Salvianolic Acid A Improves Rat Kidney Injury by Regulating MAPKs and TGF-β1/Smads Signaling Pathways
Abstract
1. Introduction
2. Results
2.1. SAA Improved Gentamicin-Induced Acute Kidney Injury
2.2. SAA Reduced KIM-1, NGAL and UP Levels in AKI Rats
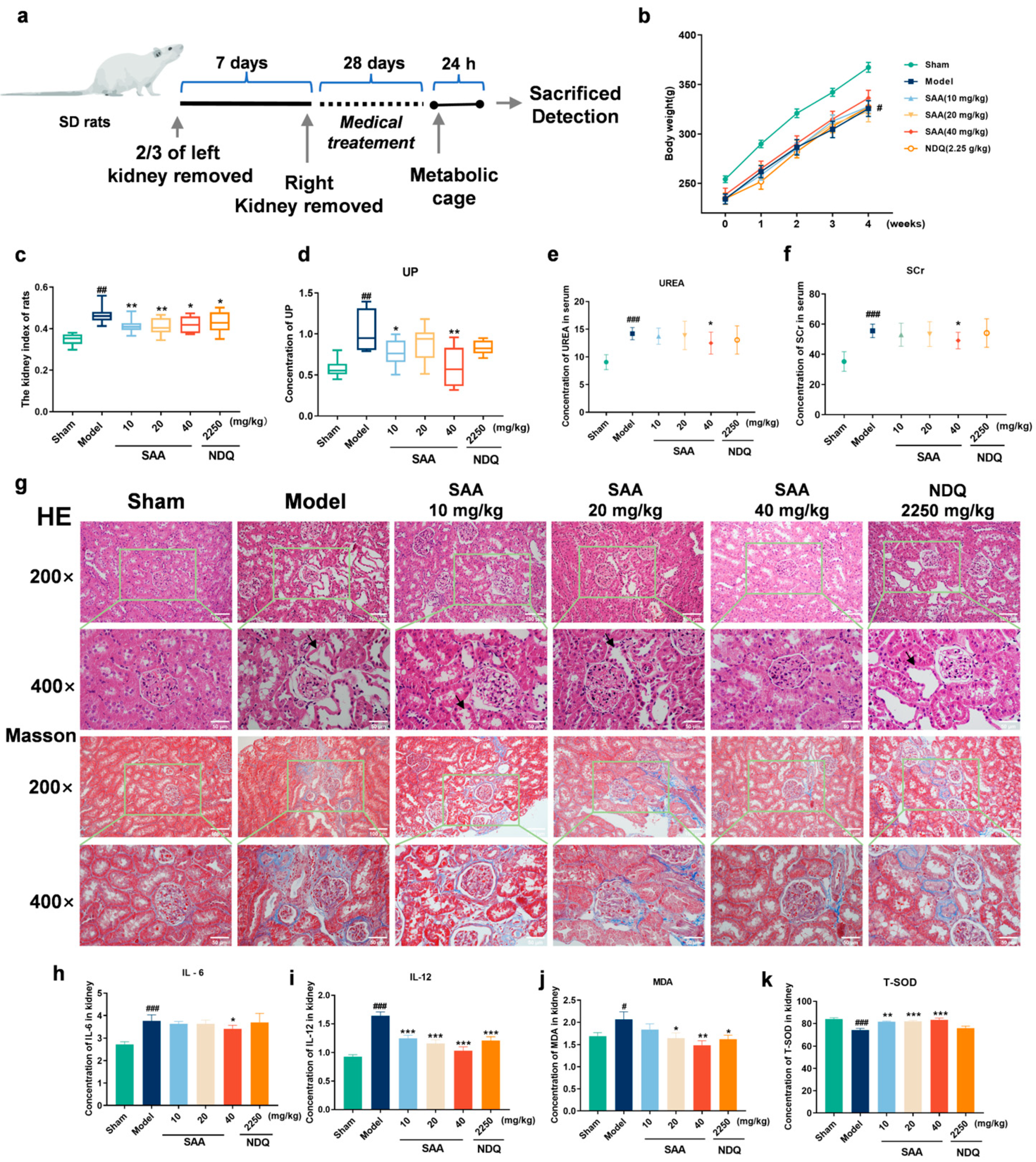
2.3. SAA Improved 5/6 Nephrectomized Model-Induced Chronic Kidney Disease
2.4. SAA Inhibited the Release of Inflammatory Cytokines and Anti-Oxidative Stress in 5/6Nx Rats
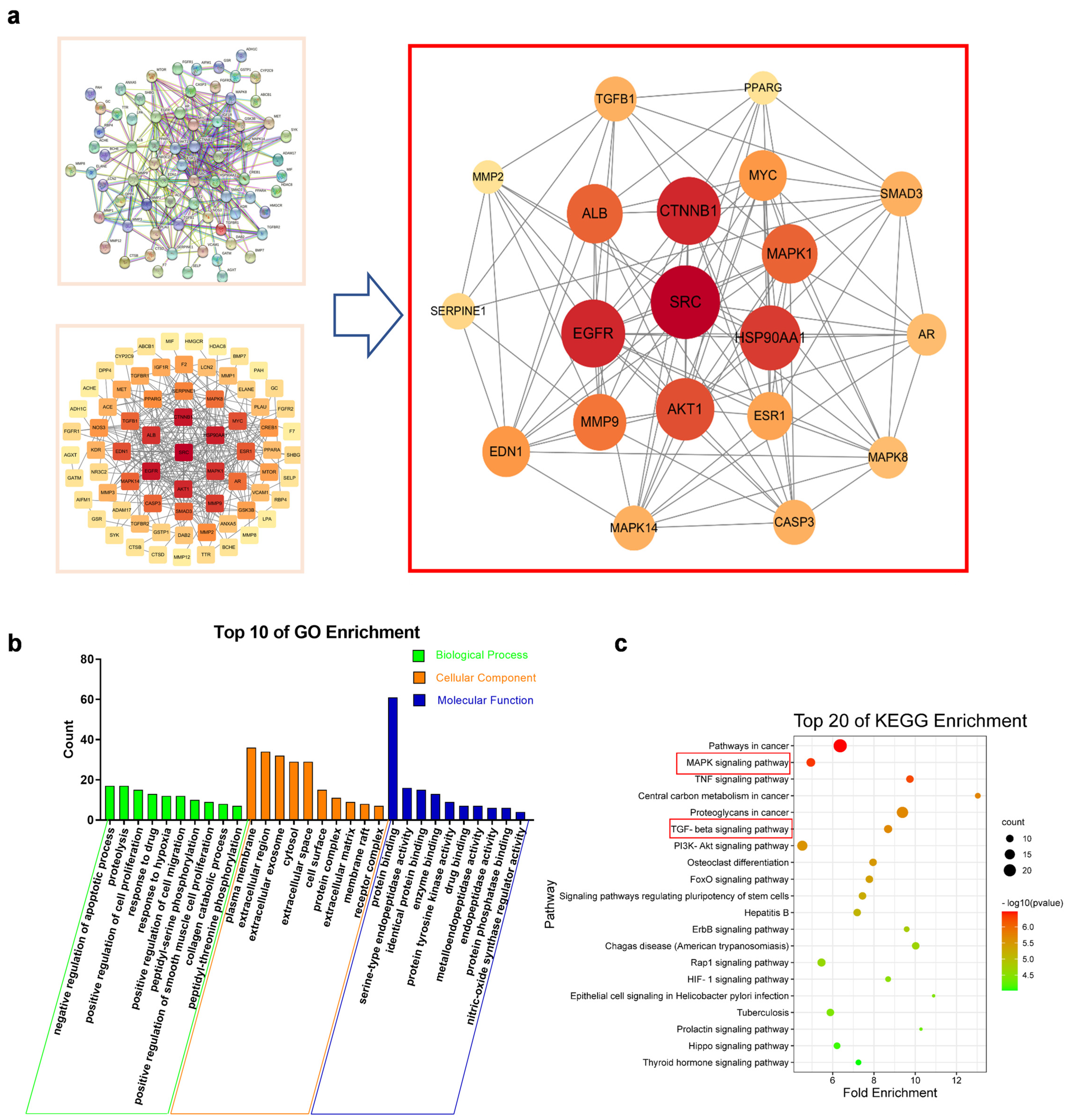
2.5. Network Pharmacology Suggested That MAPKs and TGF-β1/Smads Signaling Pathways Were Key Mechanisms of SAA in the Treatment of CKD
2.6. SAA Affected TGF-β1/smads Protein Expression in the Kidneys of CKD Rats
2.7. SAA Inhibited TLR4 Expression in the Kidneys of CKD Rats
2.8. SAA Inhibited the Activation of the MAPKs Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. AKI Model (Gentamicin-Induced Rats)
4.3. CKD Model (5/6 Nephrectomized Rats)
4.4. Kidney Index of Rats
4.5. Histopathological Analysis
4.6. Masson Staining
4.7. Biochemical Analysis
4.8. ELISA
4.9. Western Blotting
4.10. Network Pharmacology
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SAA | Salvianolic acid A |
| AKI | Acute kidney injury |
| CKD | Chronic kidney disease |
| 5/6Nx | 5/6 nephrectomized |
| KIM-1 | Kidney injury molecule-1 |
| NGAL | Neutrophil gelatinase-associated lipocalin |
| SCr | Serum creatinine |
| UREA | Urea |
| UP | Urine protein |
| IL-6 | Interleukin 6 |
| IL-12 | Interleukin 12 |
| MDA | malondialdehyde |
| T-SOD | Total superoxide dismutase |
| ERK | Extracellular regulated protein kinases |
| MAPKs | Mitogen-activated protein kinase signaling |
| TGF-β | Transforming growth factor-beta |
| PKD | Polycystic kidney disease |
| LPS | Lipopolysaccharide |
| STZ | Streptozocin |
| DEX | Dexamethasone |
| α-SMA | α-Smooth Muscle Actin |
| NDQ | Niaoduqing granules |
| MKKs | MAP kinase kinases |
References
- Crews, D.C.; Bello, A.K.; Saadi, G.; World Kidney Day Steering Committee. Burden, access, and disparities in kidney disease. Pediatr. Nephrol. 2019, 34, 541–548. [Google Scholar] [CrossRef]
- Eckardt, K.-U.; Coresh, J.; Devuyst, O.; Johnson, R.J.; Köttgen, A.; Levey, A.S.; Levin, A. Evolving importance of kidney disease: From subspecialty to global health burden. Lancet 2013, 382, 158–169. [Google Scholar] [CrossRef]
