Abstract
Fe (II)-and 2-ketoglutarate-dependent dioxygenases (Fe (II)/α-KG DOs) have been applied to catalyze hydroxylation of amino acids. However, the Fe (II)/α-KG DOs that have been developed and characterized are not sufficient. L-isoleucine dioxygenase (IDO) is an Fe (II)/α-KG DO that specifically catalyzes the formation of 4-hydroxyisoleucine (4-HIL) from L-isoleucine (L-Ile) and exhibits a substrate specificity toward L-aliphatic amino acids. To expand the substrate spectrum of IDO toward aromatic amino acids, in this study, we analyzed the regularity of the substrate spectrum of IDO using molecular dynamics (MD) simulation and found that the distance between Fe2+, C2 of α-KG and amino acid chain’s C4 may be critical for regulating the substrate specificity of the enzyme. The mutation sites (Y143, S153 and R227) were also subjected to single point saturation mutations based on polarity pockets and residue free energy contributions. It was found that Y143D, Y143I and S153A mutants exhibited catalytic L-phenylalanine activity, while Y143I, S153A, S153Q and S153Y exhibited catalytic L-homophenylalanine activity. Consequently, this study extended the substrate spectrum of IDO with aromatic amino acids and enhanced its application property.
1. Introduction
Hydroxyamino acids are unusual amino acids that are commonly found in nature. They exist as secondary metabolites and components of peptides and proteins. Many hydroxyamino acids are components of sugars, peptides and antibiotics and also play an important role in the chemical synthesis of other compounds as precursors and chiral auxiliaries [1,2,3,4,5,6].
Fe (II)- and 2-ketoglutarate-dependent dioxygenases (Fe (II)/α-KG DOs) are a diverse superfamily of proteins that catalyze a variety of oxidative transformations. Industrially, these enzymes are mainly used in the biosynthesis of hydroxylated amino acids and antibiotics [7,8,9,10,11]. Currently, there is a common understanding of the Fe (II)/α-KG DO reaction mechanism for hydroxylation activity. The Fe (II)/α-KG DO requires Fe (II), usually bound by three residues, to form a highly conserved HXD/E…H triad [12]. α-KG forms complexes with iron in a bidentate manner via its C-1 carboxylate and ketone oxygens [13]. When the substrate binds to the enzyme active site (not, however, to the Fe ion), Fe (II)-bound water is displaced, thus creating a site for binding an O2 molecules, generating an Fe (III)-superoxo intermediate. The distal oxygen atom of the Fe (III)-superoxo intermediate attacks C2 of α-KG to release CO2 and to yield Fe (IV)=O (ferryl) intermediate. The Fe (IV)=O intermediate attacks the C-H closer to the substrate to form a hydroxylation reaction. Therefore, the distance between the iron ion and the C-H of the substrate is very important [13]. However, the regulation of enzyme selectivity of the Fe (II)/α-KG DO-mediated hydroxylation remains unclear. In addition, not enough Fe (II)/α-KG DOs have been developed and identified, and only a few specific hydroxylases have been reported, such as L-proline 3-hydroxylase from Streptomyces, L-proline 3-hydroxylase from Bacillus thuringiensis, L-proline cis-4-hydroxylase from Mesorhizobium loti, L-proline cis-4-hydroxylase from Sinorhizobium meliloti, L-proline trans-4-hydroxylase from Dactylosporangium, L-proline trans-4-hydroxylase from Amycolatopsis and L-isoleucine dioxygenase (IDO) from Bacillus thuringiensis [14]. In short, these factors prevent the application of these enzymes in the production of various hydroxylated amino acids.
IDO is an Fe(II)/α-KG DO that specifically catalyzes the generation of 4-hydroxyisoleucine (4-HIL) from L-isoleucine (L-Ile) [15,16]. In addition to L-Ile as the target substrate, IDO can also act on other L-aliphatic amino acids and has a substrate spectrum toward such amino acid substrates [17,18,19]. We have previously solved the IDO structure and analyzed the catalytic orientation mechanism of its hydroxylation [20].
To further expand the substrate spectrum of IDO reactions, especially toward aromatic amino acids, we used molecular docking and molecular dynamics (MD) to simulate the enzyme dynamics of Fe2+ and α-KG, L-amino acids [L-Ile, D-isoleucine (D-Ile), L-leucine (L-Leu), L-norleucine (L-Nle), L-norvaline (L-Nva), L-phenylalanine (PHE) and L-homophenylalanine (HPHE)]. Then, single point saturation mutagenesis at selected mutation sites was carried out based on polarity pockets and residue free energy contribution, thereby expanding the IDO substrate spectrum for aromatic amino acids to enhance their application properties.
2. Results and Discussion
2.1. MD Simulation of IDO with L-Aliphatic Amino Acids
Our previous structural analysis showed that the IDO monomer (PDB: 6lnh, Chain A) has 240 amino acids (with the N-terminal purification tag removed) and contains a conserved motif H159-X1-D161-X50-H212 that chelates Fe2+, the active site of IDO [20]. Molecular dynamics simulations using Fe2+, α-KG and L-Ile as substrates showed that in the IDO active pockets, S153 and P155 formed hydrogen bonds with the amino group of L-Ile, and residues Y143 and R227 had hydrogen bonds to the carboxyl group of L-Ile (Figure 1). Residues S153, P155, Y143 and R227 formed a polar substrate binding pocket, and the formation of this pocket restricted the other end of L-Ile toward Fe2+ and α-KG, when the carbonyl group of α-KG (opposite D161 bound to Fe2+) and the C1 position carboxyl group (opposite H159, H212 bound to Fe2+) are bound to Fe2+ in a dentate bound (Figure 1). This makes the C4 of L-Ile in proximity to the Fe2+ (5.9 Å) and C2 of α-KG (4.7 Å). Such a distance may facilitate the attack of oxygen molecules (the diameter of oxygen molecule is about 3.5 Å) on C4 of L-Ile, where hydroxylation occurs [21,22]. Thus, the distance between Fe2+, C2 of α-KG and the fourth carbon of L-Ile (where hydroxylation occurs) may be crucial for the incorporation of oxygen molecules and oxidation reactions and important for substrate recognition and catalytic orientation.
