Cytotoxicity Profiles and Neuroprotective Properties of the Novel Ifenprodil Analogues as Sigma Ligands
Abstract
1. Introduction
2. Results and Discussion
2.1. Compound Design
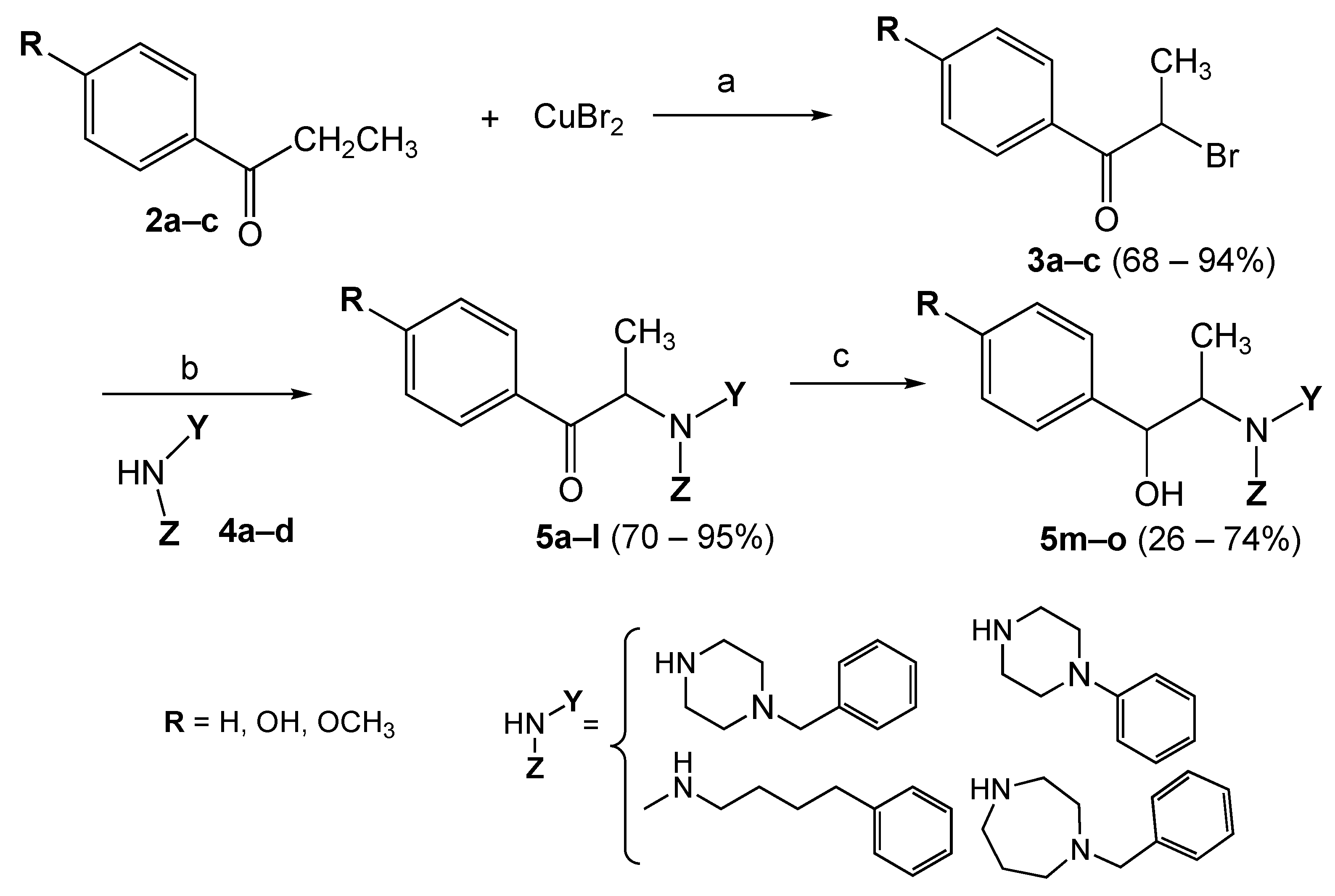
2.2. Chemistry
2.3. Biology and Computational
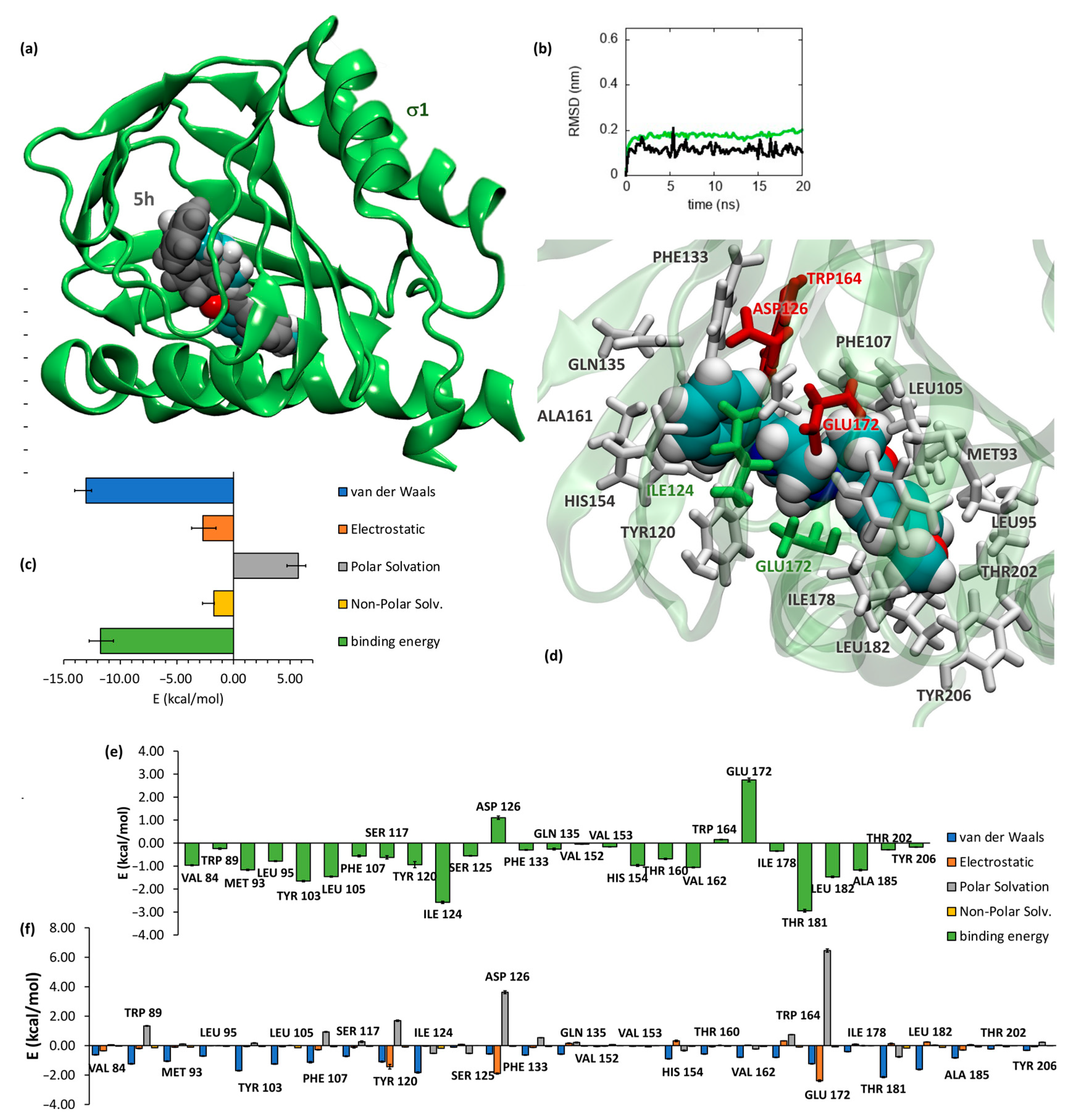
2.3.1. SR Binding Affinities, SAR Discussion, Molecular Dynamics and Docking Studies
2.3.2. Cytotoxic Profile
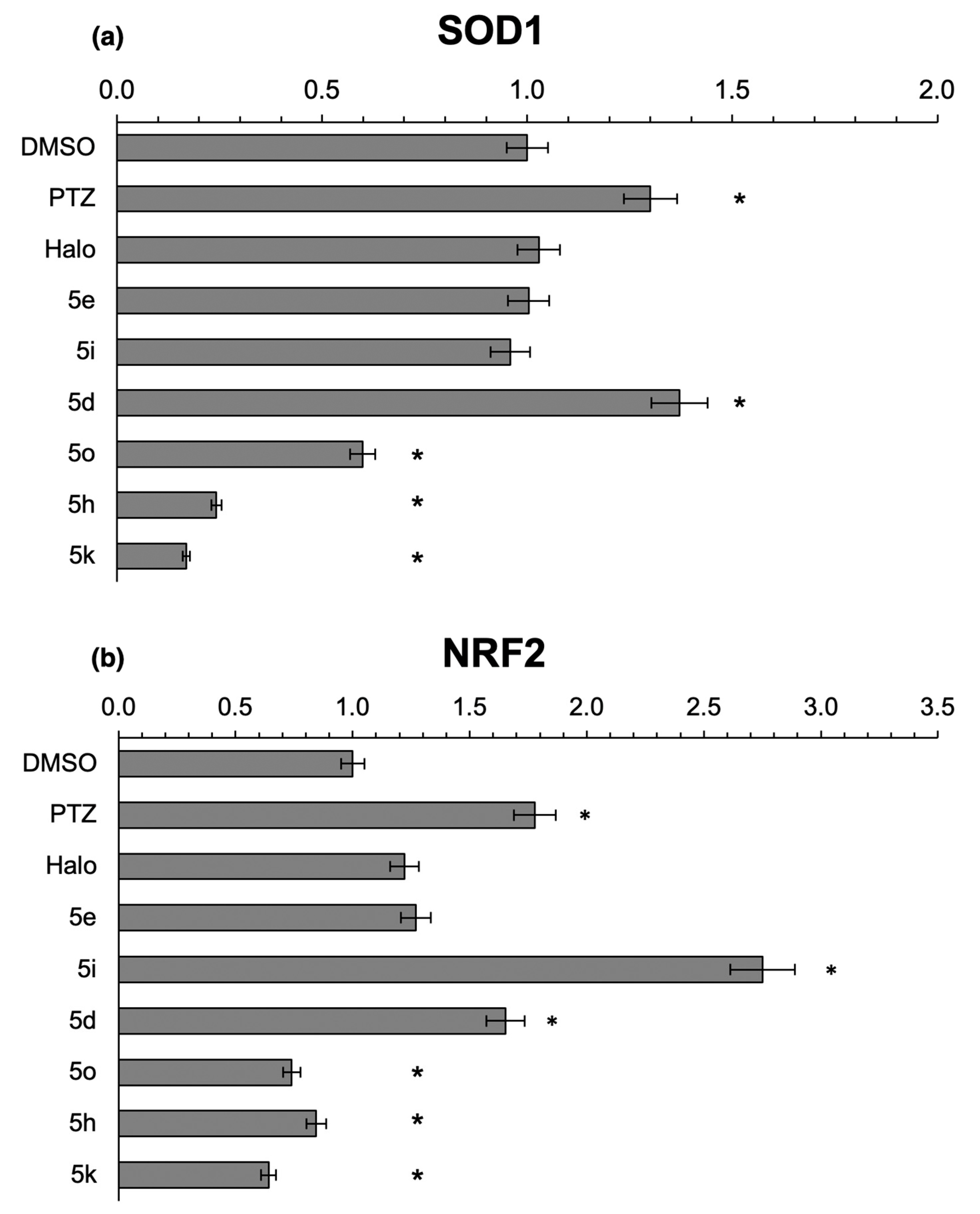
2.3.3. Effects of the Novel Ifenprodil Analogues on Antioxidant SOD1 and NRF2 in SH-SY5Y Cells
2.4. Antioxidant Activity
In Vitro Intrinsic Antioxidant Activity Evaluation
3. Materials and Methods
3.1. Chemistry
3.1.1. Chemical Reagents and Instruments
3.1.2. Synthetic Procedure
General Synthesis of Brominated Compounds 3a–c
- 2-Bromo-1-phenylpropan-1-one (3a)
- 2-Bromo-1-(4-methoxyphenyl)propan-1-one (3b)
- 2-Bromo-1-(4-hydroxyphenyl)propan-1-one (3c)
General Synthesis of the Final Compounds 5a–m
- 2-(4-Benzyl-1,4-diazepan-1-yl)-1-phenylpropan-1-one (5a)
- 2-(4-Phenylpiperazin-1-yl)-1-phenylpropan-1-one (5b)
- 2-(Methyl(4-phenylbutyl)amino)-1-phenylpropan-1-one (5c)
- 2-(4-Benzylpiperazin-1-yl)-1-phenylpropan-1-one (5d)
- 2-(4-Benzyl-1,4-diazepan-1-yl)-1-(4-methoxyphenyl)propan-1-one (5e)
- 2-(4-Phenylpiperazin-1-yl)-1-(4-methoxyphenyl)propan-1-one (5f)
- 1-(4-Methoxyphenyl)-2-(methyl(4-phenylbutyl)amino)propan-1-one (5g)
- 2-(4-Benzylpiperazin-1-yl)-1-(4-methoxyphenyl)propan-1-one (5h)
- 2-(4-Benzyl-1,4-diazepan-1-yl)-1-(4-hydroxyphenyl)propan-1-one (5i)
- 1-(4-Hydroxyphenyl)-2-(methyl(4-phenylbutyl)amino)propan-1-one (5j)