- Wonnacott, A.; Meran, S.; Amphlett, B.; Talabani, B.; Phillips, A. Epidemiology and outcomes in community-acquired vs. hospital-acquired AKI. Clin. J. Am. Soc. Nephrol. CJASN 2014, 9, 1007–1014. [Google Scholar] [CrossRef]
- Xue, J.L.; Daniels, F.; Star, R.A.; Kimmel, P.L.; Eggers, P.W.; Molitoris, B.A.; Himmelfarb, J.; Collins, A.J. Incidence and mortality of acute renal failure in Medicare beneficiaries, 1992 to 2001. J. Am. Soc. Nephrol. JASN 2006, 17, 1135–1142. [Google Scholar] [CrossRef]
- Jacob, J.; Dannenhoffer, J.; Rutter, A. Acute Kidney Injury. Prim. Care 2020, 47, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Ronco, C.; Bellomo, R.; Kellum, J.A. Acute kidney injury. Lancet 2019, 394, 1949–1964. [Google Scholar] [CrossRef]
- Heidarian, E.; Jafari-Dehkordi, E.; Valipour, P.; Ghatreh-Samani, K.; Ashrafi-Eshkaftaki, L. Nephroprotective and Anti-Inflammatory Effects of Pistacia atlantica Leaf Hydroethanolic Extract Against Gentamicin-Induced Nephrotoxicity in Rats. J. Diet. Suppl. 2017, 14, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Bledsoe, G.; Shen, B.; Yao, Y.Y.; Hagiwara, M.; Mizell, B.; Teuton, M.; Grass, D.; Chao, L.; Chao, J. Role of tissue kallikrein in prevention and recovery of gentamicin-induced renal injury. Toxicol. Sci. 2008, 102, 433–443. [Google Scholar] [CrossRef]
- Mosadegh Jabbari, Z.R. Aria Jenabi, Leila Zahedi-Shoolami, Ahmad Moorak, Simvastatin Ameliorates Gentamicin-Induced Renal Injury in Rat. Saudi J. Kidney Dis. Transplant. 2011, 22, 1181–1911. [Google Scholar]
- Udawatte, N.S.; Kang, S.W.; Wang, Y.; Arumugam, T.V.; Seneviratne, C.J. Predictive Nephrotoxicity Profiling of a Novel Antifungal Small Molecule in Comparison to Amphotericin B and Voriconazole. Front. Pharmacol. 2020, 11, 511–519. [Google Scholar] [CrossRef]
- Vaidya, V.S.; Ramirez, V.; Ichimura, T.; Bobadilla, N.A.; Bonventre, J.V. Urinary kidney injury molecule-1: A sensitive quantitative biomarker for early detection of kidney tubular injury. Am. J. Physiol. Renal. Physiol. 2006, 290, F517–F529. [Google Scholar] [CrossRef]
- Zang, X.J.; An, S.X.; Feng, Z.; Xia, Y.P.; Song, Y.; Yu, Q. In vivo mechanism study of NGAL in rat renal ischemia-reperfusion injury. Genet. Mol. Res. 2014, 13, 8740–8748. [Google Scholar] [CrossRef]