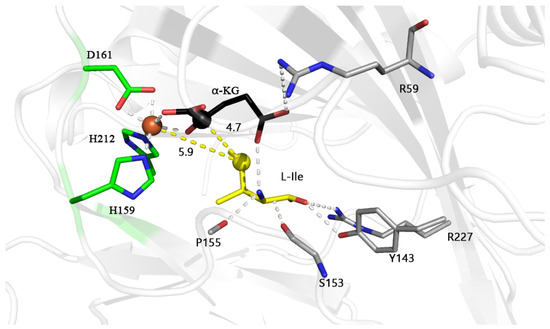
Figure 1.
Final structure of the active site of L-isoleucine dioxygenase (IDO) with L-isoleucine (L-Ile) model during molecular dynamics at 200 ns; Fe2+, C2 atom of α-KG and C4 atom of L-Ile are shown in brown, black and yellow spheres, and distance between atoms is also indicated by yellow dashed lines. Possible hydrogen bonds are indicated by white dashed lines. The unit of distance is Å.
IDO has a certain substrate spectrum for L-amino acids [17,18,19]. To explore this regularity, we chose L-aliphatic amino acids that can be catalyzed by IDO (although with lower catalytic efficiency), including L-Leu, L-Nva and L-Nle, and analyzed the reaction mechanism by MD simulations [17]. MD simulations showed that the closest distance to Fe2+ was located at C4 of the carbon chain (Fe2+ distances to L-Leu, L-Nva and L-Nle: 5.4, 5.1 and 5.2 Å, respectively) (Figure 2a–c). It is also interesting to note that the distance between C2 of α-KG and the C4 position of these L-aliphatic amino acids remains essentially around 4.4 Å (Figure 2), while it is as long as 6.2 Å in the D-Ile (L-Ile tautomer with a similar structure) which does not react with IDO (Figure 2d). Furthermore, MD simulations showed that the distance between Fe2+, C2 of α-KG and C4 position of D-Ile fluctuated greatly and was very unstable, whereas the other amino acids quickly reached a stable state (Figure 2e,f). This distance between Fe2+, C2 of α-KG and C4 of the L-aliphatic amino acid may be more favorable to IDO hydroxylated amino acid at the C4 position. Previous studies on Fe (II)/α-KG DOs suggest that α-KG binds reactive ferrous ions and displaces two water molecules to form an octahedral ligand complex [23,24,25]. During the reaction, one of the water molecules involved in the octahedral ligand complex formation is replaced by a molecule of oxygen, which, through oxidative decarboxylation, produces 1,4-butanedioic acid, CO2 and iron oxide [Fe(IV)=O], followed by high-valent iron interactions with the substrate to undergo hydroxylation [26,27,28,29]. In contrast, the distance between α-KG and the C4 position of the L-aliphatic amino acid may make the geometrical configuration of Fe-O more favorable to attack the C4 position of the amino acid for hydroxylation reactions through spatial crowding. This may be one reason why IDO has a favorable substrate spectrum for L-aliphatic amino acids.
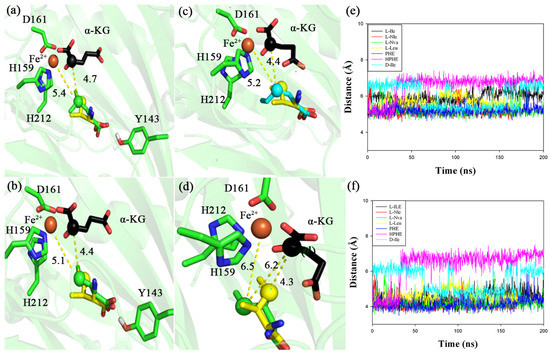
Figure 2.
Molecular structure around active site of different substrates during molecular dynamics simulation at 200 ns. IDO substrates are L-leucine (L-Leu, (a), green), L-norvaline (L-Nva, (b), green), L-norleucine (L-Nle, (c), cyan), D-isoleucine (D-Ile, (d), green) and L-isoleucine (L-Ile, (a–d), yellow), respectively. Distance between C4 of different substrates and Fe2+ (e), C2 of α-KG (f) during IDO molecular dynamics simulation. L-Ile (black), L-Nle (red), L-Nva (green), L-Leu (yellow), PHE (blue), HPHE (purple), D-Ile (cyan). The unit of distance is Å.
2.2. Analysis of the Interaction between IDO and Aromatic Amino Acids
Based on the above catalytic mechanism of IDO for L-aliphatic amino acids, MD simulations were carried out for aromatic amino acids (PHE and HPHE) to speculate whether IDO could react with them. MD simulations showed that the carbon chain orientation of PHE and HPHE was consistent with the above L-aliphatic amino acids. The distances between Fe2+ and the C4 position of PHE and HPHE were 5.1 and 6.8 Å, respectively (Figure 3a,b), and the distances between the C2 of α-KG and the C4 position of these amino acids were 4.3 and 6.9 Å, respectively (Figure 3a,b). Previous reaction patterns suggested that IDO recognition of the substrate requires hydroxylation occurs at methylene or methine carbon of the amino acid but not methyl [17]. Therefore, although C4 of PHE is in close proximity to Fe2+ and C2 of α-KG (5.1 and 4.3 Å, respectively), C4 of the carbon chain is a benzene ring, not a methylene or methine carton, and therefore presumably IDO cannot catalyze its hydroxylation. In contrast, C4 of HPHE is farther away from Fe2+ and C2 of α-KG (6.8 and 6.9 Å, respectively) and presumably cannot be hydroxylated by IDO. To further test this hypothesis, we purified the IDO protein and used IDO to catalyze the reaction with PHE and HPHE in vitro, showing no reaction between them, suggesting that IDO cannot catalyze PHE and HPHE (Figure 3c,d).
