DNA-Guided Metallization of Nanomaterials and Their Biomedical Applications
Abstract
1. Introduction
2. Strategies of DNA Functionalized Nanoseeds
3. DNA as Director for Nanomaterials Metallization
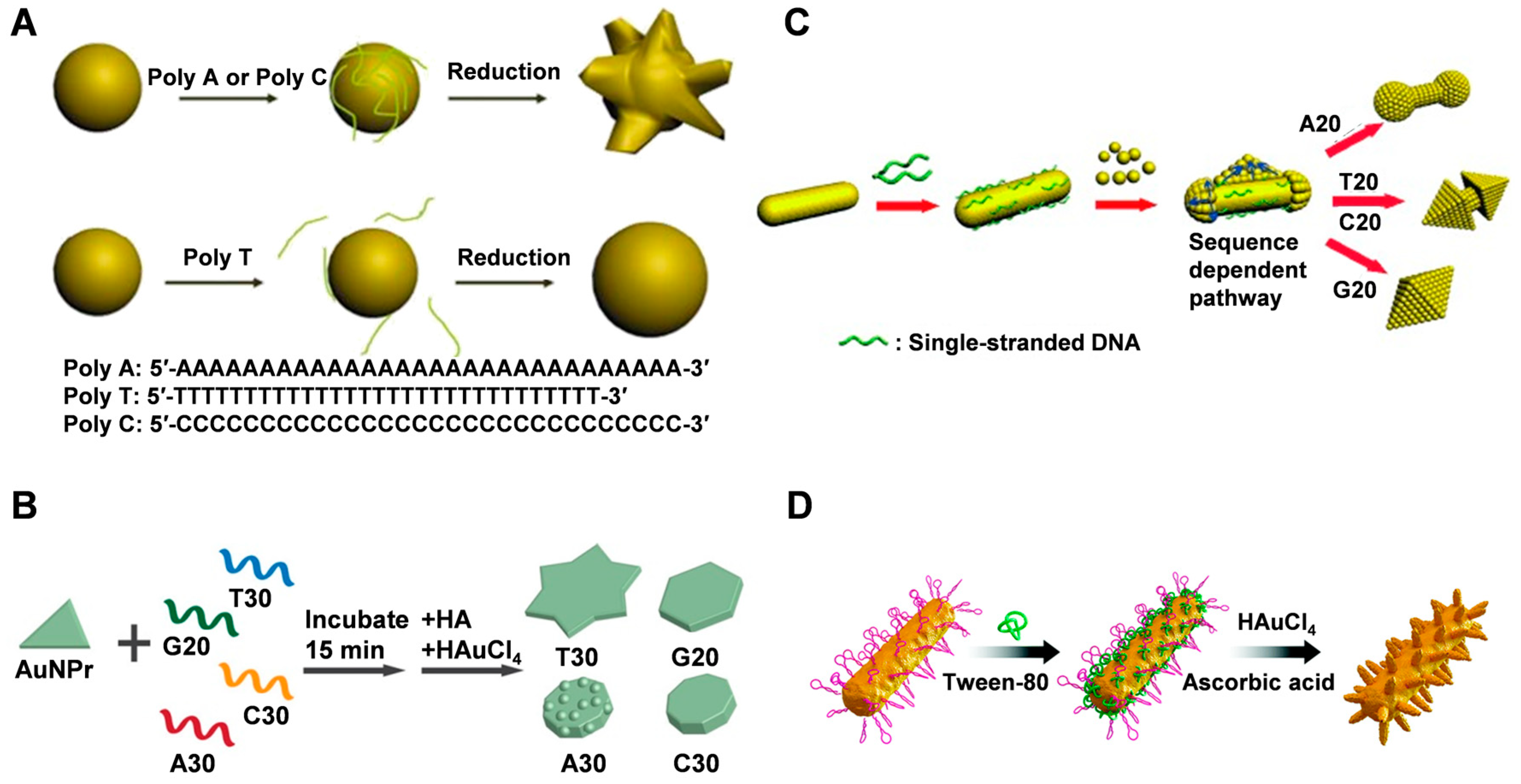
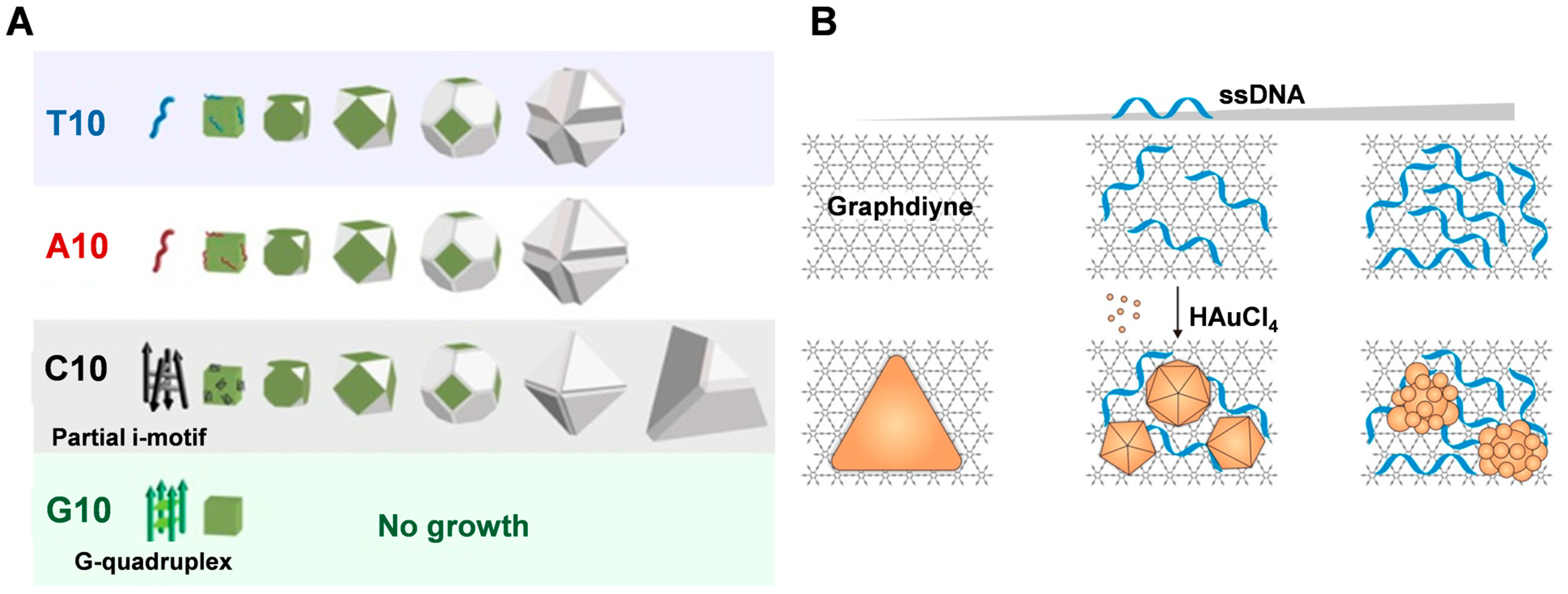
3.1. ssDNA
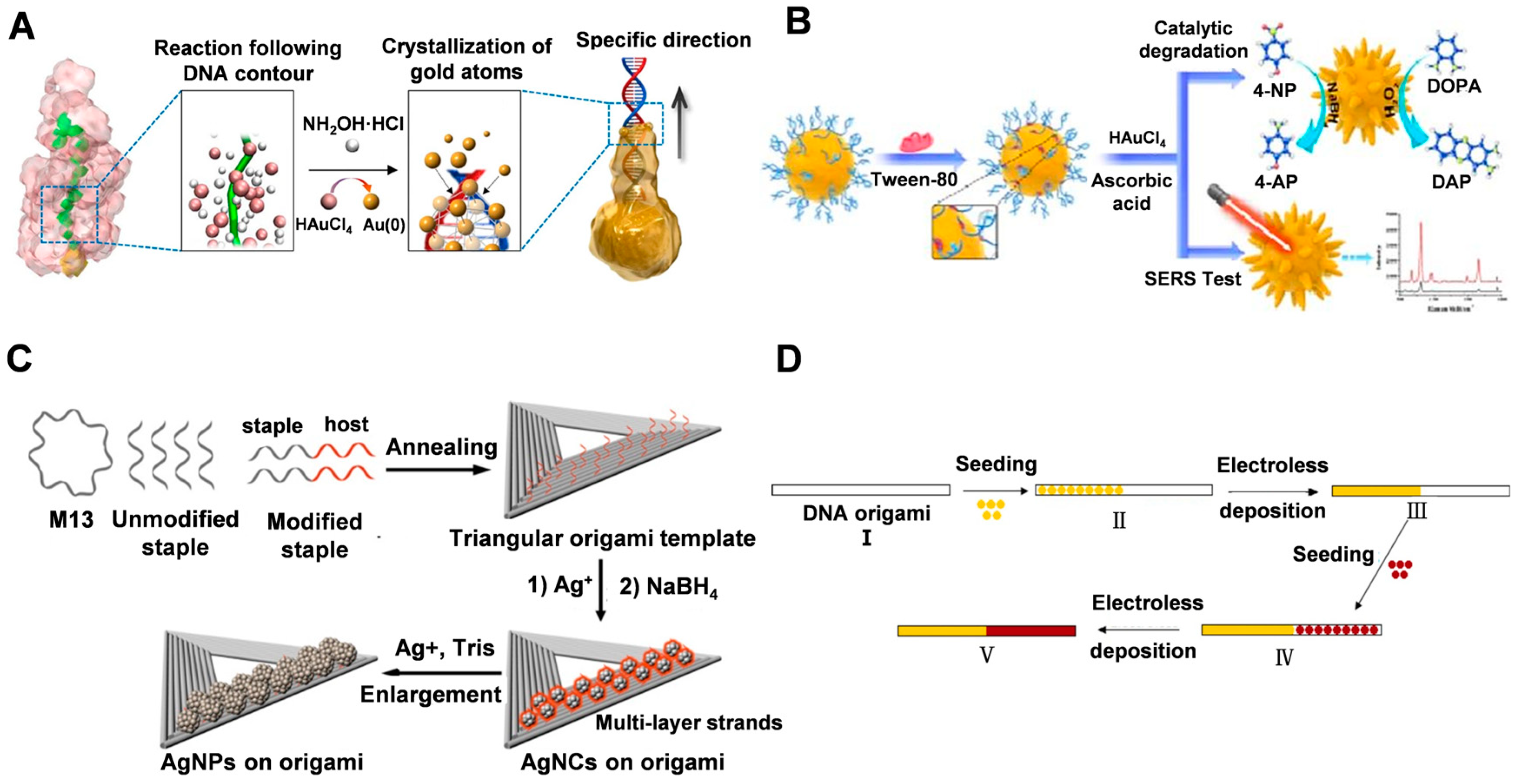
3.2. dsDNA
3.3. DNA Origami
4. Biomedical Applications
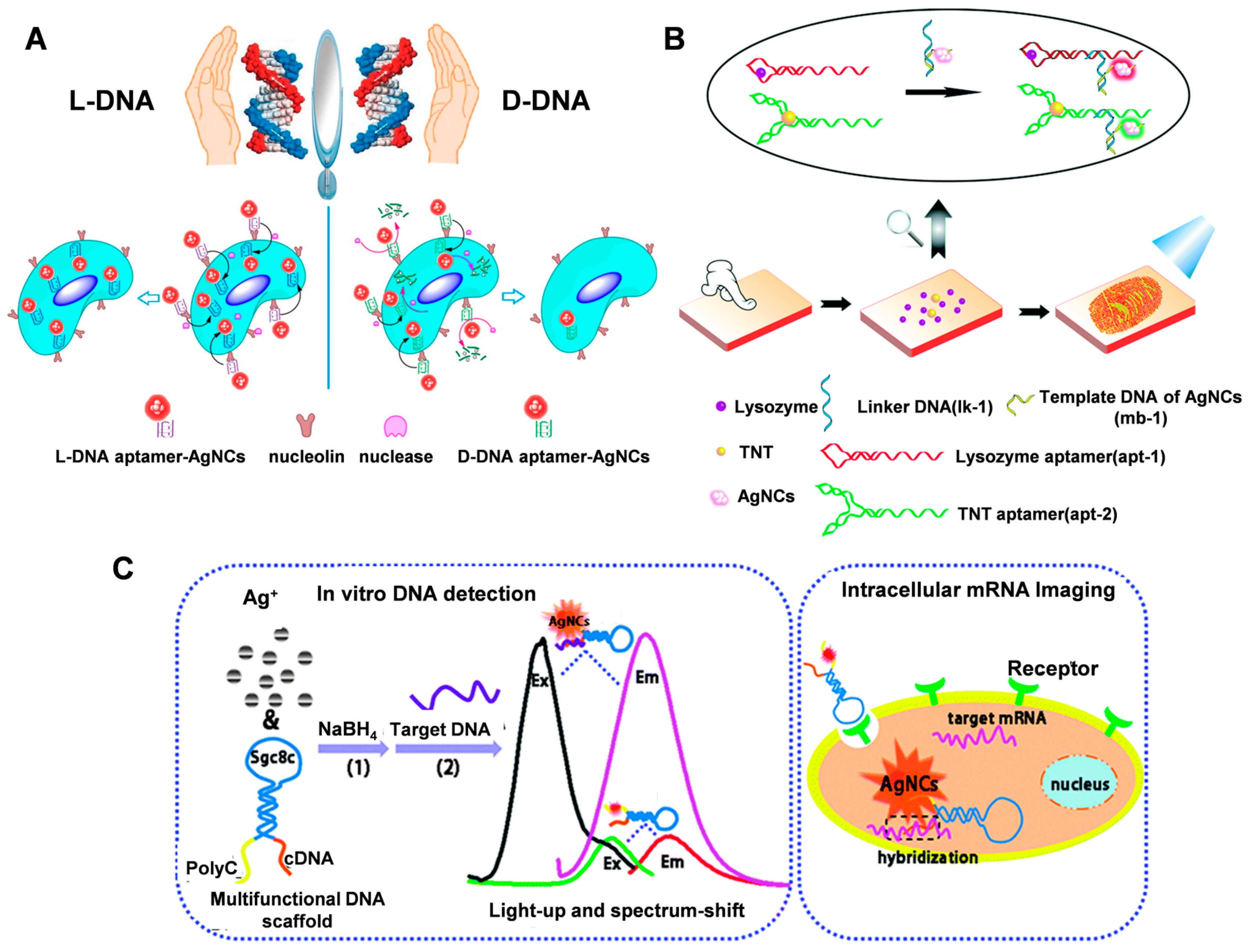
4.1. Biosensing
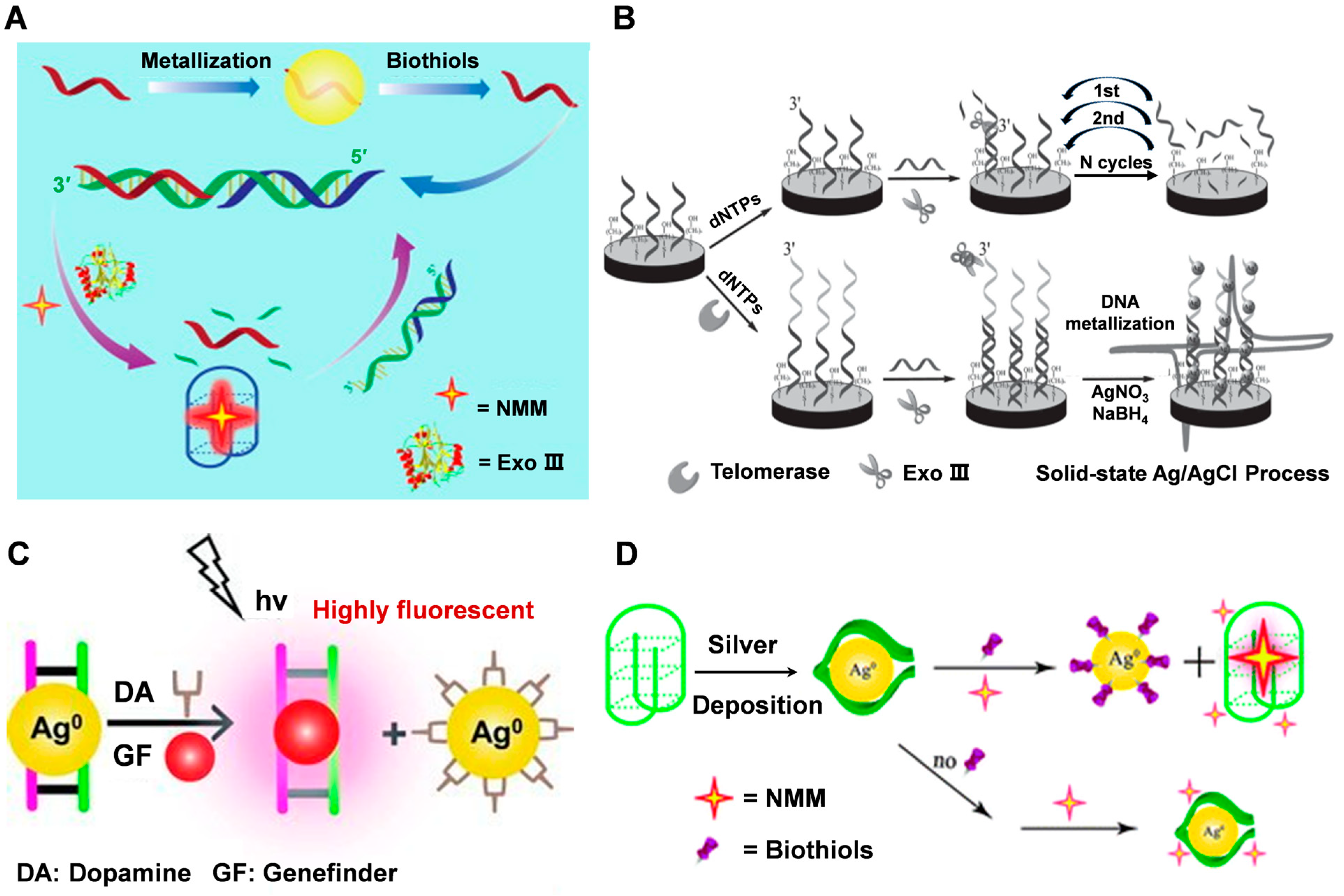
4.2. Bioimaging
4.3. Therapy
5. Advantages and Challenges
6. Prospectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, S.; Annu; Ikram, S.; Yudha S., S. Biosynthesis of gold nanoparticles: A green approach. J. Photochem. Photobiol. B Biol. 2016, 161, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fedin, I.; Zhang, H.; Talapin, D.V. Direct optical lithography of functional inorganic nanomaterials. Science 2017, 357, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.Y.; Mason, J.A.; Li, Z.; Zhou, W.; O’Brien, M.N.; Brown, K.A.; Jones, M.R.; Butun, S.; Lee, B.; Dravid, V.P.; et al. Building superlattices from individual nanoparticles via template-confined DNA-mediated assembly. Science 2018, 359, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Wang, J.; Xie, M.; Sun, J.; Liu, H.; Zhang, Y.; Chao, J.; Li, J.; Wang, L.; Lin, J.; et al. Programming DNA origami patterning with non-canonical DNA-based metallization reactions. Nat. Commun. 2019, 10, 5597. [Google Scholar] [CrossRef]
- Al-Hinai, M.N.; Hassanien, R.; Wright, N.G.; Horsfall, A.B.; Houlton, A.; Horrocks, B.R. Networks of DNA-templated palladium nanowires: Structural and electrical characterisation and their use as hydrogen gas sensors. Faraday Discuss. 2013, 164, 71–91. [Google Scholar] [CrossRef]
- Li, N.; Shang, Y.; Han, Z.; Wang, T.; Wang, Z.G.; Ding, B. Fabrication of Metal Nanostructures on DNA Templates. ACS Appl. Mater. Interfaces 2019, 11, 13835–13852. [Google Scholar] [CrossRef]
- Braun, E.; Eichen, Y.; Sivan, U.; Ben-Yoseph, G. DNA-templated assembly and electrode attachment of a conducting silver wire. Nature 1998, 391, 775–778. [Google Scholar] [CrossRef]
- Burley, G.A.; Gierlich, J.; Mofid, M.R.; Nir, H.; Tal, S.; Eichen, Y.; Carell, T. Directed DNA metallization. J. Am. Chem. Soc. 2006, 128, 1398–1399. [Google Scholar] [CrossRef]
- Keren, K.; Berman, R.S.; Braun, E. Patterned DNA Metallization by Sequence-Specific Localization of a Reducing Agent. Nano Lett. 2004, 4, 323–326. [Google Scholar] [CrossRef]
- Eidelshtein, G.; Fardian-Melamed, N.; Gutkin, V.; Basmanov, D.; Klinov, D.; Rotem, D.; Levi-Kalisman, Y.; Porath, D.; Kotlyar, A. Synthesis and Properties of Novel Silver-Containing DNA Molecules. Adv. Mater. 2016, 28, 4839–4844. [Google Scholar] [CrossRef]
- Tan, L.H.; Yue, Y.; Satyavolu, N.S.; Ali, A.S.; Wang, Z.; Wu, Y.; Lu, Y. Mechanistic Insight into DNA-Guided Control of Nanoparticle Morphologies. J. Am. Chem. Soc. 2015, 137, 14456–14464. [Google Scholar] [CrossRef]