- Chawla, L.S.; Eggers, P.W.; Star, R.A.; Kimmel, P.L. Acute kidney injury and chronic kidney disease as interconnected syndromes. New Engl. J. Med. 2014, 371, 58–66. [Google Scholar] [CrossRef]
- Chen, J.H.; Chao, C.T.; Huang, J.W.; Hung, K.Y.; Liu, S.H.; Tarng, D.C.; Chiang, C.K. Early elimination of uremic toxin ameliorates AKI-to-CKD transition. Clin. Sci. 2021, 135, 2643–2658. [Google Scholar] [CrossRef]
- Kiousi, E.; Grapsa, E. The role of an out-patient renal clinic in renal disease management. J. Transl. Intern. Med. 2015, 3, 3–7. [Google Scholar] [CrossRef]
- He, J.; Wang, Y.; Sun, S.; Yu, M.; Wang, C.; Pei, X.; Zhu, B.; Wu, J.; Zhao, W. Bone marrow stem cells-derived microvesicles protect against renal injury in the mouse remnant kidney model. Nephrology 2012, 17, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, A.; Ware, K.; Calomeni, E.; Nadasdy, T.; Forbes, R.; Satoskar, A.A.; Nadasdy, G.; Rovin, B.H.; Hebert, L.A.; Brodsky, S.V. 5/6 nephrectomy as a validated rat model mimicking human warfarin-related nephropathy. Am. J. Nephrol. 2012, 35, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Tsuprykov, O.; Ando, R.; Reichetzeder, C.; von Websky, K.; Antonenko, V.; Sharkovska, Y.; Chaykovska, L.; Rahnenfuhrer, J.; Hasan, A.A.; Tammen, H.; et al. The dipeptidyl peptidase inhibitor linagliptin and the angiotensin II receptor blocker telmisartan show renal benefit by different pathways in rats with 5/6 nephrectomy. Kidney Int. 2016, 89, 1049–1061. [Google Scholar] [CrossRef]
- Kim, K.; Anderson, E.M.; Thome, T.; Lu, G.; Salyers, Z.R.; Cort, T.A.; O’Malley, K.A.; Scali, S.T.; Ryan, T.E. Skeletal myopathy in CKD: A comparison of adenine-induced nephropathy and 5/6 nephrectomy models in mice. Am. J. Physiol. Renal. Physiol. 2021, 321, F106–F119. [Google Scholar] [CrossRef]
- Ling, Y.; Jin, L.; Ma, Q.; Huang, Y.; Yang, Q.; Chen, M.; Shou, Q. Salvianolic acid A alleviated inflammatory response mediated by microglia through inhibiting the activation of TLR2/4 in acute cerebral ischemia-reperfusion. Phytomedicine Int. J. Phytother. Phytopharm. 2021, 87, 153569. [Google Scholar] [CrossRef]
