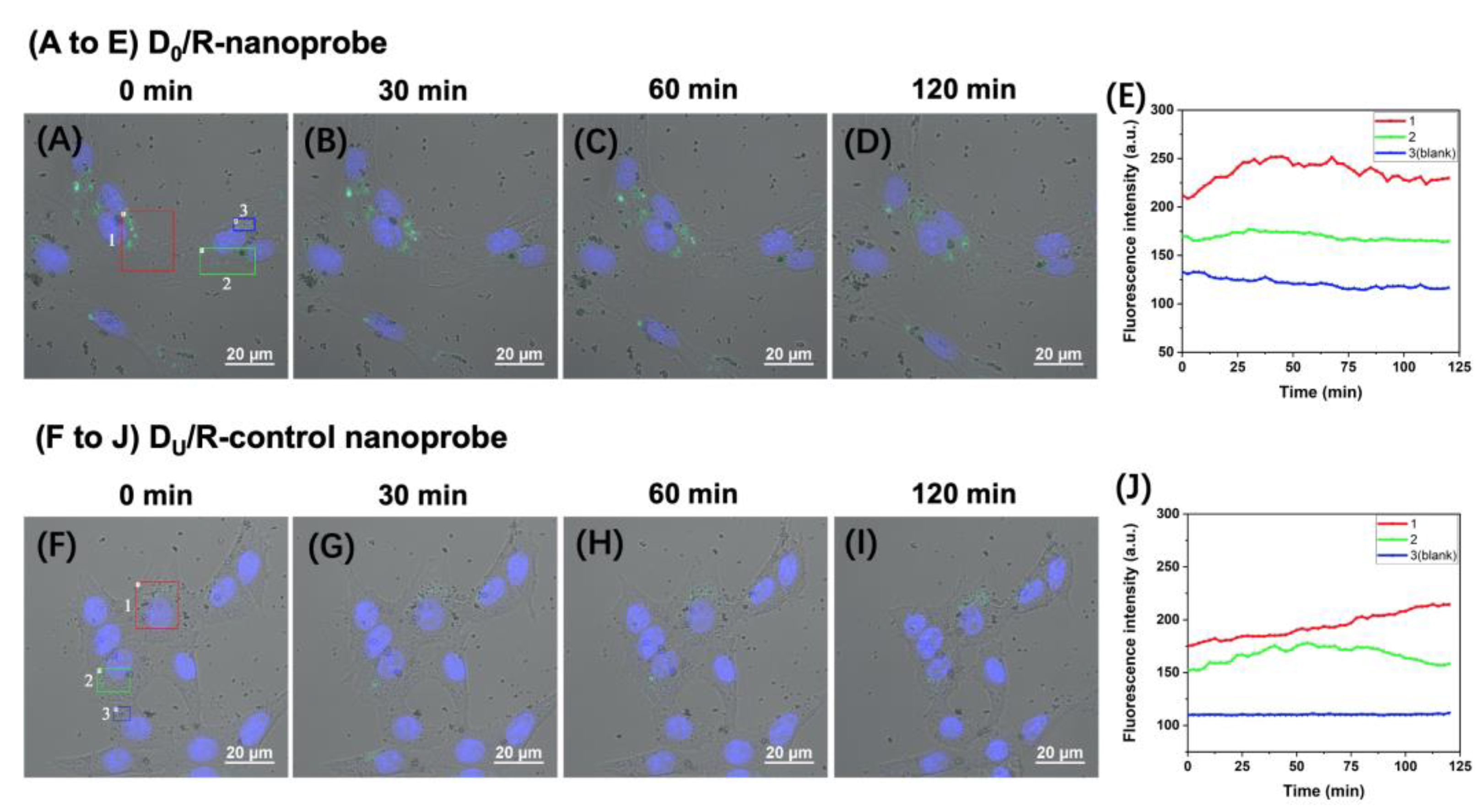
2.2. Fluorescence Imaging of APE1 Activity in Live MCF-7 and MCF-10A Cells
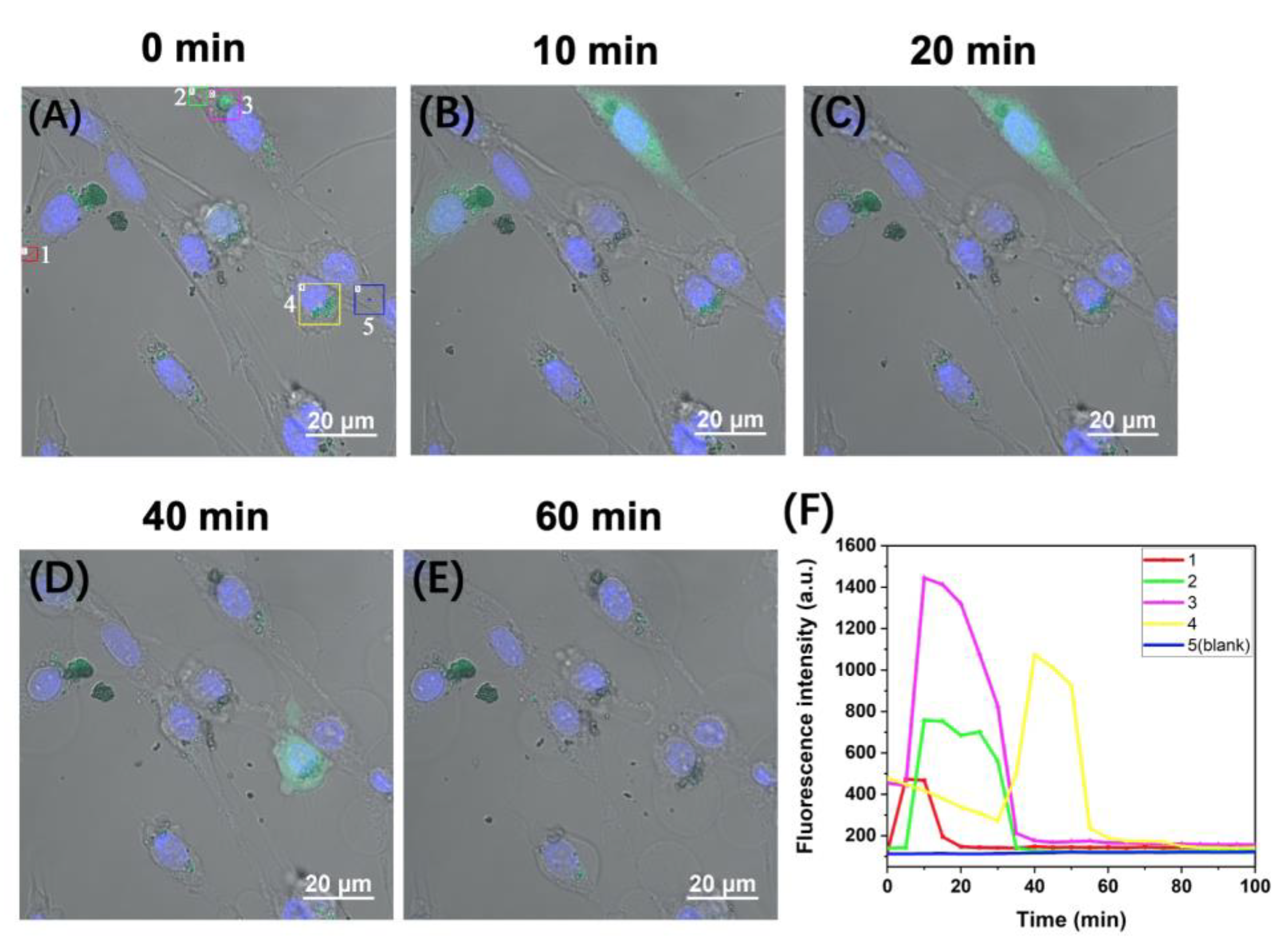
2.3. Time-Lapse Fluorescence Imaging of the APE1 Activity in PC-12 Live Cells with the D0/R-Nanoprobes
2.4. Real-Time Monitoring of the Changes of APE1 Level in PC-12 under Severe Attack by ROS
3. Materials and Methods
3.1. Chemical Reagents and Materials
3.2. Synthesis of DNA/RNA Hybrid Nanoprobes
3.3. Sensitivity and Selectivity Measurement of D0/R-Nanoprobes
3.4. Intracellular Uptake of D0/R-Nanoprobes for Fluorescence Imaging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Evans, A.R.; Limp-Foster, M.; Kelley, M.R. Going APE over ref-1. Mutat. Res.-DNA Repair 2000, 461, 83–108. [Google Scholar] [CrossRef]
- Tell, G.; Quadrifoglio, F.; Tiribelli, C.; Kelley, M.R. The Many Functions of APE1/Ref-1: Not Only a DNA Repair Enzyme. Antioxid. Redox Signal. 2009, 11, 601–619. [Google Scholar] [CrossRef]
- Dumitrache, L.C.; Shimada, M.; Downing, S.M.; Kwak, Y.D.; Li, Y.; Illuzzi, J.L.; Russell, H.R.; Wilson, D.M., 3rd; McKinnon, P.J. Apurinic endonuclease-1 preserves neural genome integrity to maintain homeostasis and thermoregulation and prevent brain tumors. Proc. Natl. Acad. Sci. USA 2018, 115, E12285–E12294. [Google Scholar] [CrossRef]
- Bobola, M.S.; Blank, A.; Berger, M.S.; Stevens, B.A.; Silber, J.R. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin. Cancer Res. 2001, 7, 3510–3518. [Google Scholar]
- Silva, L.P.; Santana, T.; Sedassari, B.T.; de Sousa, S.M.; Sobral, A.P.V.; Freitas, R.A.; Barboza, C.A.G.; de Souza, L.B. Apurinic/apyrimidinic endonuclease 1 (APE1) is overexpressed in malignant transformation of salivary gland pleomorphic adenoma. Eur. Arch. Oto-Rhino-Laryngol. 2017, 274, 3203–3209. [Google Scholar] [CrossRef]