- Tan, L.H.; Xing, H.; Lu, Y. DNA as a Powerful Tool for Morphology Control, Spatial Positioning, and Dynamic Assembly of Nanoparticles. Acc. Chem. Res. 2014, 47, 1881–1890. [Google Scholar] [CrossRef]
- Shen, X.T.; Xu, W.; Ouyang, J.; Na, N. Fluorescence resonance energy transfer-based nanomaterials for the sensing in biological systems. Chin. Chem. Lett. 2022, 33, 4505–4516. [Google Scholar] [CrossRef]
- Zhou, X.; Yao, D.B.; Hua, W.Q.; Huang, N.D.; Chen, X.W.; Li, L.B.; He, M.; Zhang, Y.H.; Guo, Y.J.; Xiao, S.Y.; et al. Programming colloidal bonding using DNA strand-displacement circuitry. Proc. Natl. Acad. Sci. USA 2020, 117, 5617–5623. [Google Scholar] [CrossRef]
- Lee, W.-J.; Kim, K.-J.; Hossain, M.K.; Cho, H.-Y.; Choi, J.-W. DNA–Gold Nanoparticle Conjugates for Intracellular miRNA Detection Using Surface-Enhanced Raman Spectroscopy. BioChip J. 2022, 16, 33–40. [Google Scholar] [CrossRef]
- Hu, Y.Q.; Zhang, Z.; Zhang, W.; Hu, M.H.; Xiao, X.J.; Wu, T.B. Probing and modulating the interactions of the DNAzyme with DNA-functionalized nanoparticles. Chin. Chem. Lett. 2022, 33, 3026–3030. [Google Scholar] [CrossRef]
- Lee, J.S.; Lytton-Jean, A.K.R.; Hurst, S.J.; Mirkin, C.A. Silver nanoparticle-oligonucleotide conjugates based on DNA with triple cyclic disulfide moieties. Nano Lett. 2007, 7, 2112–2115. [Google Scholar] [CrossRef] [PubMed]
- Dougan, J.A.; Karlsson, C.; Smith, W.E.; Graham, D. Enhanced oligonucleotide-nanoparticle conjugate stability using thioctic acid modified oligonucleotides. Nucleic Acids Res. 2007, 35, 3668–3675. [Google Scholar] [CrossRef]
- Nguyen, L.; Dass, M.; Ober, M.F.; Besteiro, L.V.; Wang, Z.M.; Nickel, B.; Govorov, A.O.; Liedl, T.; Heuer-Jungemann, A. Chiral Assembly of Gold-Silver Core-Shell Plasmonic Nanorods on DNA Origami with Strong Optical Activity. ACS Nano 2020, 14, 7454–7461. [Google Scholar] [CrossRef]
- Zhang, X.; Servos, M.R.; Liu, J. Surface science of DNA adsorption onto citrate-capped gold nanoparticles. Langmuir 2012, 28, 3896–3902. [Google Scholar] [CrossRef] [PubMed]
- Opdahl, A.; Petrovykh, D.Y.; Kimura-Suda, H.; Tarlov, M.J.; Whitman, L.J. Independent control of grafting density and conformation of single-stranded DNA brushes. Proc. Natl. Acad. Sci. USA 2007, 104, 9–14. [Google Scholar] [CrossRef]
- de Izarra, A.; Jang, Y.H.; Lansac, Y. DNA-assisted assembly of cationic gold nanoparticles: Monte Carlo simulation. Soft Matter 2021, 17, 9315–9325. [Google Scholar] [CrossRef] [PubMed]
- Zinchenko, A.A.; Sakaue, T.; Araki, S.; Yoshikawa, K.; Baigl, D. Single-chain compaction of long duplex DNA by cationic nanoparticles: Modes of interaction and comparison with chromatin. J. Phys. Chem. B 2007, 111, 3019–3031. [Google Scholar] [CrossRef] [PubMed]
- Warner, M.G.; Hutchison, J.E. Linear assemblies of nanoparticles electrostatically organized on DNA scaffolds. Nat. Mater. 2003, 2, 272–277. [Google Scholar] [CrossRef]