- Qin, T.; Rasul, A.; Sarfraz, A.; Sarfraz, I.; Hussain, G.; Anwar, H.; Riaz, A.; Liu, S.; Wei, W.; Li, J.; et al. Salvianolic acid A & B: Potential cytotoxic polyphenols in battle against cancer via targeting multiple signaling pathways. Int. J. Biol. Sci. 2019, 15, 2256–2264. [Google Scholar]
- Qiu, J.M.; Qin, C.F.; Wu, S.G.; Ji, T.Y.; Tang, G.T.; Lei, X.Y.; Cao, X.; Xie, Z.Z. A novel salvianolic acid A analog with resveratrol structure and its antioxidant activities in vitro and in vivo. Drug Dev. Res. 2021, 82, 108–114. [Google Scholar] [CrossRef]
- Wang, R.; Song, F.; Li, S.; Wu, B.; Gu, Y.; Yuan, Y. Salvianolic acid A attenuates CCl(4)-induced liver fibrosis by regulating the PI3K/AKT/mTOR, Bcl-2/Bax and caspase-3/cleaved caspase-3 signaling pathways. Drug Des. Dev. Ther. 2019, 13, 1889–1900. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.F.; Wang, J.H.; Wang, Y.L.; Gao, C.; Gu, Y.T.; Huang, J.; Wang, J.H.; Zhang, Z. Salvianolic Acid A Protects the Kidney against Oxidative Stress by Activating the Akt/GSK-3β/Nrf2 Signaling Pathway and Inhibiting the NF-κB Signaling Pathway in 5/6 Nephrectomized Rats. Oxidative Med. Cell. Longev. 2019, 2019, 2853534. [Google Scholar] [CrossRef]
- Zeng, X.; Chen, X.; Qin, H.; Han, Y.; Chen, X.; Han, Z.; Zhao, W. Preventive effects of a natural anti-inflammatory agent Salvianolic acid A on acute kidney injury in mice. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2020, 135, 110901. [Google Scholar] [CrossRef]
- Song, Y.; Liu, W.; Ding, Y.; Jia, Y.; Zhao, J.; Wang, F.; Bai, J.; Cheng, L.; Gao, K.; Liu, M.; et al. Salvianolic acid A ameliorates renal ischemia/reperfusion injury by activating Akt/mTOR/4EBP1 signaling pathway. Am. J. Physiol. Renal. Physiol. 2018, 315, F254–F262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Cui, G.; Tong, S.; Cao, Q. Salvianolic acid A alleviates the renal damage in rats with chronic renal failure1. Acta Cir. Bras. 2019, 34, e201900204. [Google Scholar] [CrossRef]
- Yao, L.; Zhao, R.; He, S.; Feng, Q.; Qiao, Y.; Wang, P.; Li, J. Effects of salvianolic acid A and salvianolic acid B in renal interstitial fibrosis via PDGF-C/PDGFR-alpha signaling pathway. Phytomedicine 2022, 106, 154414. [Google Scholar] [CrossRef]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H.; et al. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef]
