Anti-Inflammatory Effect of Izalpinin Derived from Chromolaena leivensis: λ-Carrageenan-Induced Paw Edema and In Silico Model
Abstract
1. Introduction
2. Results and Discussion
2.1. Structural Determination of Izalpinin
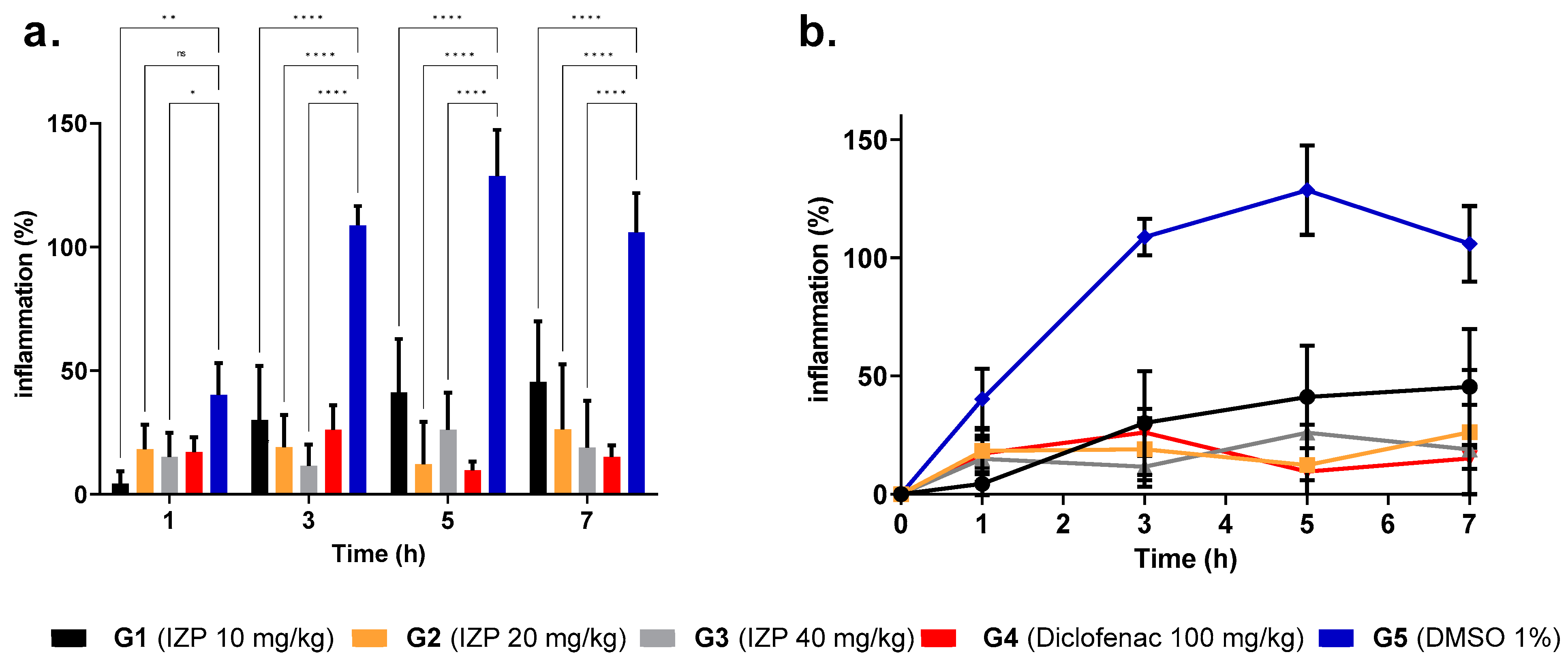
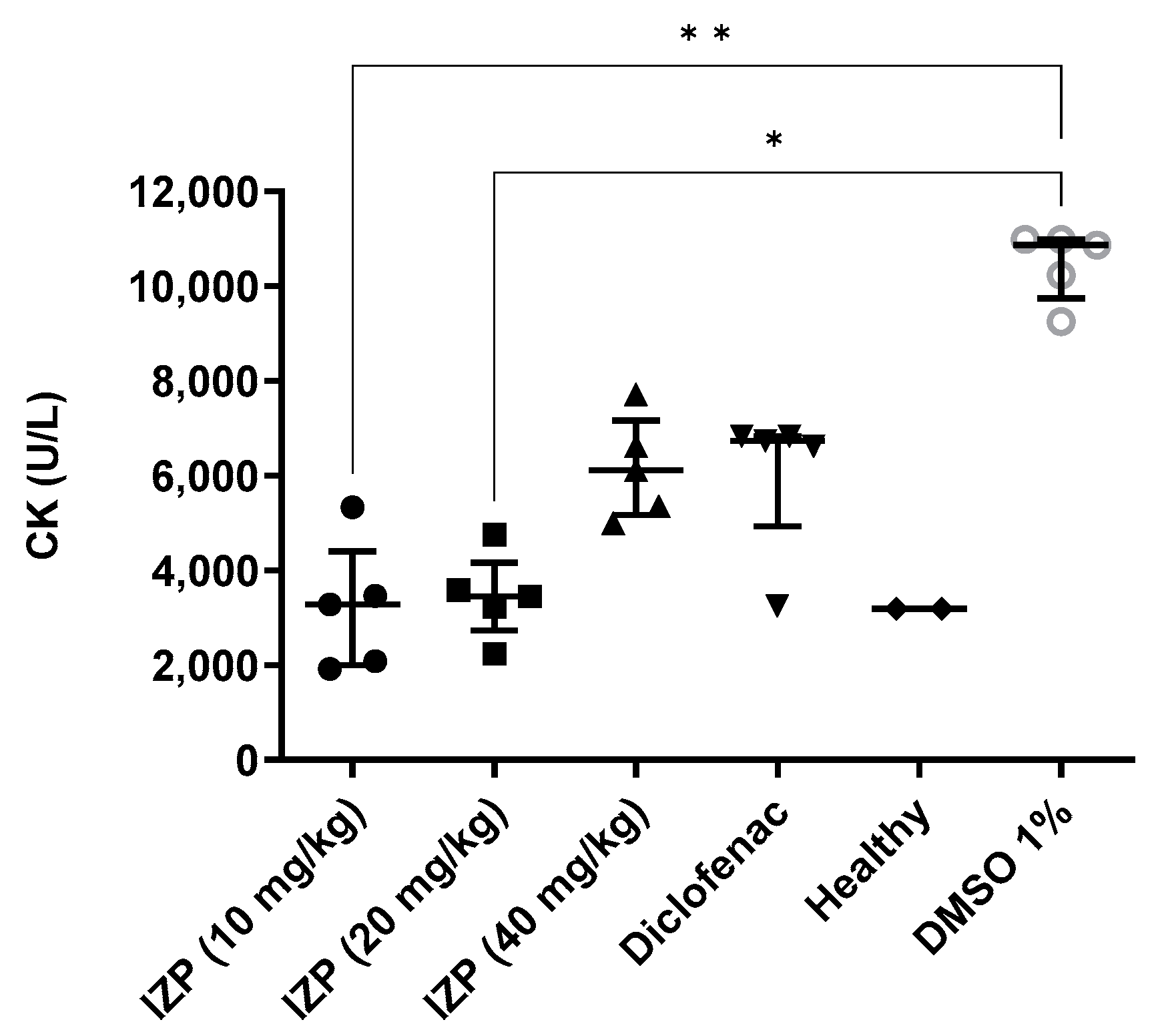
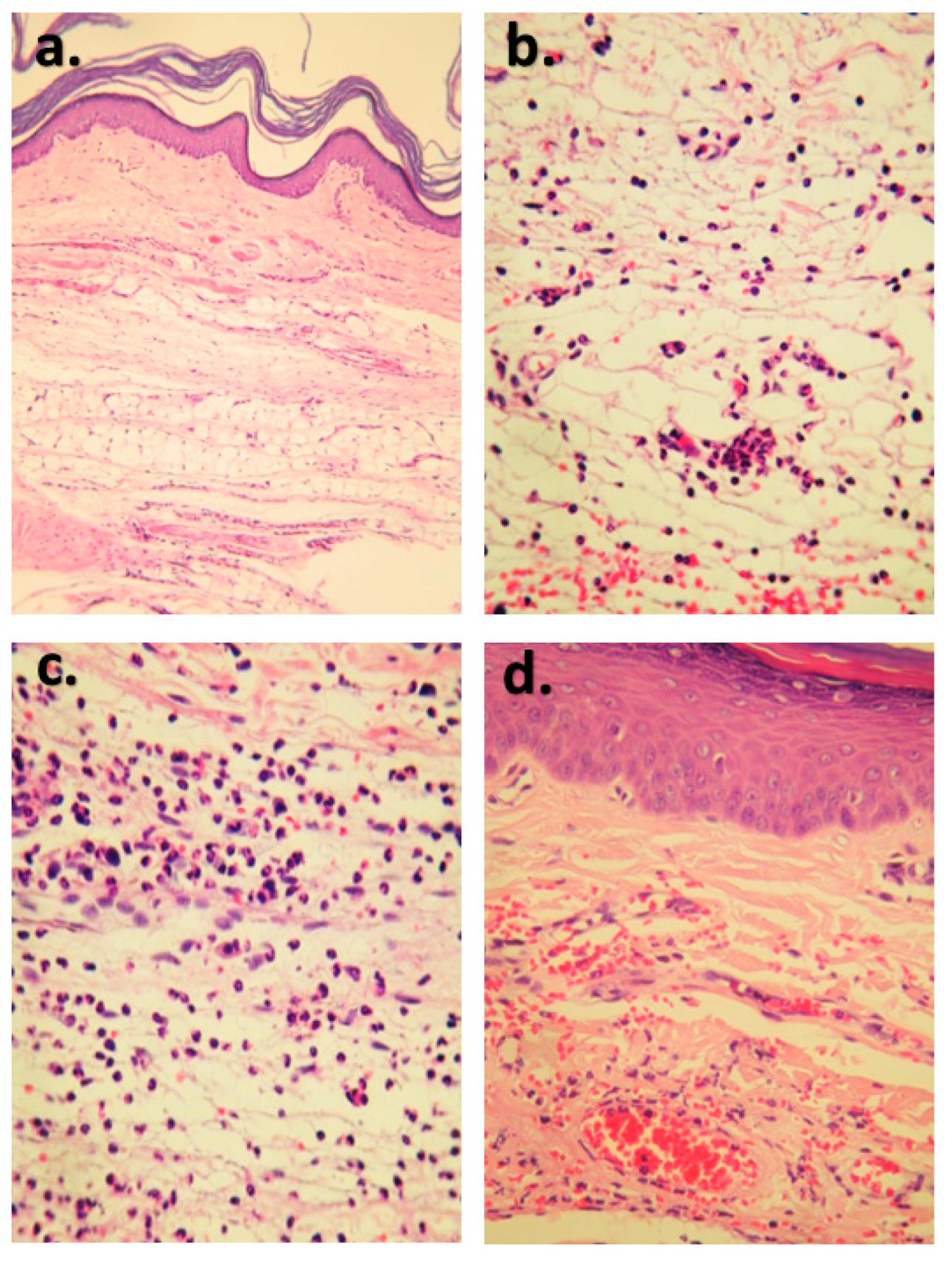
2.2. Anti-Inflammatory Activity of Izalpinin
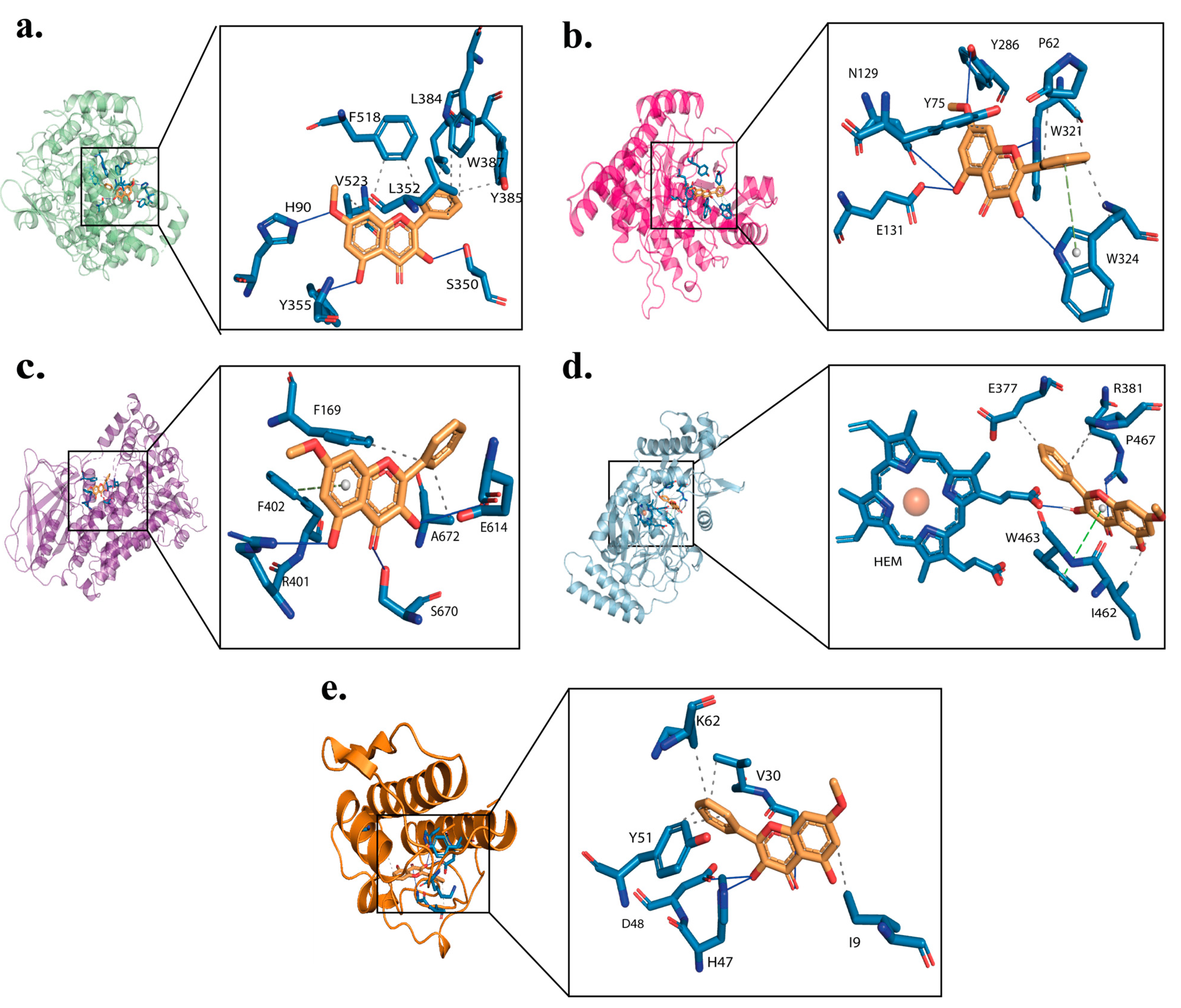
2.3. Binding Interaction of Izalpinin with Key Targets
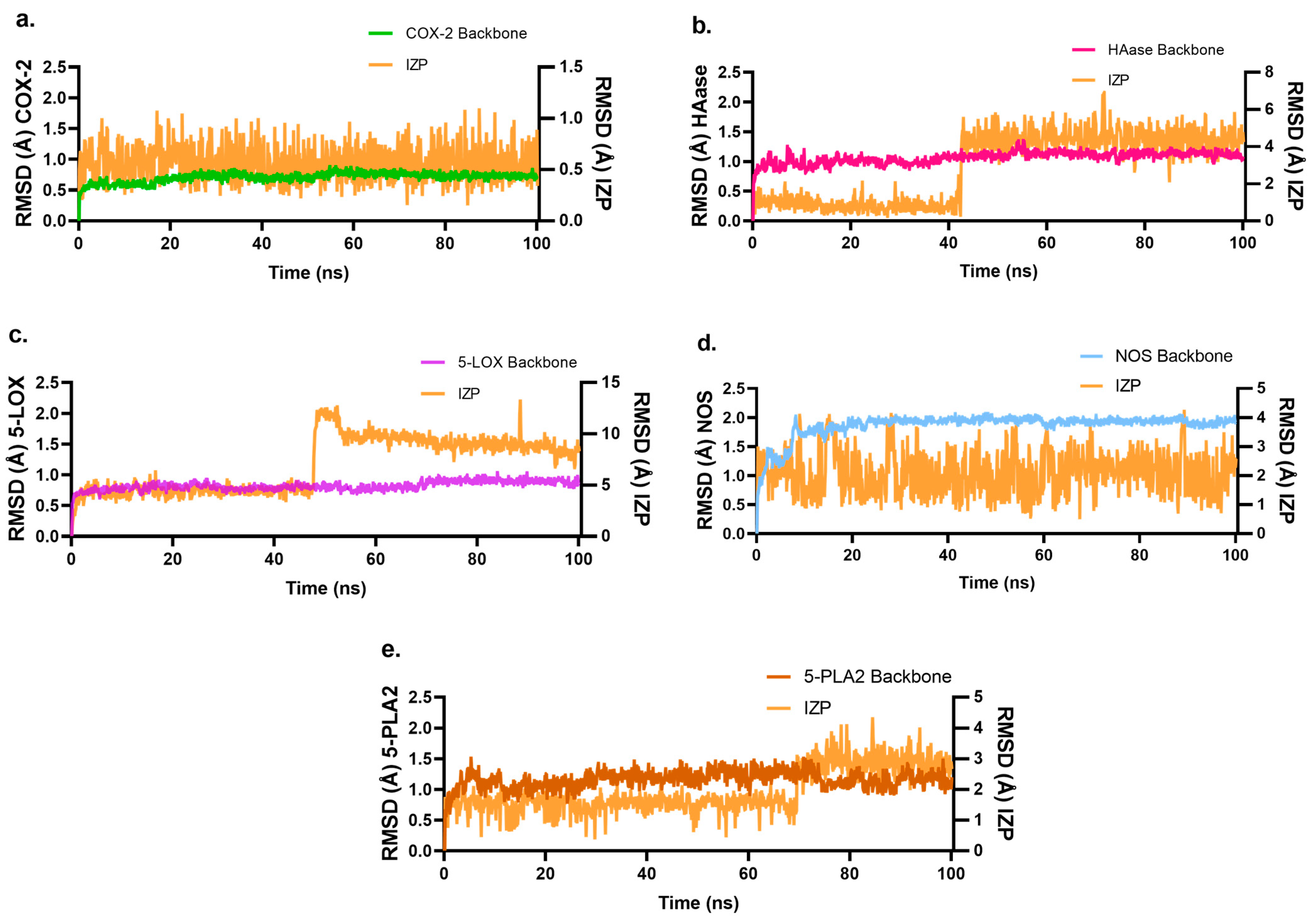
2.4. Molecular Dynamics Studies of IZP Interacting with Key Enzymes COX-2, HAase, 5-LOX, NOS, and 5-PLA2 Involved in Inflammation
3. Materials and Methods
3.1. General Information
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Anti-inflammatory Assay
3.4.1. Experimental Animals
3.4.2. Evaluation of the in Vivo Anti-Inflammatory in Rat Paw Edema Induced by λ-Carrageenan
3.4.3. Creatine Kinase (CK) Assay and Histopathological Study
3.4.4. Statistical Analysis
3.5. In Silico Study
3.5.1. Protein Selection Preparation and Grid Generation
3.5.2. Structural Optimization of IZP
3.5.3. Molecular Docking and Binding Free Energy Calculation
3.5.4. Molecular Dynamics Simulations (MDs)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Ansar, W.; Ghosh, S. Inflammation and Inflammatory Diseases, Markers, and Mediators: Role of CRP in Some Inflammatory Diseases. In Biology of C Reactive Protein in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2016; pp. 67–107. ISBN 9788132226802. [Google Scholar]