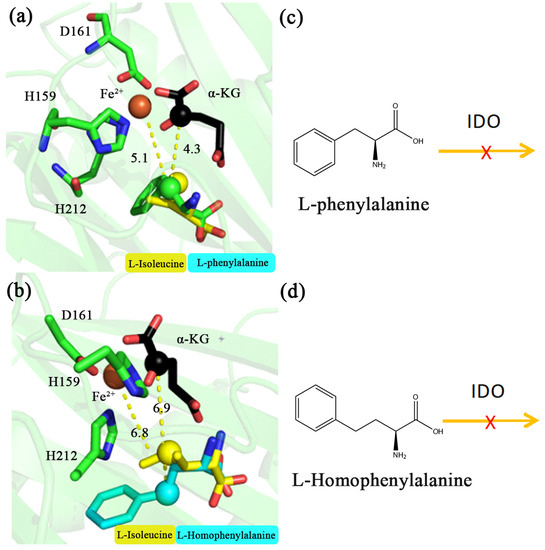
Figure 3.
Molecular structure around active site of L-phenylalanine and L-homophenylalanine during molecular dynamics simulation at 200 ns. IDO substrates are L-phenylalanine (a), green, L-homophenylalanine (b), cyan and L-isoleucine (a,b), yellow, respectively. Pattern of L-phenylalanine (c) and L-homophenylalanine (d) that cannot be catalyzed by IDO. The unit of distance is Å.
2.3. Structure-Guided Prediction of IDO Mutation Sites
To extend the reaction of IDO toward aromatic amino acids, we used MD simulations for the selection of IDO mutation sites. There was an energy difference between the free and the bound states of the ligand and the receptor of a protein, which can be used to measure the affinity of their binding [30,31,32]. Analysis of the residue contribution using binding free energy calculations would be helpful to understanding the catalytic mechanism and thus to designing enzyme modification at the molecular level [32]. To further investigate the contribution of IDO residues to the reaction, the binding energies of the IDO, Fe2+, α-KG and L-Ile complex systems were simulated and calculated by molecular dynamics, with a binding energy of −34.11 kcal/mol for the IDO complex system, and the top ten residues contributing to the selectivity were labeled by decomposing their contribution to the binding energy (Table 1), among which the top ranked residues are mostly hydrophilic residues, indicating that hydrophilicity is a key factor affecting IDO catalysis. The residues with an absolute value of binding energy contribution greater than 1 kcal/mol were displayed at 200 ns, and it can be seen that these residues are distributed near the binding cavity substrate (Figure 4). Based on the residue contribution and polarity pockets (S153, P155, Y143 and R227), we selected Y143, S153 and R227, which contribute more than others, as the mutation sites.

Table 1.
Energy contribution of IDO key residues.
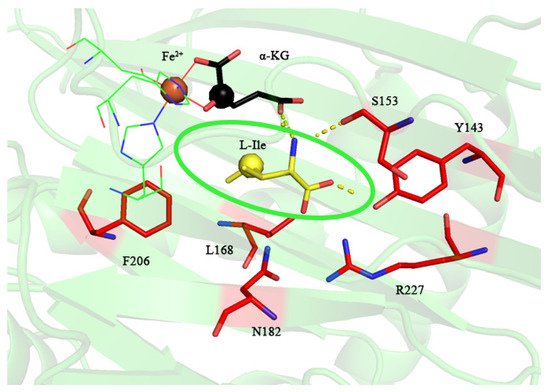
Figure 4.
Display diagram in energy contribution of IDO key residues. The green elliptical line represents the substrate molecule L-Ile.
2.4. Construction, Expression and Purification of IDO Mutant
The single point saturation mutation of residues Y143, S153 and R227 were performed by gene synthesis, respectively, and 5′(NcoI), 5′UTR (AA) and 3′(SalI) were added at both ends of the sequence and cloned into the vector pET-28a (+). The constructed pET-28a (+)-ido mutant vector was confirmed by EcoRV and Xhol digestion and sequencing. Different IDO mutants were expressed in E. coli BL21 (DE3), and then the proteins were extracted and purified by ultrasound and a Ni column (Figure 5). The single point saturation mutation at residue Y143, S153 or R227 did not have an obvious influence on protein expression.
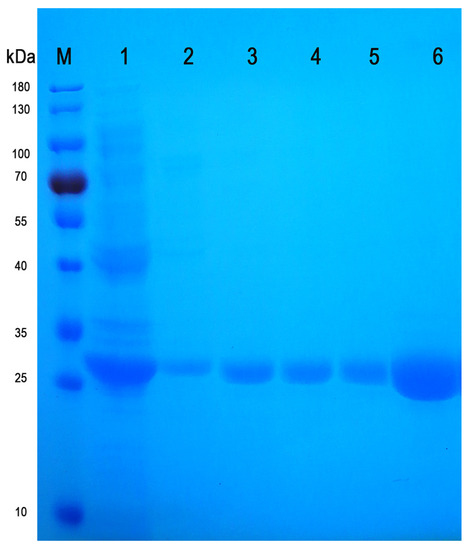
Figure 5.
SDS-PAGE electrophoresis diagram of mutant IDO and IDO-WT. Protein marker (M), representative total protein expression (line 1), representative purified mutant IDO (lines 2–5), purified IDO-WT (line 6).