- Pinero, J.; Bravo, A.; Queralt-Rosinach, N.; Gutierrez-Sacristan, A.; Deu-Pons, J.; Centeno, E.; Garcia-Garcia, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Li, X.; Liao, J.; Su, X.; Li, W.; Bi, Z.; Wang, J.; Su, Q.; Huang, H.; Wei, Y.; Gao, Y.; et al. Human urine-derived stem cells protect against renal ischemia/reperfusion injury in a rat model via exosomal miR-146a-5p which targets IRAK1. Theranostics 2020, 10, 9561–9578. [Google Scholar] [CrossRef]
- Liu, X.; Luo, D.; Huang, S.; Liu, S.; Zhang, B.; Wang, F.; Lu, J.; Chen, J.; Li, S. Impaired Nicotinamide Adenine Dinucleotide Biosynthesis in the Kidney of Chronic Kidney Disease. Front. Physiol. 2021, 12, 723690. [Google Scholar] [CrossRef]
- Li, F.; Chen, Y.; Li, Y.; Huang, M.; Zhao, W. Geniposide alleviates diabetic nephropathy of mice through AMPK/SIRT1/NF-kappaB pathway. Eur. J. Pharmacol. 2020, 886, 173449. [Google Scholar] [CrossRef] [PubMed]
- Ying, T.; Clayton, P.; Naresh, C.; Chadban, S. Predictive value of spot versus 24-hour measures of proteinuria for death, end-stage kidney disease or chronic kidney disease progression. BMC Nephrol. 2018, 19, 55–63. [Google Scholar] [CrossRef]
- Cui, S.; Wu, L.; Feng, X.; Su, H.; Zhou, Z.; Luo, W.; Su, C.; Li, Y.; Shi, M.; Yang, Z.; et al. Urinary angiotensinogen predicts progressive chronic kidney disease after an episode of experimental acute kidney injury. Clin. Sci. 2018, 132, 2121–2133. [Google Scholar] [CrossRef]
- Wallbach, M.; Tampe, B.; Dihazi, H.; Koziolek, M.J. Acute kidney injury: From creatinine to KIM1? Internist 2019, 60, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.; Wang, Z. The Update of NGAL in Acute Kidney Injury. Curr. Protein. Pept. Sci. 2017, 18, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Marcello, M.; Virzi, G.M.; Mucino-Bermejo, M.J.; Milan Manani, S.; Giavarina, D.; Salvador, L.; Ronco, C.; Zanella, M. Subclinical AKI and Clinical Outcomes in Elderly Patients Undergoing Cardiac Surgery: Diagnostic Utility of NGAL versus Standard Creatinine Increase Criteria. Cardiorenal. Med. 2022, 12, 94–105. [Google Scholar] [CrossRef]