- 2-(4-Benzylpiperazin-1-yl)-1-(4-hydroxyphenyl)propan-1-one (5k)
- 2-(4-Phenylpiperazin-1-yl)-1-(4-hydroxyphenyl)propan-1-one (5l)
General Synthesis for the Reduced Compounds 5m–o
- 2-(Methyl(4-phenylbutyl)amino)-1-phenylpropan-1-ol (5m)
- 1-(4-Methoxyphenyl)-2-(methyl(4-phenylbutyl)amino)propan-1-ol (5n)
- 2-(4-Benzyl-1,4-diazepan-1-yl)-1-(4-methoxyphenyl)propan-1-ol (5o)
3.2. Computational
3.2.1. Docking
3.2.2. Molecular Dynamics
3.3. Hydrogen Peroxide Radical Scavenging Activity
3.4. Biology
3.4.1. S1R and S2R Binding Assays
Materials
Preparation of the Test Compounds
Preparation of the Membranes from Pig Brain
Preparation of the Membranes from Rat Liver
S2R Ligand Binding Assay
S2R Ligand Binding Assay
Data Analysis
3.4.2. Cytotoxicity Studies
Cell Culture
Cell Viability Test
Data Analysis
Real-Time PCR to Test Neuroprotective Properties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Giacalone, M.; Di Sacco, F.; Traupe, I.; Pagnucci, N.; Forfori, F.; Giunta, F. Chapter 2: Blueberry Polyphenols and Neuroprotection. In Bioactive Nutraceuticals and Dietary Supplements in Neurological and Brain Disease; Academic Press: Cambridge, MA, USA, 2015; pp. 17–28. [Google Scholar]
- Bogar, F.; Fulop, L.; Penke, B. Novel Therapeutic Target for Prevention of Neurodegenerative Diseases: Modulation of Neuroinflammation with Sig-1R Ligands. Biomolecule 2022, 12, 363. [Google Scholar] [CrossRef] [PubMed]
- GBD 2016 Neurology Collaborators. Global, regional, and national burden of neurological disorders, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 459–480. [Google Scholar] [CrossRef] [PubMed]