2.5. Enzymatic Activity Assay and Kinetic Characterization of IDO-WT and IDO Mutants
We performed enzyme activity assays on IDO-WT and IDO mutants. The enzyme activity results showed that IDO-WT had high enzyme activity (0.55 ± 0.15 μmol/min/mg) on the optimal substrate L-Ile, but no enzyme activity on PHE and HPHE was detected. This indicated that IDO could not catalyze PHE and HPHE. We unexpectedly found that mutants Y143D, Y143I and S153A showed catalytic activity of PHE, and their enzyme activities were 0.056 ± 0.008, 0.025 ± 0.003 and 0.012 ± 0.002 μmol/min/mg (Table 2), respectively. We also found that the mutants Y143I, S153A, S153Q and S153Y were active toward HPHE with enzymatic activities of 0.014 ± 0.002, 0.035 ± 0.005, 0.063 ± 0.003 and 0.028 ± 0.004 μmol/min/mg (Table 2), respectively. To further investigate the enzyme’s ability to bind the substrate and catalyze the reaction rate, the enzyme kinetic parameters were examined, and the results are shown in Table 3. For PHE, among all mutants, the kcat/Km value of IDO-Y143I was the highest at 2.00 ± 0.01 s−1mM−1, indicating that the IDO-Y143I had the highest catalytic efficiency. For HPHE, among the various mutants, the kcat/Km value of IDO-S153Q was the highest at 3.50 ± 0.10 s−1mM−1, indicating that IDO-S153Q had the highest catalytic efficiency.

Table 2.
Specific activity of IDO mutants.

Table 3.
Kinetic parameters of IDO-WT and mutants.
To identify whether the product of the catalytic reaction was hydroxylated, LC-MS was performed. The results obtained from LC-MS analyses showed the presence of mass spectrometric peaks consistent with the molecular weight of the hydroxylated products, thereby confirming the formation of 2-amino-3-hydroxy-3-phenylpropanoic acid after hydroxylation of PHE catalyzed by mutant Y143D, Y143I and S153A (Figures S1–S3). Our findings verified the potential of IDO mutants to catalyze the hydroxylation of PHE.
These results indicated that the modification of the IDO molecule realizes its possibility to catalyze aromatic amino acids, although their catalytic activity is low. This will provide an idea to further improve the activity of IDO catalyzed aromatic amino acids.
2.6. Molecular Docking Analysis
The previous analysis has shown that because the C4 position of the PHE side chain is a benzene ring but not methylene or methine carbon, IDO cannot hydroxylate with it, although the C4 of the carbon chain is close to Fe2+ and C2 of α-KG [17]. However, these mutants showed the activity of catalyzing PHE, and we speculated that the methylene at position C3 was hydroxylated [17]. Molecular docking results showed that the distance between C3 of PHE and Fe2+ in mutants Y143D, Y143I and S153A was 6.9, 6.7 and 6.7 Å (Figure 6b–d), respectively, and the distances to C2 of α-KG was 4.8, 4.6 and 4.8 Å (Figure 6b–d), respectively, which was closer than that of d (Fe2+-PHEC3) (7.5 Å), d (α-KGC2-PHEC3) (5.3 Å) in IDO-WT (Figure 6a). Moreover, the binding free energies of these mutants to PHE were decreased (−6.2, −6.5, −6.4 and −6.5 kcal/mol for IDO-WT, Y143D, Y143I and S153A, respectively) (Table 4), which may favor the hydroxylation of Fe-O attacking C3 of PHE to produce the non-essential amino acid 2-amino-3-hydroxy-3-phenylpropanoic acid. Of course, this does not exclude the possibility of the positive mutant also acting on the benzene ring [33,34,35,36]. In the future, we should accurately detect the reaction site in the tested amino acid substrate.
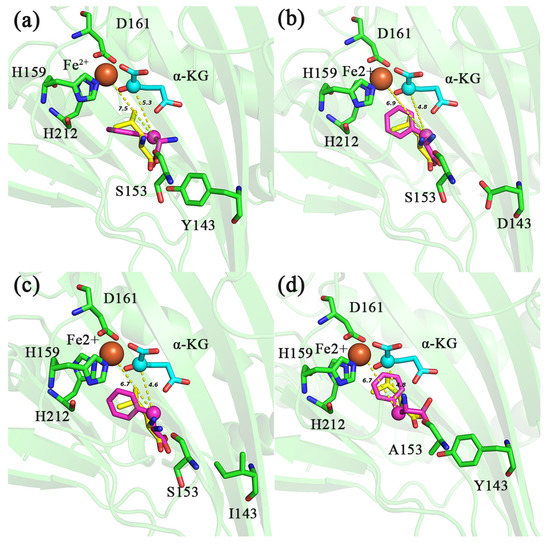
Figure 6.
Molecular docking diagram of different IDO mutants (Y143D-IDO: (b); Y143I-IDO: (c); S153A-IDO: (d)) and IDO-WT (a) with PHE and L-Ile. Distance between C2 of α-KG, Fe2+ and C4 of different substrates is indicated by yellow dashed lines. PHE (purple), L-Ile (yellow). The unit of distance is Å.

Table 4.
Receptor–ligand affinity.
The molecular docking results showed that the distance of C4 of HPHE from Fe2+ and C2 of α-KG in IDO-WT was 6.9 and 5.8 Å, respectively (Figure 7a), while the distance of C4 of HPHE from Fe2+ in mutants Y143I, S153A, S153Q and S153Y were 6.0, 6.5, 6.2 and 6.2 Å, respectively (Figure 7b–e), and the distances to C2 of α-KG were 4.7, 4.8, 5.0 and 4.9 Å (Figure 7b–e), respectively. This indicated that C4 of HPHE in the mutant was closer to Fe2+ and C2 of α-KG than that in the wild type, which would be more favorable for C4 of HPHE to be attacked. Moreover, the binding free energy analysis showed that the binding free energies of Y143I, S153A, S153Q and S153Y to HPHE (−6.6, −6.5, −6.4 and −6.5 kcal/mol) were slightly lower compared to IDO-WT (−6.3 kcal/mol) (Table 4). Taken together, these site mutations may be more favorable for hydroxylation on C4 of PHE.