- Soto, K.; Papoila, A.L.; Coelho, S.; Bennett, M.; Ma, Q.; Rodrigues, B.; Fidalgo, P.; Frade, F.; Devarajan, P. Plasma NGAL for the diagnosis of AKI in patients admitted from the emergency department setting. Clin. J. Am. Soc. Nephrol. 2013, 8, 2053–2063. [Google Scholar] [CrossRef]
- Dase, J.; Rasyid, H.; Masadah, R.; Cangara, M.H.; Bukhari, A.; Dwiyanti, R.; Hatta, M. Analysis of mRNA and protein kidney injury Molecule-1 (KIM-1) expression in a kidney model during the initiation phase of ischemia reperfusion injury. Ann. Med. Surg. 2022, 75, 103373. [Google Scholar] [CrossRef]
- Ichimura, T.; Bonventre, J.V.; Bailly, V.; Wei, H.; Hession, C.A.; Cate, R.L.; Sanicola, M. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J. Biol. Chem. 1998, 273, 4135–4142. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.H.; Chen, M.L.; Sun, F.J.; Chen, Z.L.; Li, M.Y.; Zeng, W.; Gong, L.; Cheng, A.C.; Peng, X.; Fang, J.; et al. KIM-1 and NGAL as biomarkers of nephrotoxicity induced by gentamicin in rats. Mol. Cell. Biochem. 2014, 397, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, S.; Pergola, P.E.; Zager, R.A.; Vaziri, N.D. Targeting the transcription factor Nrf2 to ameliorate oxidative stress and inflammation in chronic kidney disease. Kidney Int. 2013, 83, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Mizoguchi, I.; Chiba, Y.; Ohashi, M.; Xu, M.; Yoshimoto, T. Expanding Diversity in Molecular Structures and Functions of the IL-6/IL-12 Heterodimeric Cytokine Family. Front. Immunol. 2016, 7, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Minciullo, P.L.; Catalano, A.; Mandraffino, G.; Casciaro, M.; Crucitti, A.; Maltese, G.; Morabito, N.; Lasco, A.; Gangemi, S.; Basile, G. Inflammaging and Anti-Inflammaging: The Role of Cytokines in Extreme Longevity. Arch. Immunol. Ther. Exp. 2016, 64, 111–126. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-beta and the TGF-beta Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb. Perspect. Biol. 2016, 8, 5. [Google Scholar] [CrossRef]
- Voisin, A.; Damon-Soubeyrand, C.; Bravard, S.; Saez, F.; Drevet, J.R.; Guiton, R. Differential expression and localisation of TGF-beta isoforms and receptors in the murine epididymis. Sci. Rep. 2020, 10, 995–1003. [Google Scholar] [CrossRef]
- Tang, P.C.; Chan, A.S.; Zhang, C.B.; Garcia Cordoba, C.A.; Zhang, Y.Y.; To, K.F.; Leung, K.T.; Lan, H.Y.; Tang, P.M. TGF-beta1 Signaling: Immune Dynamics of Chronic Kidney Diseases. Front. Med. 2021, 8, 628519. [Google Scholar] [CrossRef]