- Li, F.; Pei, H.; Wang, L.; Lu, J.; Gao, J.; Jiang, B.; Zhao, X.; Fan, C. Nanomaterial-Based Fluorescent DNA Analysis: A Comparative Study of the Quenching Effects of Graphene Oxide, Carbon Nanotubes, and Gold Nanoparticles. Adv. Funct. Mater. 2013, 23, 4140–4148. [Google Scholar] [CrossRef]
- Bera, S.C.; Sanyal, K.; Senapati, D.; Mishra, P.P. Conformational Changes Followed by Complete Unzipping of DNA Double Helix by Charge-Tuned Gold Nanoparticles. J. Phys. Chem. B 2016, 120, 4213–4220. [Google Scholar] [CrossRef]
- Karthick, K.; Anantharaj, S.; Ede, S.R.; Sankar, S.S.; Kumaravel, S.; Karmakar, A.; Kundu, S. Developments in DNA metallization strategies for water splitting electrocatalysis: A review. Adv. Colloid Interface Sci. 2020, 282, 102205. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.; Wang, K.; Huitink, D.; Liang, H. Photoinduced Formation of Electrically Conductive Thin Palladium Nanowires on DNA Scaffolds. Langmuir 2009, 25, 10146–10152. [Google Scholar] [CrossRef]
- Berti, L.; Alessandrini, A.; Facci, P. DNA-templated photoinduced silver deposition. J. Am. Chem. Soc. 2005, 127, 11216–11217. [Google Scholar] [CrossRef]
- Watson, S.M.D.; Mohamed, H.D.A.; Horrocks, B.R.; Houlton, A. Electrically conductive magnetic nanowires using an electrochemical DNA-templating route. Nanoscale 2013, 5, 5349–5359. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, H.D.A.; Watson, S.M.D.; Horrocks, B.R.; Houlton, A. Chemical and electrochemical routes to DNA-templated rhodium nanowires. J. Mater. Chem. C 2015, 3, 438–446. [Google Scholar] [CrossRef]
- Zhu, D.; Pei, H.; Chao, J.; Su, S.; Aldalbahi, A.; Rahaman, M.; Wang, L.; Wang, L.; Huang, W.; Fan, C.; et al. Poly-adenine-based programmable engineering of gold nanoparticles for highly regulated spherical DNAzymes. Nanoscale 2015, 7, 18671–18676. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.; He, G.; Wang, C.; Li, S.; Zhao, X.; Xu, Y.; Mi, X. Poly-adenine-mediated spherical nucleic acid probes for live cell fluorescence imaging of tumor-related microRNAs. Mol. Biol. Rep. 2022, 49, 3705–3712. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luan, B.Q.; Yang, Z.Y.; Zhang, X.Y.; Ritzo, B.; Gates, K.; Gu, L.Q. Single Molecule Investigation of Ag+ Interactions with Single Cytosine-, Methylcytosine- and Hydroxymethylcytosine-Cytosine Mismatches in a Nanopore. Sci. Rep. 2014, 4, 5883. [Google Scholar] [CrossRef]
- Zhu, D.; Chao, J.; Pei, H.; Zuo, X.L.; Huang, Q.; Wang, L.H.; Huang, W.; Fan, C.H. Coordination-Mediated Programmable Assembly of Unmodified Oligonucleotides on Plasmonic Silver Nanoparticles. ACS Appl. Mater. Interfaces 2015, 7, 11047–11052. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, Q.; Ye, X.; Cao, Y.; Hu, C.; Liu, R.; Liu, Y. Spectrophotometric determination of single-stranded DNA with self-assembly hairpin DNA and silver nanoparticles. Instrum. Sci. Technol. 2021, 49, 81–90. [Google Scholar] [CrossRef]