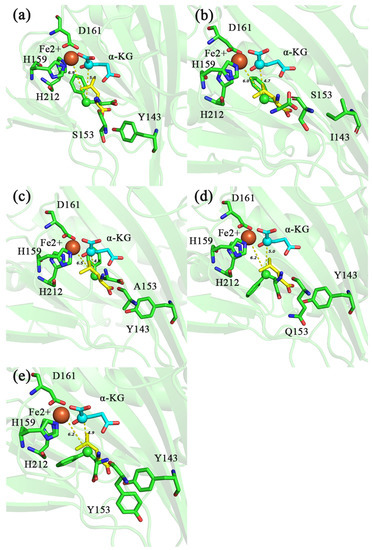
Figure 7.
Molecular docking diagram of different IDO mutants (Y143I-IDO: (b); S153A-IDO: (c); S153Q-IDO: (d); S153Y-IDO: (e)) and IDO-WT (a) with HPHE and L-Ile. Distance between C2 of α-KG, Fe2+ and C4 of different substrates is indicated by yellow dashed lines. HPHE (green), L-Ile (yellow). The unit of distance is Å.
However, the saturated mutant at residue R227 showed no catalytic activity for the aromatic amino acids (PHE and HPHE) (Table 2). This may be due to the influence of R227 on the conformation of the amino group of the amino acid, thus affecting the orientation of the substrate, which cannot keep an appropriate distance between the C3 or C4 of PHE and HPHE chains from Fe2+ and C2 of α-KG.
3. Materials and Methods
3.1. Materials
The bacterial strains and plasmids used in this study are listed in Table 5. E. coli DH5α was used as a cloning host. E. coli BL21(DE3) was used for expression of ido and mutant genes. The chemicals, including α-KG, L-Ile, L-Leu, L-Nva, L-Nle, D-Ile, PHE, HPHE, NaH2PO4, FeSO₄·7H₂O, NaCl, kanamycin, Isopropyl β-D-thiogalactoside (IPTG), imidazole and LB medium, were purchased from Sigma-Aldrich.

Table 5.
Strains and plasmids.
3.2. Mutagenesis Vector Construction
For saturation mutagenesis at each site, the ido mutant genes were designed by replacing the codon encoding the predicted residue with those encoding other 19 residues separately. A total of 57 ido mutant genes were synthesized by Suzhou GENEWIZ Biotechnology Co., Ltd. (Suzhou, China), with added 5′(NcoI), 5′UTR ([AA]) and 3′(SalI). The gene was cloned into expression vector pET-28a (+) (Kanamycin) by 5′ NcoI and 3′ SalI to construct pET-28a (+)-ido mutant, and then the plasmid was transformed into E. coli BL21 (DE3) for subsequent expression of the target protein [37].
3.3. Protein Expression and Purification
The plasmid, pET-28a (+)-ido involving wild-type ido gene or pET-28a (+)-ido mutant involving ido mutant gene, was transformed into E. coli BL21 (DE3) and smeared on an LB plate containing 50 μg/mL kanamycin. Single colonies were selected and identified by enzyme digestion. The single colonies of the recombinant strain were inoculated in 5 mL LB medium containing 50 μg/mL kanamycin and cultured overnight at 37 °C with 200 r/min shaking. The 5 mL seed liquid was transferred to 500 mL liquid medium containing 50 μg/mL kanamycin in LB medium. At 37 °C, the medium was shaken at 200 r/min and cultured to OD600 of about 0.6–0.8. IPTG with a final concentration of 1 mM was added to the medium and induced and cultured overnight at 25 °C and 200 r/min [37].
The precipitate was collected by centrifugation at 4000× g for 20 min and suspended in a lysis solution (50 mM NaH2PO4, 300 mM NaCl, pH 8.0). The cells resuspended in the lysate above were sonicated in an ice bath to break the bacteria. Sonication conditions: 360 w, 2 s of work, 3 s of interval, 15 min and 4 °C. The supernatant was collected by 12,000× g/min centrifugation for 20 min. The supernatant was filtered through a 0.22 μm membrane, and the crude enzyme solution was used for further purification.
The crude enzyme solution was then purified by applying a Ni-Charged Resin FF column (GenScript, Nanjing, China). Purification conditions: Wash buffer 1 (50 mM NaH2PO4, 300 mM NaCl, 150 mM imidazole, pH 8.0) and wash buffer 2 (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0) were used to elute non-target proteins. The target proteins were finally eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole, pH 8.0). A 12% SDS-PAGE was performed to determine the size of the target protein molecule and the degree of purification [38].
3.4. Enzyme Activity Determination
In this work, the enzyme activity of IDO and its variants was assayed by measuring the yield of succinic acid, which was generated accompanying the IDO-catalyzed hydroxylation. One unit of enzyme activity was defined as the amount of enzyme required to produce 1 μmol succinate per minute in a standard enzyme assay system. The catalytic reaction system configuration was as follows: 10 mM substrate, 10 mM α-KG, 0.5 mM FeSO4·7H2O, 10 mM ascorbic acid, 50 mM Bis-Tris (pH 6.0) and 0.2 mg/mL purified IDO or IDO mutants in 1 mL; reaction conditions: 25 °C, 10 min. The supernatant was collected by centrifugation at 4000× g for 10 min at 4 °C. The supernatant was assayed using the Succinic Acid Assay Kit (MAK184, Sigma-Aldrich, Inc., Shanghai, China). All assays were performed in triplicate.