- Gopi, S.; Amalraj, A.; Kunnumakkara, A.; Thomas, S. Inflammation and Natural Products; Elsevier: Amsterdam, The Netherlands, 2021; ISBN 9780128192184. [Google Scholar]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory Responses and Inflammation-Associated Diseases in Organs. Oncotarget 2018, 9, 7204–7218. [Google Scholar] [CrossRef]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic Inflammation in the Etiology of Disease across the Life Span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, R.; Cheng, Z.; Song, Z.; Wang, Z.; Duan, H.; Wu, X.; Ni, T. Comparative Study on the Interaction between Flavonoids with Different Core Structures and Hyaluronidase. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2021, 262, 120079. [Google Scholar] [CrossRef]
- Atrahimovich, D.; Avni, D.; Khatib, S. Flavonoids-macromolecules Interactions in Human Diseases with Focus on Alzheimer, Atherosclerosis and Cancer. Antioxidants 2021, 10, 423. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.L.; Fan, M.X.; Wu, J.L.; Li, N.; Guo, M.Q. Antioxidant and Anti-Inflammatory Properties of Flavonoids from Lotus Plumule. Food Chem. 2019, 277, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Catarino, M.D.; Talhi, O.; Rabahi, A.; Silva, A.M.S.; Cardoso, S.M. The Antiinflammatory Potential of Flavonoids: Mechanistic Aspects, 1st ed.; Elsevier B.V.: Amsterdam, The Netherlands, 2016; Volume 48, ISBN 9780444636027. [Google Scholar]
- Moulishankar, A.; Lakshmanan, K. Data on Molecular Docking of Naturally Occurring Flavonoids with Biologically Important Targets. Data Br. 2020, 29, 105243. [Google Scholar] [CrossRef]