- Coskun, E.; Jaruga, P.; Reddy, P.T.; Dizdaroglu, M. Extreme Expression of DNA Repair Protein Apurinic/Apyrimidinic Endonuclease 1 (APE1) in Human Breast Cancer As Measured by Liquid Chromatography and Isotope Dilution Tandem Mass Spectrometry. Biochemistry 2015, 54, 5787–5790. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Irani, K.; Heffron, S.E.; Jurnak, F.; Meyskens, F.L., Jr. Alterations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE/Ref-1) in human melanoma and identification of the therapeutic potential of resveratrol as an APE/Ref-1 inhibitor. Mol. Cancer Ther. 2005, 4, 1923–1935. [Google Scholar] [CrossRef] [PubMed]
- Kelley, M.R.; Logsdon, D.; Fishel, M.L. Targeting DNA repair pathways for cancer treatment: What’s new? Future Oncol. 2014, 10, 1215–1237. [Google Scholar] [CrossRef]
- Qing, Y.; Li, Q.; Ren, T.; Xia, W.; Peng, Y.; Liu, G.L.; Luo, H.; Yang, Y.X.; Dai, X.Y.; Zhou, S.F.; et al. Upregulation of PD-LI and APEI is associated with tumorigenesis and poor prognosis of gastric cancer. Drug Des. Dev. Ther. 2015, 9, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Kakolyris, S.; Kaklamanis, L.; Engels, K.; Fox, S.B.; Taylor, M.; Hickson, I.D.; Gatter, K.C.; Harris, A.L. Human AP endonuclease 1 (HAP1) protein expression in breast cancer correlates with lymph node status and angiogenesis. Br. J. Cancer 1998, 77, 1169–1173. [Google Scholar] [CrossRef] [PubMed]
- Bobola, M.S.; Finn, L.S.; Ellenbogen, R.G.; Geyer, J.R.; Berger, M.S.; Braga, J.M.; Meade, E.H.; Gross, M.E.; Silber, J.R. Apurinic/apyrimidinic endonuclease activity is associated with response to radiation and chemotherapy in medulloblastoma and primitive neuroectodermal tumors. Clin. Cancer Res. 2005, 11, 7405–7414. [Google Scholar] [CrossRef]
- Vasko, M.R.; Guo, C.; Kelley, M.R. The multifunctional DNA repair/redox enzyme Ape1/Ref-1 promotes survival of neurons after oxidative stress. DNA Repair 2005, 4, 367–379. [Google Scholar] [CrossRef]