3.5. Determination of Kinetic Parameters
The kinetic parameters were determined using 1 mL of the assay mixture. For PHE and HPHE, the standard conditions for the hydroxylation reaction and the activity assay with the Succinic Acid Assay method were used, and the concentration of PHE and HPHE was 0.01–5 mM. The assays were carried out in triplicate, and the Michaelis–Menten model was fitted using GraphPad Prism 9 (GraphPad, Inc., San Diego, CA, USA).
3.6. Analysis of the Catalytic Product
The products of the amino acid hydroxylation were analyzed using the Waters ACQUITY UPLC-MS/MS system. A Waters ACQUITY UPLC HSS C18 column (1.8 μm) was used. Gradient elution was performed for mobile phase A consisting of 20 mM NH4Ac-HAc buffer (pH 4.2) and mobile phase B consisting of acetonitrile. The flow rate was 200 μL/min, the column temperature was 25 °C, and the sample size was 5 μL.
3.7. Molecular Docking and Molecular Dynamics (MD) Simulation Analysis
The IDO monomer A (PDB: 6lnh, Chain A) containing an Fe ion was used as the initial structure of the enzyme, and the Fe ion was used as the reference site of the active center. α-KG and amino acids (L-Ile, L-leu, L-Nva, L-Nle, D-Ile, PHE and HPHE) were carefully aligned to the active site by AutoDock Vina. The protein atom was described by an AMBER18 ff14SB force field, and the substrate and 2OG were parameterized by the general AMBER force field (GAFF) [39,40]. The metal central bonding model was applied to this system. The whole modeling process was completed under the guidance of the Metal Center Parameter Builder (MCPB) program, which included the calculation of force constants, geometric optimization of active center residues and restrained electrostatic potential charge (RESP charge) using Gaussian09 [41]. After obtaining the force field parameters, AMBER further minimized the system to obtain the correct molecular structure. At 200 ns, the C1-N1-C4 atoms of other amino acid substrates were aligned with the L-Ile model structure, and a good docking structure was obtained.
The systems were then solvated in a rectangular box of TIP3P water and were neutralize with sodium ions. The total number of systems was about 32,000. The positions of water were then optimized with 800 steps of the steepest descent (SD) method and 9200 steps of the conjugate gradient (CD) method followed by 1800 steps of steepest descent (SD) method and 38,200 steps of the conjugate gradient (CD) method to the entire system. After that, the systems were gradually heated from 0 to 300 K over 50 ps in the NVT ensemble. Then, the systems were equilibrated for 75 ps under 1 atm of pressure. Finally, a molecular simulation of 200 ns was performed for each system, during which the Verlet algorithm with a time step of 2 fs was employed and the SHAKE algorithm adopted for the constraint of bond stretching of those covalent bonds involving hydrogen atoms [42,43].
To understand the interaction of IDO mutants with the substrates PHE and HPHE, we selected IDO-WT and mutants to dock PHE and HPHE, respectively. Molecular docking was performed by AutoDock Vina, based on the substrate L-Ile of the original IDO and its surrounding residues as the active pocket. At the end of docking, the scored optimal conformation was selected to merge with the protein and saved as a complex. The small molecule-receptor binding site and minimum binding energy data were obtained by Pymol and AutoDock Vina, respectively.
4. Conclusions
In this study, the regularity of the wider substrate spectrum of IDO was explored using our previously obtained IDO structure in combination with molecular docking and MD simulations, i.e., the distance of C4 on the amino acid chain to Fe2+ and C2 of α-KG may be critical. The selection of mutation sites for single point saturation mutagenesis based on polarity pockets and residue free energy contribution revealed mutations in sites Y143 and S153 that expanded the substrate spectrum of IDO toward aromatic amino acids, despite their low catalytic activity. In the future, the Y143 and S153 sites can be mutated simultaneously with IDO sites around the benzene ring to further improve their catalytic activity for aromatic side-chain amino acids, thus expanding the reaction of IDO with aromatic amino acids to enhance its application performance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28093750/s1, Figure S1: LC-MS analysis of products catalyzed by Y143D with substrate PHE; Figure S2: LC-MS analysis of products catalyzed by Y143I with substrate PHE; Figure S3: LC-MS analysis of products catalyzed by S153A with substrate PHE.
Author Contributions
Conceptualization, Y.N.; methodology, J.A.; software, J.A.; formal analysis, J.A. and J.G.; writing—original draft preparation, J.A.; writing—review and editing, J.A. and Y.N.; funding acquisition, Y.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Key R&D Program of China (grant number 2021YFC2102000), National Natural Science Foundation of China (grant numbers 22178147, 31872891), Program of Introducing Talents of Discipline to Universities (grant number 111-2-06), High-End Foreign Experts Recruitment Program (grant number G2021144005L), National Program for Support of Top-Notch Young Professionals, Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, Top-Notch Academic Programs Project of Jiangsu Higher Education Institutions, Jiangsu Province “Collaborative Innovation Center for Advanced Industrial Fermentation” Industry Development Program and National First-Class Discipline Program of Light Industry Technology and Engineering (grant number LITE2018-09).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The experimental data provided in this work are available in articles.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
IDO mutants are available from the authors.