- Xiao, H.; Lü, F.; Stewart, D.; Zhang, Y. Mechanisms Underlying Chemopreventive Effects of Flavonoids via Multiple Signaling Nodes within Nrf2-ARE and AhR-XRE Gene Regulatory Networks. Curr. Chem. Biol. 2013, 7, 151–176. [Google Scholar] [CrossRef]
- Cho, H.; Yun, C.W.; Park, W.K.; Kong, J.Y.; Kim, K.S.; Park, Y.; Lee, S.; Kim, B.K. Modulation of the Activity of Pro-Inflammatory Enzymes, COX-2 and INOS, by Chrysin Derivatives. Pharmacol. Res. 2004, 49, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Moussa, N.; Hassan, A.; Gharaghani, S. Pharmacophore Model, Docking, QSAR, and Molecular Dynamics Simulation Studies of Substituted Cyclic Imides and Herbal Medicines as COX-2 Inhibitors. Heliyon 2021, 7, e06605. [Google Scholar] [CrossRef]
- Torrenegra, R.D.; Rodríguez, J.; Rodríguez, O.; Palau, V.E.; Méndez, G.M. Antiproliferative Activity of 3,5,7-Trihydroxy-6-Methoxy Flavone Obtained from Chromolaena Leivensis (Hieron) on Cancer Cell Lines of Breast, Prostate, Lung, Colon and Cervix. Pharmacol. OnLine 2016, 1, 7–11. [Google Scholar]
- Torrenegra, R.; Rodríguez, O. Chemical and Biological Activity of Leaf Extracts of Chromolaena Leivensis. Nat. Prod. Commun. 2011, 6, 947–950. [Google Scholar] [PubMed]
- Solis-Vasquez, L.; Tillack, A.F.; Santos-Martins, D.; Koch, A.; LeGrand, S.; Forli, S. Benchmarking the Performance of Irregular Computations in AutoDock-GPU Molecular Docking. Parallel Comput. 2022, 109, 102861. [Google Scholar] [CrossRef]
- Manivannan, A.; Soundararajan, P.; Park, Y.G.; Sakkiah, S.; Jeong, B.R. Binding Mode Investigation of Polyphenols from Scrophularia Targeting Human Aldose Reductase Using Molecular Docking and Molecular Dynamics Simulations. J. Chem. 2015, 2015, 434256. [Google Scholar] [CrossRef]