- Jay, T.R.; Bemiller, S.M.; Neilson, L.E.; Cheng-Hathaway, P.J.; Lamb, B.T. Neuroinflammation and Neurodegenerative Diseases; Oxford University Press: Oxford, UK, 2016; Volume 1. [Google Scholar]
- MacPherson, K.P.; de Sousa Rodrigues, M.E.; Cintron, A.F.; Tansey, M.G. Neuroinflammation in Age-Related Neurodegenerative Diseases. In The Molecular and Cellular Basis of Neurodegenerative Diseases; Elsevier: Amsterdam, The Netherlands, 2018; pp. 477–507. ISBN 978-0-12-811304-2. [Google Scholar]
- Di Sabato, D.J.; Quan, N.; Godbout, J.P. Neuroinflammation: The devil is in the details. J. Neurochem. 2016, 139, 136–153. [Google Scholar] [CrossRef] [PubMed]
- Korovesis, D.; Rubio-Tomás, T.; Tavernarakis, N. Oxidative Stress in Age-Related Neurodegenerative Diseases: An Overview of Recent Tools and Findings. Antioxidants 2023, 12, 131. [Google Scholar] [CrossRef]
- Uttara, B.; Singh, A.V.; Zamboni, P.; Mahajan, R.T. Oxidative stress and neurodegenerative diseases: A review of upstream and downstream antioxidant therapeutic options. Curr. Neuropharmacol. 2009, 7, 65–74. [Google Scholar] [CrossRef]
- Khan, T.A.; Hassan, I.; Ahmad, A.; Perveen, A.; Aman, S.; Quddusi, S.; Alhazza, I.; Ashraf, G.M.; Aliev, G. Recent Updates on the Dynamic Association Between Oxidative Stress and Neurodegenerative Disorders. CNS Neurol. Disord. Drug Targets 2016, 15, 310–320. [Google Scholar] [CrossRef]