References
- Ciabatti, R.; Kettenring, J.K.; Winters, G.; Tuan, G.; Zerilli, L.; Cavalleri, B. Ramoplanin (A-16686), a new glycolipodepsipeptide antibiotic. III. Structure elucidation. J. Antibiot. 1989, 42, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Matsunaga, S.; Yano, G.; Fusetani, N. Polytheonamides A and B, highly cytotoxic, linear polypeptides with unprecedented structural features, from the marine sponge, Theonella swinhoei. J. Am. Chem. Soc. 2005, 127, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Van Soest, R.W.M.; Ford, P.W.; Gustafson, K.R.; Mc Kee, T.C. Papuamides A–D, HIV-inhibitory and cytotoxic depsipeptides from the sponges Theonella mirabilis and Theonella swinhoei, collected in Papua New Guinea. J. Am. Chem. Soc. 1999, 121, 8632. [Google Scholar]
- Gelse, K.; Pöschl, E.; Aigner, T. Collagens—Structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef]
- Shen, Y.-P.; Niu, F.-X.; Yan, Z.-B.; Fong, L.S.; Huang, Y.-B.; Liu, J.-Z. Recent Advances in Metabolically Engineered Microorganisms for the Production of Aromatic Chemicals Derived From Aromatic Amino Acids. Front. Bioeng. Biotechnol. 2020, 8, 407. [Google Scholar] [CrossRef]
- Luo, H.; Yang, L.; Kim, S.H.; Wulff, T.; Feist, A.M.; Herrgard, M.; Palsson, B.Ø. Directed Metabolic Pathway Evolution Enables Functional Pterin-Dependent Aromatic-Amino-Acid Hydroxylation in Escherichia coli. ACS Synth. Biol. 2020, 9, 494–499. [Google Scholar] [CrossRef]
- Nazor, J.; Osborne, R.; Liang, J.; Vroom, J.; Zhang, X.; Entwistle, D.; Voladri, R.; Garcia, R.D.; Moore, J.C.; Grosser, S. Biocatalysts and Methods for Hydroxylation of Chemical Compounds. EP2847327 A2. U.S. Patent 10,184,117, 22 January 2019. [Google Scholar]
- Miyake, R.; Dekishima, Y. Method for Manufacturing Cis-5-hydroxy-l-pipecolic Acid. 15/526031. U.S. Patent 10,087,473, 2 October 2018. [Google Scholar]
- Ozaki, A.; Mori, H.; Shibasaki, T.; Ando, K.; Ochiai, K.; Chiba, S.; Uosaki, Y.J.E. Process for Producing Cis-3-hydroxy-L-proline. U.S. Patent 6413748, 6 December 1999. [Google Scholar]
- Kino, K.; Hara, R. L-proline Cis-4-hydroxylase and Use Thereof to Produce Cis-4-hydroxy-L-proline. U.S. Patent 8,541,209, 24 September 2013. [Google Scholar]
- Eng, J.G.M.; Shahsavarani, M.; Smith, D.P.; Hájíček, J.; De Luca, V.; Qu, Y. A Catharanthus roseus Fe(II)/α-ketoglutarate-dependent dioxygenase catalyzes a redox-neutral reaction responsible for vindolinine biosynthesis. Nat. Commun. 2022, 13, 3335. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Leissing, T.M.; Chowdhury, R.; Hopkinson, R.J.; Schofield, C.J. 2-Oxoglutarate-Dependent Oxygenases. Annu. Rev. Biochem. 2018, 87, 585–620. [Google Scholar] [CrossRef]
- Martinez, S.; Hausinger, R.P. Catalytic Mechanisms of Fe(II)- and 2-Oxoglutarate-dependent Oxygenases. J. Biol. Chem. 2015, 290, 20702–20711. [Google Scholar] [CrossRef]
- Peters, C.; Buller, R.M. Industrial Application of 2-Oxoglutarate-Dependent Oxygenases. Catalysts 2019, 9, 221. [Google Scholar] [CrossRef]
- Kodera, T.; Smirnov, S.V.; Samsonova, N.N.; Kozlov, Y.I.; Koyama, R.; Hibi, M.; Ogawa, J.; Yokozeki, K.; Shimizu, S. A novel l-isoleucine hydroxylating enzyme, l-isoleucine dioxygenase from Bacillus thuringiensis, produces (2S,3R,4S)-4-hydroxyisoleucine. Biochem. Biophys. Res. Commun. 2009, 390, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Fowden, L.; Pratt, H.M.; Smith, A.J.P. 4-Hydroxyisoleucine from seed of Trigonella foenum-graecum. Phytochemistry 1973, 12, 1707–1711. [Google Scholar] [CrossRef]
- Hibi, M.; Kawashima, T.; Kodera, T.; Smirnov, S.V.; Sokolov, P.M.; Sugiyama, M.; Shimizu, S.; Yokozeki, K.; Ogawa, J. Characterization of Bacillus thuringiensis L-Isoleucine Dioxygenase for Production of Useful Amino Acids. Appl. Environ. Microbiol. 2011, 77, 6926–6930. [Google Scholar] [CrossRef]
- Sun, D.; Gao, D.; Liu, X.; Zhu, M.; Li, C.; Chen, Y.; Zhu, Z.; Lu, F.; Qin, H.-M. Redesign and engineering of a dioxygenase targeting biocatalytic synthesis of 5-hydroxyl leucine. Catal. Sci. Technol. 2019, 9, 1825–1834. [Google Scholar] [CrossRef]
- Jing, X.; Liu, H.; Nie, Y.; Xu, Y. Advances in Fe(II)/2-ketoglutarate-dependent dioxygenase-mediated C–H bond oxidation for regioselective and stereoselective hydroxyl amino acid synthesis: From structural insights into practical applications. Syst. Microbiol. Biomanuf. 2021, 1, 275–290. [Google Scholar] [CrossRef]
- Wu, L.; An, J.; Jing, X.; Chen, C.-C.; Dai, L.; Xu, Y.; Liu, W.; Guo, R.-T.; Nie, Y. Molecular Insights into the Regioselectivity of the Fe(II)/2-Ketoglutarate-Dependent Dioxygenase-Catalyzed C–H Hydroxylation of Amino Acids. ACS Catal. 2022, 12, 11586–11596. [Google Scholar] [CrossRef]