- Ye, H.; Sun, L.; Li, J.; Wang, Y.; Bai, J.; Wu, L.; Han, Q.; Yang, Z.; Li, L. Sesamin Attenuates Carrageenan-Induced Lung Inflammation through Upregulation of A20 and TAX1BP1 in Rats. Int. Immunopharmacol. 2020, 88, 107009. [Google Scholar] [CrossRef]
- Eddouks, M.; Chattopadhyay, D.; Zeggwagh, N.A. Animal Models as Tools to Investigate Antidiabetic and Anti-Inflammatory Plants. Evid.-Based Complement. Altern. Med. 2012, 2012, 14. [Google Scholar] [CrossRef] [PubMed]
- Vencovský, J.; Alexanderson, H.; Lundberg, I.E. Idiopathic Inflammatory Myopathies. Rheum. Dis. Clin. N. Am. 2019, 45, 569–581. [Google Scholar] [CrossRef]
- Gunawardena, D.; Govindaraghavan, S.; Münch, G. Anti-Inflammatory Properties of Cinnamon Polyphenols and Their Monomeric Precursors. In Polyphenols in Human Health and Disease; Elsevier: Amsterdam, The Netherlands, 2014; pp. 409–425. ISBN 9780123984562. [Google Scholar]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef]
- Arming, S.; Strobl, B.; Wechselberger, C.; Kreil, G. In Vitro Mutagenesis of PH-20 Hyaluronidase from Human Sperm. Eur. J. Biochem. 1997, 247, 810–814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Bharadwaj, A.G.; Casper, A.; Barkley, J.; Barycki, J.J.; Simpson, M.A. Hyaluronidase Activity of Human Hyal1 Requires Active Site Acidic and Tyrosine Residues. J. Biol. Chem. 2009, 284, 9433–9442. [Google Scholar] [CrossRef] [PubMed]