- Korinek, M.; Kapras, V.; Vyklicky, V.; Adamusova, E.; Borovska, J.; Vales, K.; Stuchlik, A.; Horak, M.; Chodounska, H.; Vyklicky, L., Jr. Neurosteroid modulation of N-methyl-d-aspartate receptors: Molecular mechanism and behavioral effects. Steroids 2011, 13, 1409–1418. [Google Scholar] [CrossRef]
- Clemente, A.S.; Nicoll, R.A.; Roche, K.W. Diversity in NMDA receptor composition: Many regulators, many consequences. Neuroscientists 2013, 19, 62–75. [Google Scholar] [CrossRef]
- Danysz, W.; Parsons, C.G. Glycine and N-Methyl-D-Aspartate Receptors: Physiological Significance and Possible Therapeutic Applications. Pharmacol. Rev. 1998, 50, 597–664. [Google Scholar]
- Reynolds, I.J.; Miller, R.J. Ifenprodil is a novel type of N-methyl-D-aspartate receptor antagonist: Interaction with polyamines. Mol. Pharmacol. 1989, 36, 758–765. [Google Scholar]
- Maurice, T.; Lockhart, B.P. Neuroprotective and anti-amnesic potentials of sigma (s) receptor ligands. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 1997, 21, 69–102. [Google Scholar]
- King, M.; Pan, Y.X.; Mei, J.; Chang, A.; Xu, J.; Pasternak, G.W. Enhanced kappa-opioid receptor-mediated analgesia by antisense targeting the sigma1 receptor. Eur. J. Pharmacol. 1997, 331, R5–R6. [Google Scholar] [CrossRef] [PubMed]
- Modell, S.; Nober, D.; Holzbach, R. Efficacy and safety of an opiate sigma receptor antagonist (SL 82.0715) in schizophrenic patients with negative symptoms: An open dose-range study. Pharmacopsychiatry 1996, 29, 63–66. [Google Scholar] [CrossRef]