- Meng, X.M.; Chung, A.C.; Lan, H.Y. Role of the TGF-beta/BMP-7/Smad pathways in renal diseases. Clin. Sci. 2013, 124, 243–254. [Google Scholar] [CrossRef]
- Derynck, R.; Ying, E. Zhang. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef]
- Stempien-Otero, A.; Kim, D.H.; Davis, J. Molecular networks underlying myofibroblast fate and fibrosis. J. Mol. Cell. Cardiol. 2016, 97, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Xu, W.; Wang, J.; Yan, J.; Shi, Y.; Zhang, C.; Ge, W.; Wu, J.; Du, P.; Chen, Y. Boosting mTOR-dependent autophagy via upstream TLR4-MyD88-MAPK signalling and downstream NF-kappaB pathway quenches intestinal inflammation and oxidative stress injury. EBioMedicine 2018, 35, 345–360. [Google Scholar] [CrossRef]
- Kuroki, Y.; Tsuchida, K.; Go, I.; Aoyama, M.; Naganuma, T.; Takemoto, Y.; Nakatani, T. A study of innate immunity in patients with end-stage renal disease: Special reference to toll-like receptor-2 and -4 expression in peripheral blood monocytes of hemodialysis patients. Int. J. Mol. Med. 2007, 19, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Lepenies, J.; Eardley, K.S.; Kienitz, T.; Hewison, M.; Ihl, T.; Stewart, P.M.; Cockwell, P.; Quinkler, M. Renal TLR4 mRNA expression correlates with inflammatory marker MCP-1 and profibrotic molecule TGF-beta(1) in patients with chronic kidney disease. Nephron. Clin. Pract. 2011, 119, c97–c104. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Minden, A.; Martinetto, H.; Claret, F.-X.; Lange-Carter, C.; Mercurio, F.; Johnson, G.L.; Karin, M. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science 1995, 268, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.E.; McDonnell, M.A.; Law, B.K.; Moses, H.L. Interdependent SMAD and JNK signaling in transforming growth factor-beta-mediated transcription. J. Biol. Chem. 1999, 274, 37413–37420. [Google Scholar] [CrossRef] [PubMed]