- Meka, B.; Ravada, S.R.; Muthyala, M.K.K.; Kurre, P.N.; Golakoti, T. Synthesis, in Vitro and in Silico Evaluation of Diaryl Heptanones as Potential 5LOX Enzyme Inhibitors. Bioorg. Chem. 2018, 80, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Baul, H.S.; Rajiniraja, M. Molecular Docking Studies of Selected Flavonoids on Inducible Nitric Oxide Synthase (INOs) in Parkinson’s Disease. Res. J. Pharm. Technol. 2018, 11, 3685–3688. [Google Scholar] [CrossRef]
- Lättig, J.; Böhl, M.; Fischer, P.; Tischer, S.; Tietböhl, C.; Menschikowski, M.; Gutzeit, H.O.; Metz, P.; Pisabarro, M.T. Mechanism of Inhibition of Human Secretory Phospholipase A2 by Flavonoids: Rationale for Lead Design. J. Comput. Aided. Mol. Des. 2007, 21, 473–483. [Google Scholar] [CrossRef] [PubMed]
- OECD. Test No. 404: Acute Dermal Irritation/Corrosion; OECD Publishing: Paris, France, 2015. [Google Scholar]
- Winter, C.A.; Risley, E.A.; Nuss, G.W. Carrageenin-Induced Edema in Hind Paw. Exp. Biol. Med. 1962, 111, 544–547. [Google Scholar] [CrossRef]
- Morris, C.J. Carrageenan-Induced Paw Edema in the Rat and Mouse. In Inflammation Protocols. Methods in Molecular Biology; Winyard, P.G., Willoughby, D.A., Eds.; Humana Press: Totowa, NJ, USA, 2003; pp. 115–121. ISBN 978-1-59259-374-3. [Google Scholar]
- Goicoechea, M.; Cía, F.; San José, C.; Asensio, A.; Emparanza, J.I.; Gil, A.G.; López De Cerain, A.; Aldazabal, P.; Azpitarte, M.; Otaegui, D.; et al. Minimizing Creatine Kinase Variability in Rats for Neuromuscular Research Purposes. Lab. Anim. 2008, 42, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, H.; Wang, T.; Chong, Y.; Yu, P.; Lu, C.; Xue, Y.; Fu, F.; Zhang, L. Anti-Inflammatory Activity of Yanshu Spraying Agent in Animal Models. Exp. Ther. Med. 2013, 5, 73–76. [Google Scholar] [CrossRef]