- Gür, F.N.; Schwarz, F.W.; Ye, J.; Diez, S.; Schmidt, T.L. Toward Self-Assembled Plasmonic Devices: High-Yield Arrangement of Gold Nanoparticles on DNA Origami Templates. ACS Nano 2016, 10, 5374–5382. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.; Santiago, I.; Ardavan, A.; Turberfield, A.J. Ordering Gold Nanoparticles with DNA Origami Nanoflowers. ACS Nano 2016, 10, 7303–7306. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhang, J.; Chen, X.; Huang, T.; Duan, X.; Li, W.; Wang, J. Silver Nanomaterials Regulated by Structural Competition of G-/C-Rich Oligonucleotides. J. Phys. Chem. C 2011, 115, 10370–10379. [Google Scholar] [CrossRef]
- Wu, L.; Wang, J.; Ren, J.; Qu, X. Ultrasensitive Telomerase Activity Detection in Circulating Tumor Cells Based on DNA Metallization and Sharp Solid-State Electrochemical Techniques. Adv. Funct. Mater. 2014, 24, 2727–2733. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, J.; Yu, Z.; Li, G. Synthesis of tunable DNA-directed trepang-like Au nanocrystals for imaging application. Nanoscale 2019, 11, 18099–18108. [Google Scholar] [CrossRef]
- Zhao, Y.; Dai, X.; Wang, F.; Zhang, X.; Fan, C.; Liu, X. Nanofabrication based on DNA nanotechnology. Nano Today 2019, 26, 123–148. [Google Scholar] [CrossRef]
- Eichhorn, G.L. Inorganic Biochemistry; Elsevier Scientific Publishing Company: Amsterdam, The Netherlands, 1973; Volume 2. [Google Scholar]
- Stoltenberg, R.M.; Woolley, A.T. DNA-Templated Nanowire Fabrication. Biomed. Microdev. 2004, 6, 105–111. [Google Scholar] [CrossRef]
- Ritchie, C.M.; Johnsen, K.R.; Kiser, J.R.; Antoku, Y.; Dickson, R.M.; Petty, J.T. Ag Nanocluster Formation Using a Cytosine Oligonucleotide Template. J. Phys. Chem. C 2007, 111, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, N.; Prozorov, T. Direct Observation of Early Stages of Growth of Multilayered DNA-Templated Au-Pd-Au Core-Shell Nanoparticles in Liquid Phase. Front. Bioeng. Biotechnol. 2019, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.Z.; Wang, X.; Xing, Y.K.; Ren, S.K.; Teng, N.; Wang, J.; Chao, J.; Wang, L.H. DNA origami-templated assembly of plasmonic nanostructures with enhanced Raman scattering. Nucl. Sci. Tech. 2018, 29, 6. [Google Scholar] [CrossRef]
- Thacker, V.V.; Herrmann, L.O.; Sigle, D.O.; Zhang, T.; Liedl, T.; Baumberg, J.J.; Keyser, U.F. DNA origami based assembly of gold nanoparticle dimers for surface-enhanced Raman scattering. Nat. Commun. 2014, 5, 3448. [Google Scholar] [CrossRef] [PubMed]
- Kumaravel, S.; Karthick, K.; Sankar, S.S.; Karmakar, A.; Madhu, R.; Kundu, S. Prospects in interfaces of biomolecule DNA and nanomaterials as an effective way for improvising surface enhanced Raman scattering: A review. Adv. Colloid Interface Sci. 2021, 291, 102399. [Google Scholar] [CrossRef]
- Zhang, Q.; Han, L.; Jing, H.; Blom, D.A.; Lin, Y.; Xin, H.L.; Wang, H. Facet Control of Gold Nanorods. ACS Nano 2016, 10, 2960–2974. [Google Scholar] [CrossRef]