- Obata, K.; Katsura, H.; Mizushima, T.; Sakurai, J.; Kobayashi, K.; Yamanaka, H.; Dai, Y.; Fukuoka, T.; Noguchi, K. Roles of extracellular signal-regulated protein kinases 5 in spinal microglia and primary sensory neurons for neuropathic pain. J. Neurochem. 2007, 102, 1569–1584. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Campanale, N.V.; Liang, R.J.; Deane, J.A.; Bertram, J.F.; Ricardo, S.D. Inhibition of p38 mitogen-activated protein kinase and transforming growth factor-beta1/Smad signaling pathways modulates the development of fibrosis in adriamycin-induced nephropathy. Am. J. Pathol. 2006, 169, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Andrikopoulos, P.; Kieswich, J.; Pacheco, S.; Nadarajah, L.; Harwood, S.M.; O’Riordan, C.E.; Thiemermann, C.; Yaqoob, M.M. The MEK Inhibitor Trametinib Ameliorates Kidney Fibrosis by Suppressing ERK1/2 and mTORC1 Signaling. J. Am. Soc. Nephrol. 2019, 30, 33–49. [Google Scholar] [CrossRef]
- Alcorn, J.F.; van der Velden, J.; Brown, A.L.; McElhinney, B.; Irvin, C.G.; Janssen-Heininger, Y.M. c-Jun N-terminal kinase 1 is required for the development of pulmonary fibrosis. Am. J. Respir. Cell. Mol. Biol. 2009, 40, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Liang, L.; Liu, Z.; Wang, Y.; Dong, X.; Qu, L.; Gou, R.; Wang, Y.; Wang, Q.; Liu, Z.; et al. Inhibition of IRE1/JNK pathway in HK-2 cells subjected to hypoxia-reoxygenation attenuates mesangial cells-derived extracellular matrix production. J. Cell. Mol. Med. 2020, 24, 13408–13420. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diao, H.-Y.; Zhu, W.; Liu, J.; Yin, S.; Wang, J.-H.; Li, C.-L. Salvianolic Acid A Improves Rat Kidney Injury by Regulating MAPKs and TGF-β1/Smads Signaling Pathways. Molecules 2023, 28, 3630. https://doi.org/10.3390/molecules28083630
Diao H-Y, Zhu W, Liu J, Yin S, Wang J-H, Li C-L. Salvianolic Acid A Improves Rat Kidney Injury by Regulating MAPKs and TGF-β1/Smads Signaling Pathways. Molecules. 2023; 28(8):3630. https://doi.org/10.3390/molecules28083630
Chicago/Turabian StyleDiao, Hai-Yang, Wei Zhu, Jie Liu, Sheng Yin, Jin-Hui Wang, and Chun-Li Li. 2023. "Salvianolic Acid A Improves Rat Kidney Injury by Regulating MAPKs and TGF-β1/Smads Signaling Pathways" Molecules 28, no. 8: 3630. https://doi.org/10.3390/molecules28083630
APA StyleDiao, H.-Y., Zhu, W., Liu, J., Yin, S., Wang, J.-H., & Li, C.-L. (2023). Salvianolic Acid A Improves Rat Kidney Injury by Regulating MAPKs and TGF-β1/Smads Signaling Pathways. Molecules, 28(8), 3630. https://doi.org/10.3390/molecules28083630