- Santos, P.; Lopez-Vallejo, F.; Ramírez, D.; Caballero, J.; Mata Espinosa, D.; Hernández-Pando, R.; Soto, C.Y. Identification of Mycobacterium Tuberculosis CtpF as a Target for Designing New Antituberculous Compounds. Bioorganic Med. Chem. 2020, 28, 115256. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, M.K.; Sharma, P.; Tripathi, A.; Tripathi, P.N.; Srivastava, P.; Seth, A.; Shrivastava, S.K. Computational Exploration and Experimental Validation to Identify a Dual Inhibitor of Cholinesterase and Amyloid-Beta for the Treatment of Alzheimer’s Disease. J. Comput. Aided. Mol. Des. 2020, 34, 983–1002. [Google Scholar] [CrossRef]
- Borrego-Muñoz, P.; Becerra, L.D.; Ospina, F.; Coy-Barrera, E.; Quiroga, D. Synthesis (Z) vs (E) Selectivity, Antifungal Activity against Fusarium Oxysporum, and Structure-Based Virtual Screening of Novel Schiff Bases Derived from L-Tryptophan. ACS Omega 2022, 7, 24714–24726. [Google Scholar] [CrossRef]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the Scope of the Protein-Ligand Interaction Profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W. PyMOL: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Cryst. 2002, 40, 82–92. [Google Scholar]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006. [Google Scholar]
- Cheng, A.; Merz, K.M. Application of the Nosé-Hoover Chain Algorithm to the Study of Protein Dynamics. J. Phys. Chem. 1996, 100, 1927–1937. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Humphrey, W.; Andrew, D.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]