- Talamini, L.; Violatto, M.B.; Cai, Q.; Monopoli, M.P.; Kantner, K.; Krpetić, Ž.; Perez-Potti, A.; Cookman, J.; Garry, D.; Silveira, C.P.; et al. Influence of Size and Shape on the Anatomical Distribution of Endotoxin-Free Gold Nanoparticles. ACS Nano 2017, 11, 5519–5529. [Google Scholar] [CrossRef]
- De Silva Indrasekara, A.S.; Johnson, S.F.; Odion, R.A.; Vo-Dinh, T. Manipulation of the Geometry and Modulation of the Optical Response of Surfactant-Free Gold Nanostars: A Systematic Bottom-Up Synthesis. ACS Omega 2018, 3, 2202–2210. [Google Scholar] [CrossRef]
- Gao, J.; Huang, L.; Zhang, Z.; Li, G. Synthesis of sea urchin-shaped Au nanocrystals by double-strand diblock oligonucleotides for surface-enhanced Raman scattering and catalytic application. Nanotechnology 2021, 32, 175501. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Wang, J.; Song, C.; Li, Q.; Yang, Y.; Teng, N.; Su, S.; Zhu, D.; Huang, W.; Chao, J.; et al. Single-Step Organization of Plasmonic Gold Metamaterials with Self-Assembled DNA Nanostructures. Research 2019, 2019, 7403580. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; He, F.; Fang, W.; Shen, J.; Liu, X.; Xue, Y.; Liu, H.; Li, J.; Wang, L.; Li, Y.; et al. DNA-Guided Room-Temperature Synthesis of Single-Crystalline Gold Nanostructures on Graphdiyne Substrates. ACS Cent. Sci. 2020, 6, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Uprety, B.; Gates, E.P.; Geng, Y.; Woolley, A.T.; Harb, J.N. Site-Specific Metallization of Multiple Metals on a Single DNA Origami Template. Langmuir 2014, 30, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Tan, L.H.; Hwang, K.; Xing, H.; Wu, P.; Li, W.; Lu, Y. DNA sequence-dependent morphological evolution of silver nanoparticles and their optical and hybridization properties. J. Am. Chem. Soc. 2014, 136, 15195–15202. [Google Scholar] [CrossRef]
- Wang, Z.G.; Liu, Q.; Li, N.; Ding, B.Q. DNA-Based Nanotemplate Directed In Situ Synthesis of Silver Nanoclusters with Specific Fluorescent Emission: Surface-Guided Chemical Reactions. Chem. Mater. 2016, 28, 8834–8841. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, L.; Zhao, A.; Zhang, Z.; Wang, Z.; Lin, Y.; Ren, J.; Qu, X. Coupling exonuclease III with DNA metallization for amplified detection of biothiols at picomolar concentration. Biosens. Bioelectron. 2014, 58, 214–218. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, J.; Ekman, J.M.; Kenis, P.J.A.; Lu, Y. DNA-Mediated Control of Metal Nanoparticle Shape: One-Pot Synthesis and Cellular Uptake of Highly Stable and Functional Gold Nanoflowers. Nano Lett. 2010, 10, 1886–1891. [Google Scholar] [CrossRef]
- Song, T.; Tang, L.; Tan, L.H.; Wang, X.; Satyavolu, N.S.; Xing, H.; Wang, Z.; Li, J.; Liang, H.; Lu, Y. DNA-Encoded Tuning of Geometric and Plasmonic Properties of Nanoparticles Growing from Gold Nanorod Seeds. Angew. Chem. Int. Ed. 2015, 54, 8114–8118. [Google Scholar] [CrossRef]