- Peng, Z.; Jiang, Q.; Peng, P.; Li, F.-F. NH3-activated Fullerene Derivative Hierarchical Microstructures to Porous Fe3O4/N-C for Oxygen Reduction Reaction and Zn-air Battery. Eng. Sci. 2021, 14, 27–38. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, N.; Narváez Villarrubia, C.W.; Huang, X.; Xie, L.; Wang, X.; Kong, X.; Xu, H.; Wu, G.; Zeng, J.; et al. Single Fe atoms anchored by short-range ordered nanographene boost oxygen reduction reaction in acidic media. Nano Energy 2019, 66, 104164. [Google Scholar] [CrossRef]
- Dai, L.; Zhang, X.; Hu, Y.; Shen, J.; Zhang, Q.; Zhang, L.; Min, J.; Chen, C.-C.; Liu, Y.; Huang, J.-W.; et al. Structural and Functional Insights into a Nonheme Iron- and α-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates. Appl. Environ. Microbiol. 2022, 88, e02497-21. [Google Scholar] [CrossRef]
- Tao, H.; Ushimaru, R.; Awakawa, T.; Mori, T.; Uchiyama, M.; Abe, I. Stereoselectivity and Substrate Specificity of the Fe(II)/α-Ketoglutarate-Dependent Oxygenase TqaL. J. Am. Chem. Soc. 2022, 144, 21512–21520. [Google Scholar] [CrossRef]
- Tao, H.; Abe, I. Oxidative modification of free-standing amino acids by Fe(II)/αKG-dependent oxygenases. Eng. Microbiol. 2023, 3, 100062. [Google Scholar] [CrossRef]
- Roach, P.L.; Clifton, I.J.; Fülöp, V.; Harlos, K.; Barton, G.J.; Hajdu, J.; Andersson, I.; Schofield, C.J.; Baldwin, J.E. Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes. Nature 1995, 375, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Clifton, I.J.; McDonough, M.A.; Ehrismann, D.; Kershaw, N.J.; Granatino, N.; Schofield, C.J. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J. Inorg. Biochem. 2006, 100, 644–669. [Google Scholar] [CrossRef] [PubMed]
- Markolovic, S.; Leissing, T.M.; Chowdhury, R.; Wilkins, S.E.; Lu, X.; Schofield, C.J. Structure–function relationships of human JmjC oxygenases—Demethylases versus hydroxylases. Curr. Opin. Struct. Biol. 2016, 41, 62–72. [Google Scholar] [CrossRef]
- Mcdonough, M.A.; Loenarz, C.; Chowdhury, R.; Clifton, I.J.; Schofield, C.J. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr. Opin. Struct. Biol. 2010, 20, 659–672. [Google Scholar] [CrossRef]
- He, X.; Man, V.H.; Ji, B.; Xie, X.-Q.; Wang, J. Calculate protein–ligand binding affinities with the extended linear interaction energy method: Application on the Cathepsin S set in the D3R Grand Challenge 3. J. Comput. Aided Mol. Des. 2019, 33, 105–117. [Google Scholar] [CrossRef]
- Oshima, H.; Re, S.; Sugita, Y. Prediction of Protein–Ligand Binding Pose and Affinity Using the gREST+FEP Method. J. Chem. Inf. Model. 2020, 60, 5382–5394. [Google Scholar] [CrossRef]
- Siebenmorgen, T.; Zacharias, M. Computational prediction of protein–protein binding affinities. WIREs Comput. Mol. Sci. 2020, 10, e1448. [Google Scholar] [CrossRef]
- Masferrer-Rius, E.; Borrell, M.; Lutz, M.; Costas, M.; Klein Gebbink, R.J.M. Aromatic C−H Hydroxylation Reactions with Hydrogen Peroxide Catalyzed by Bulky Manganese Complexes. Adv. Synth. Catal. 2021, 363, 3783–3795. [Google Scholar] [CrossRef]
- Regoutz, A.; Wolinska, M.S.; Fernando, N.K.; Ratcliff, L.E. A combined density functional theory and x-ray photoelectron spectroscopy study of the aromatic amino acids. Electron. Struct. 2020, 2, 044005. [Google Scholar] [CrossRef]
- Chenprakhon, P.; Wongnate, T.; Chaiyen, P. Monooxygenation of aromatic compounds by flavin-dependent monooxygenases. Protein Sci. 2019, 28, 8–29. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, P.F. Mechanism of Aromatic Amino Acid Hydroxylation. Biochemistry 2003, 42, 14083–14091. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Zhang, W.; Jing, X.; Nie, Y.; Xu, Y. Reconstitution of TCA cycle involving l-isoleucine dioxygenase for hydroxylation of l-isoleucine in Escherichia coli using CRISPR-Cas9. 3 Biotech 2020, 10, 167. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Wang, X.; Zhang, W.; An, J.; Luo, P.; Nie, Y.; Xu, Y. Highly Regioselective and Stereoselective Hydroxylation of Free Amino Acids by a 2-Oxoglutarate-Dependent Dioxygenase from Kutzneria albida. ACS Omega 2019, 4, 8350–8358. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Li, P.; Merz, K.M., Jr. MCPB.py: A Python Based Metal Center Parameter Builder. J. Chem. Inf. Model. 2016, 56, 599–604. [Google Scholar] [CrossRef]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Grubmüller, H.; Heller, H.; Windemuth, A.; Schulten, K. Generalized Verlet Algorithm for Efficient Molecular Dynamics Simulations with Long-range Interactions. Mol. Simul. 1991, 6, 121–142. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).