| Position | δH | δc |
|---|---|---|
| 2 | - | 146.1 |
| 3 | 9.8(s, OH) | 137.4 |
| 4 | - | 176.4 |
| 5 | 12.3 (s, OH) | 160.4 |
| 6 | 6.37 (d, J = 1.8 Hz, 1H) | 97.6 |
| 7 | - | 165.1 |
| 8 | 6.77 (s, J = 1.8 Hz, 1H) | 92.1 |
| 9 | - | 156.3 |
| 10 | - | 104.2 |
| 1′ | - | 130.8 |
| 2′ | 8.19 (d, J = 7 Hz, 1H) | 127.5 |
| 3′ | 7.55 (d, J = 7 Hz, 2H) | 128.5 |
| 4′ | 7.53 (m, J = 7 Hz, 2H) | 130.0 |
| 5′ | 7.52 (m, J = 7 Hz, 2H) | 128.5 |
| 6′ | 8.16 (d, J = 7 Hz, 1H) | 127.5 |
| 7-OCH3 | 3.85 (s,3H) | 56.07 |
| Target | PDB Code | GlideScore a | ΔGBind b | Interacting Residues c |
|---|---|---|---|---|
| COX-2 | 5KIR | −7.33 | −66.07 | H90 1, R120 6, Q192 1, Y348 3, V349 3, L352 3, S353 1, Y355 3, F381 3, L384 3, Y385 3, W387 3, R513 6, F518 3, I517 3, A516 3, M522 3, V523 3, A527 3, S530 1 |
| HAase | 2PE4 | −5.39 | −52.14 | N37 1, N39 1, P62 3, I73 3, Y75 3, V127 3, E131 6, Y202 3, Y247 3, W321 3, V322 3, S323 1, W324 3 |
| 5-LOX | 3V99 | −4.29 | −40.03 | S14 1, Q15 1, A18 3, Y81 3, F169 3, A398 3, R401 6, F402 3, I406 3, N554 3, F555 3, F610 3, E614 6, L615 3, E622 6, P668 3, N669 1, S670 1, A672 3. |
| iNOS | 2NSI | −3.18 | −32.96 | M120 3, I201 3, M374 3, T376 1, E377 6, Arg381 6, W461 3, I462 3, W463 3, V465 3, P466 3, P467 3, HEM550 2 |
| PLA2-IIA | 1KQU | −6.89 | −57.41 | L2 3, F5 3,5, H6 1, I9 3, A17 3, A18 3, Y21 3, H27 1, C28 3, V30 3, C44 3, H47 4,5,6, D48 2, Y51 3,5, K62 6, F98 3. |
| Groups (G) | Treatment |
|---|---|
| Izalpinin (IZP) | |
| 1 | IZP (10 mg/Kg) |
| 2 | IZP (20 mg/Kg) |
| 3 | IZP (40 mg/Kg) |