- Wang, Z.; Tang, L.; Tan, L.H.; Li, J.; Lu, Y. Discovery of the DNA “Genetic Code” for Abiological Gold Nanoparticle Morphologies. Angew. Chem. Int. Ed. 2012, 51, 9078–9082. [Google Scholar] [CrossRef]
- Liu, G.; Shao, Y.; Peng, J.; Dai, W.; Liu, L.; Xu, S.; Wu, F.; Wu, X. Highly thymine-dependent formation of fluorescent copper nanoparticles templated by ss-DNA. Nanotechnology 2013, 24, 345502. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, J.; Fang, Z.; Zeng, L. Random dsDNA-templated formation of copper nanoparticles as novel fluorescence probes for label-free lead ions detection. Chem. Commun. 2012, 48, 1057–1059. [Google Scholar] [CrossRef]
- Diao, W.; Tang, M.; Ding, S.; Li, X.; Cheng, W.; Mo, F.; Yan, X.; Ma, H.; Yan, Y. Highly sensitive surface plasmon resonance biosensor for the detection of HIV-related DNA based on dynamic and structural DNA nanodevices. Biosens. Bioelectron. 2018, 100, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Huh, J.; Park, W.; Lee, L.P.; Kwon, Y.J.; Sim, S.J. Gold nanocrystals with DNA-directed morphologies. Nat. Commun. 2016, 7, 12873. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Wang, M.; Dong, J.; Zhou, C.; Wang, Q. Modular Assembly of Plasmonic Nanoparticles Assisted by DNA Origami. Langmuir 2018, 34, 14963–14968. [Google Scholar] [CrossRef]
- Tian, C.; Cordeiro, M.A.L.; Lhermitte, J.; Xin, H.L.L.; Shani, L.; Liu, M.Z.; Ma, C.L.; Yeshurun, Y.; DiMarzio, D.; Gang, O. Supra-Nanoparticle Functional Assemblies through Programmable Stacking. ACS Nano 2017, 11, 7036–7048. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Su, Z.M.; Zhou, Y.D.; Meyer, T.; Ke, Y.G.; Wang, Q.B.; Chiu, W.; Liu, N.; Zou, S.L.; Yan, H.; et al. Programmable Supra-Assembly of a DNA Surface Adapter for Tunable Chiral Directional Self-Assembly of Gold Nanorods. Angew. Chem. Int. Ed. 2017, 56, 14632–14636. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, C.; Cao, F.; Ren, J.; Qu, X. DNA metallization: Principles, methods, structures, and applications. Chem. Soc. Rev. 2018, 47, 4017–4072. [Google Scholar] [CrossRef]
- Sharma, J.; Rocha, R.C.; Phipps, M.L.; Yeh, H.C.; Balatsky, K.A.; Vu, D.M.; Shreve, A.P.; Werner, J.H.; Martinez, J.S. A DNA-templated fluorescent silver nanocluster with enhanced stability. Nanoscale 2012, 4, 4107–4110. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yin, M.; Pu, F.; Ren, J.; Qu, X. DNA-Templated Silver Nanoparticles as a Platform for Highly Sensitive and Selective Fluorescence Turn-On Detection of Dopamine. Small 2011, 7, 1557–1561. [Google Scholar] [CrossRef]
- Hao, L.; Zhao, L.; Li, G.; Li, Y.; Ma, L.; Liu, Y.; Wang, W.; Kong, J. Ultrasensitive detection of CYFRA 21-1 DNA via SI-RAFT based in-situ metallization signal amplification. Microchem. J. 2020, 158, 105216. [Google Scholar] [CrossRef]
- Gong, L.; Kuai, H.; Ren, S.; Zhao, X.-H.; Huan, S.-Y.; Zhang, X.-B.; Tan, W. Ag nanocluster-based label-free catalytic and molecular beacons for amplified biosensing. Chem. Commun. 2015, 51, 12095–12098. [Google Scholar] [CrossRef]