- Kirkali, G.; Jaruga, P.; Reddy, P.T.; Tona, A.; Nelson, B.C.; Li, M.; Wilson, D.M., 3rd; Dizdaroglu, M. Identification and quantification of DNA repair protein apurinic/apyrimidinic endonuclease 1 (APE1) in human cells by liquid chromatography/isotope-dilution tandem mass spectrometry. PLoS ONE 2013, 8, e69894. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Cao, X.; Zheng, J.; Zhu, C.; Zhang, R.; Sun, Y.; Yang, Z.; Tang, Z.; Wang, J.; Zhao, M. A DNA/RNA hybrid fluorescent probe for high-throughput quantification of the activity of human apurinic/apyrimidinic endonuclease 1 in subcellular extracts. Biosens. Bioelectron. X 2023, 14, 100329. [Google Scholar] [CrossRef]
- Zhai, J.; Liu, Y.; Huang, S.; Fang, S.; Zhao, M. A specific DNA-nanoprobe for tracking the activities of human apurinic/apyrimidinic endonuclease 1 in living cells. Nucleic Acids Res. 2017, 45, e45. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, D.B. Intracellular uptake, transport, and processing of gold nanostructures. Mol. Membr. Biol. 2010, 27, 299–311. [Google Scholar] [CrossRef]
- Hinde, E.; Thammasiraphop, K.; Duong, H.T.T.; Yeow, J.; Karagoz, B.; Boyer, C.; Gooding, J.J.; Gaus, K. Pair correlation rnicroscopy reveals the role of nanoparticle shape in intracellular transport and site of drug release. Nat. Nanotechnol. 2017, 12, 81–89. [Google Scholar] [CrossRef]
- Hu, R.; Cao, Q.; Sun, Z.; Chen, J.; Zheng, Q.; Xiao, F. A novel method of neural differentiation of PC12 cells by using Opti-MEM as a basic induction medium. Int. J. Mol. Med. 2018, 41, 195–201. [Google Scholar] [CrossRef]
- Greene, L.A.; Tischler, A.S. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA 1976, 73, 2424–2428. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Edwards, S.K.; Wang, S.; Jiang, W.; Kool, E.T. Monitoring eukaryotic and bacterial UDG repair activity with DNA-multifluorophore sensors. Nucleic Acids Res. 2013, 41, e127. [Google Scholar] [CrossRef]