| 4 | Positive control (Diclofenac: 100 mg/Kg) |
| 5 | Negative control (DMSO 1%) |
| 6 | Control group (2) Healthy animals |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mancipe, J.C.; Vargas-Pinto, P.; Rodríguez, O.E.; Borrego-Muñoz, P.; Castellanos Londoño, I.; Ramírez, D.; Piñeros, L.G.; Mejía, M.C.; Pombo, L.M. Anti-Inflammatory Effect of Izalpinin Derived from Chromolaena leivensis: λ-Carrageenan-Induced Paw Edema and In Silico Model. Molecules 2023, 28, 3722. https://doi.org/10.3390/molecules28093722
Mancipe JC, Vargas-Pinto P, Rodríguez OE, Borrego-Muñoz P, Castellanos Londoño I, Ramírez D, Piñeros LG, Mejía MC, Pombo LM. Anti-Inflammatory Effect of Izalpinin Derived from Chromolaena leivensis: λ-Carrageenan-Induced Paw Edema and In Silico Model. Molecules. 2023; 28(9):3722. https://doi.org/10.3390/molecules28093722
Chicago/Turabian StyleMancipe, Juan C., Pedro Vargas-Pinto, Oscar E. Rodríguez, Paola Borrego-Muñoz, Iovana Castellanos Londoño, David Ramírez, Luis G. Piñeros, María Camila Mejía, and Luis M. Pombo. 2023. "Anti-Inflammatory Effect of Izalpinin Derived from Chromolaena leivensis: λ-Carrageenan-Induced Paw Edema and In Silico Model" Molecules 28, no. 9: 3722. https://doi.org/10.3390/molecules28093722
APA StyleMancipe, J. C., Vargas-Pinto, P., Rodríguez, O. E., Borrego-Muñoz, P., Castellanos Londoño, I., Ramírez, D., Piñeros, L. G., Mejía, M. C., & Pombo, L. M. (2023). Anti-Inflammatory Effect of Izalpinin Derived from Chromolaena leivensis: λ-Carrageenan-Induced Paw Edema and In Silico Model. Molecules, 28(9), 3722. https://doi.org/10.3390/molecules28093722