- Chen, Z.; Lin, Y.; Zhao, C.; Ren, J.; Qu, X. Silver metallization engineered conformational switch of G-quadruplex for fluorescence turn-on detection of biothiols. Chem. Commun. 2012, 48, 11428–11430. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Xu, G.; Sun, Y.; Zheng, W.; Zhu, X.; Wang, B.; Zhang, X.; Wang, G. A “turn-on” silver nanocluster based fluorescent sensor for folate receptor detection and cancer cell imaging under visual analysis. Chem. Commun. 2015, 51, 11810–11813. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Yu, J.; Patel, S.A.; Tzeng, Y.L.; Dickson, R.M. Tailoring silver nanodots for intracellular staining. Photochem. Photobiol. Sci. 2011, 10, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.C.; Yu, Y.; Jin, G.; Li, K.; Lu, Y.; Xie, J.; Tan, Y.N. Establishing empirical design rules of nucleic acid templates for the synthesis of silver nanoclusters with tunable photoluminescence and functionalities towards targeted bioimaging applications. Nanoscale Adv. 2020, 2, 3921–3932. [Google Scholar] [CrossRef]
- Han, G.-M.; Jia, Z.-Z.; Zhu, Y.-J.; Jiao, J.-J.; Kong, D.-M.; Feng, X.-Z. Biostable L-DNA-Templated Aptamer-Silver Nanoclusters for Cell-Type-Specific Imaging at Physiological Temperature. Anal. Chem. 2016, 88, 10800–10804. [Google Scholar] [CrossRef]
- Ran, X.; Wang, Z.; Zhang, Z.; Pu, F.; Ren, J.; Qu, X. Nucleic-acid-programmed Ag-nanoclusters as a generic platform for visualization of latent fingerprints and exogenous substances. Chem. Commun. 2016, 52, 557–560. [Google Scholar] [CrossRef]
- Li, J.; You, J.; Zhuang, Y.; Han, C.; Hu, J.; Wang, A.; Xu, K.; Zhu, J.J. A “light-up” and “spectrum-shift” response of aptamer-functionalized silver nanoclusters for intracellular mRNA imaging. Chem. Commun. 2014, 50, 7107–7110. [Google Scholar] [CrossRef]
- Hamner, K.L.; Alexander, C.M.; Coopersmith, K.; Reishofer, D.; Provenza, C.; Maye, M.M. Using temperature-sensitive smart polymers to regulate DNA-mediated nanoassembly and encoded nanocarrier drug release. ACS Nano 2013, 7, 7011–7020. [Google Scholar] [CrossRef]
- Tao, Y.; Ju, E.; Ren, J.; Qu, X. Metallization of plasmid DNA for efficient gene delivery. Chem. Commun. 2013, 49, 9791–9793. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, C.; Cheng, L.; Lee, S.T.; Liu, Z. Noble metal coated single-walled carbon nanotubes for applications in surface enhanced Raman scattering imaging and photothermal therapy. J. Am. Chem. Soc. 2012, 134, 7414–7422. [Google Scholar] [CrossRef]
- Liu, C.; Qing, Z.; Zheng, J.; Deng, L.; Ma, C.; Li, J.; Li, Y.; Yang, S.; Yang, J.; Wang, J.; et al. DNA-templated in situ growth of silver nanoparticles on mesoporous silica nanospheres for smart intracellular GSH-controlled release. Chem. Commun. 2015, 51, 6544–6547. [Google Scholar] [CrossRef] [PubMed]
- Laramy, C.R.; O’Brien, M.N.; Mirkin, C.A. Crystal engineering with DNA. Nat. Rev. Mater. 2019, 4, 201–224. [Google Scholar] [CrossRef]