- Martin, C.; Martinez, R.; Navarro, R.; Ruiz-Sanz, J.I.; Lacort, M.; Ruiz-Larrea, M.B. tert-Butyl hydroperoxide-induced lipid signaling in hepatocytes: Involvement of glutathione and free radicals. Biochem. Pharmacol. 2001, 62, 705–712. [Google Scholar] [PubMed]
- Rush, G.F.; Gorski, J.R.; Ripple, M.G.; Sowinski, J.; Bugelski, P.; Hewitt, W.R. Organic hydroperoxide-induced lipid peroxidation and cell death in isolated hepatocytes. Toxicol. Appl. Pharmacol. 1985, 78, 473–483. [Google Scholar] [CrossRef]
- Zhao, W.; Feng, H.; Sun, W.; Liu, K.; Lu, J.J.; Chen, X. Tert-butyl hydroperoxide (t-BHP) induced apoptosis and necroptosis in endothelial cells: Roles of NOX4 and mitochondrion. Redox Biol. 2017, 11, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Guo, C.; Vasko, M.R.; Kelley, M.R. Implications of apurinic/apyrimidinic endonuclease in reactive oxygen signaling response after cisplatin treatment of dorsal root ganglion neurons. Cancer Res. 2008, 68, 6425–6434. [Google Scholar] [CrossRef] [PubMed]
- Juhnke, M.; Heumann, A.; Chirico, V.; Hoflmayer, D.; Menz, A.; Hinsch, A.; Hube-Magg, C.; Kluth, M.; Lang, D.S.; Moller-Koop, C.; et al. Apurinic/apyrimidinic endonuclease 1 (APE1/Ref-1) overexpression is an independent prognostic marker in prostate cancer without TMPRSS2:ERG fusion. Mol. Carcinog. 2017, 56, 2135–2145. [Google Scholar] [CrossRef]
- Londero, A.P.; Orsaria, M.; Tell, G.; Marzinotto, S.; Capodicasa, V.; Poletto, M.; Vascotto, C.; Sacco, C.; Mariuzzi, L. Expression and prognostic significance of APE1/Ref-1 and NPM1 proteins in high-grade ovarian serous cancer. Am. J. Clin. Pathol. 2014, 141, 404–414. [Google Scholar] [CrossRef]