- Xie, M.; Fang, W.; Qu, Z.; Hu, Y.; Zhang, Y.; Chao, J.; Shi, J.; Wang, L.; Wang, L.; Tian, Y.; et al. High-entropy alloy nanopatterns by prescribed metallization of DNA origami templates. Nat. Commun. 2023, 14, 1745. [Google Scholar] [CrossRef]
- He, M.-Q.; Yu, Y.-L.; Wang, J.-H. Biomolecule-tailored assembly and morphology of gold nanoparticles for LSPR applications. Nano Today 2020, 35, 101005. [Google Scholar] [CrossRef]
- Fu, J.T.; Zhang, Z.M.; Li, G.K. Progress on the development of DNA-mediated metal nanomaterials for environmental and biological analysis. Chin. Chem. Lett. 2019, 30, 285–291. [Google Scholar] [CrossRef]
- Xie, M.; Hu, Y.; Yin, J.; Zhao, Z.; Chen, J.; Chao, J. DNA Nanotechnology-Enabled Fabrication of Metal Nanomorphology. Research 2022, 2022, 9840131. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.D.; Xu, Y.W.; Hu, P.; Yao, C.; Yang, D.Y. Construction and applications of DNA-based nanomaterials in cancer therapy. Chin. Chem. Lett. 2022, 33, 1131–1140. [Google Scholar] [CrossRef]

| Seeds | Type of DNA Templates | Shape of Nanomaterials | References |
|---|---|---|---|
| AuNPs/AuNRs functionalized with thiol-DNA | Ribbon-like DNA origami nanostructures | 1D AuNPs/AuNRs lines or 2D AuNPs lattices | [54] |
| 30 nm AuNPs | Rectangular DNA origami | AuNP chains | [47] |
| Graphdiyne and HAuCl4 | ssDNAs (A20) | Decahedrons, icosahedrons, or flower-like Au nanoparticles | [55] |
| Gold nanorods | ssDNA, dsDNA, hpDNA, and taDNA | Trepang-like AuNCs | [41] |
| AuNPs and Cu | Bar-shaped DNA origami | Cu-Au metal junction | [56] |
| Ag Nanocube | 10-mer oligo-A, -T, -C | Edge-truncated octahedral, and truncated tetrahedral AgNPs | [57] |
| AgNO3 | G-/C-Rich oligonucleotides | Silver clusters | [39] |
| AgNO3 | Triangular DNA origami template | Ag nanoclusters | [58] |
| AgNO3 | target DNA, its complementary sequence parts, and probe DNA G4) | Colloidal silver solution | [59] |
| AgNO3 | Telomerase primer oligonucleotides | Silver NPs | [40] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Liu, Y.; Lou, B.; Tan, Y.; Chen, L.; Liu, Z. DNA-Guided Metallization of Nanomaterials and Their Biomedical Applications. Molecules 2023, 28, 3922. https://doi.org/10.3390/molecules28093922
Li K, Liu Y, Lou B, Tan Y, Chen L, Liu Z. DNA-Guided Metallization of Nanomaterials and Their Biomedical Applications. Molecules. 2023; 28(9):3922. https://doi.org/10.3390/molecules28093922
Chicago/Turabian StyleLi, Ke, Yanfei Liu, Beibei Lou, Yifu Tan, Liwei Chen, and Zhenbao Liu. 2023. "DNA-Guided Metallization of Nanomaterials and Their Biomedical Applications" Molecules 28, no. 9: 3922. https://doi.org/10.3390/molecules28093922
APA StyleLi, K., Liu, Y., Lou, B., Tan, Y., Chen, L., & Liu, Z. (2023). DNA-Guided Metallization of Nanomaterials and Their Biomedical Applications. Molecules, 28(9), 3922. https://doi.org/10.3390/molecules28093922