- Wang, D.; Luo, M.; Kelley, M.R. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: Enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol. Cancer Ther. 2004, 3, 679–686. [Google Scholar] [CrossRef]
- Wang, D.; Xiang, D.B.; Yang, X.Q.; Chen, L.S.; Li, M.X.; Zhong, Z.Y.; Zhang, Y.S. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer 2009, 66, 298–304. [Google Scholar] [CrossRef]
- Wilson, D.M., 3rd; Simeonov, A. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell. Mol. Life Sci. 2010, 67, 3621–3631. [Google Scholar] [CrossRef]
- O’Connor, M.J. Targeting the DNA Damage Response in Cancer. Mol. Cell 2015, 60, 547–560. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, P.; Cao, X.; Zheng, J.; Sun, Y.; Tang, Z.; Zhao, M. Visualization and Comparison of the Level of Apurinic/Apyrimidinic Endonuclease 1 in Live Normal/Cancerous and Neuron Cells with a Fluorescent Nanoprobe. Molecules 2023, 28, 3935. https://doi.org/10.3390/molecules28093935
Lu P, Cao X, Zheng J, Sun Y, Tang Z, Zhao M. Visualization and Comparison of the Level of Apurinic/Apyrimidinic Endonuclease 1 in Live Normal/Cancerous and Neuron Cells with a Fluorescent Nanoprobe. Molecules. 2023; 28(9):3935. https://doi.org/10.3390/molecules28093935
Chicago/Turabian StyleLu, Peng, Xiangjian Cao, Jinghui Zheng, Ying Sun, Ziyu Tang, and Meiping Zhao. 2023. "Visualization and Comparison of the Level of Apurinic/Apyrimidinic Endonuclease 1 in Live Normal/Cancerous and Neuron Cells with a Fluorescent Nanoprobe" Molecules 28, no. 9: 3935. https://doi.org/10.3390/molecules28093935
APA StyleLu, P., Cao, X., Zheng, J., Sun, Y., Tang, Z., & Zhao, M. (2023). Visualization and Comparison of the Level of Apurinic/Apyrimidinic Endonuclease 1 in Live Normal/Cancerous and Neuron Cells with a Fluorescent Nanoprobe. Molecules, 28(9), 3935. https://doi.org/10.3390/molecules28093935