- Huber, M.T.; Gotthardt, U.; Schreiber, W.; Krieg, J.C. Efficacy and safety of the sigma receptor ligand EMD 57445 (panamesine) in patients with schizophrenia: An open clinical trial. Pharmacopsychiatry 1999, 32, 68–72. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.A.; Bowen, W.D.; Walker, F.O.; De Costa, B.; Matsumoto, R.R. Two novel sigma receptor ligands, BD1047 and LR172, attenuate cocaine-induced toxicity and locomotor activity. Eur. J. Pharmacol. 1999, 370, 225–232. [Google Scholar] [CrossRef]
- Kato, K.; Hayako, H.; Ishihara, Y.; Marui, S.; Iwane, M.; Miyamoto, M. TAK-147, an acetylcholinesterase inhibitor, increases choline acetyltransferase activity in cultured rat septal cholinergic neurons. Neurosci. Lett. 1999, 260, 5–8. [Google Scholar] [CrossRef]
- Vecchio, L.; Sorrentino, A.; Paoletti, R.; Marra, M.; Arbitrio, M. The State of The Art on Acetylcholinesterase Inhibitors in the Treatment of Alzheimer’s Disease. J. Cent. Nerv. Syst. Dis. 2021, 13, 11795735211029112. [Google Scholar] [CrossRef] [PubMed]
- Zampieri, D.; Fortuna, S.; Cassina, A.; Amata, E.; Dichiara, M.; Marrazzo, A.; Calabretti, A.; Mamolo, M.G. New Ifenprodil analogues as neuroprotective agents. In Proceedings of the XXVII National Meeting in Medicinal Chemistry, Bari, Italy, 11–14 September 2022. [Google Scholar]
- Zampieri, D.; Fortuna, S.; Calabretti, A.; Romano, M.; Menegazzi, R.; Schepmann, D.; Wünsch, B.; Collina, S.; Zanon, D.; Mamolo, M.G. Discovery of new potent dual sigma receptor/GluN2b ligands with antioxidant property as neuroprotective agents. Eur. J. Med. Chem. 2019, 180, 268–282. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- King, L.C.; Ostrum, G.K. Selective bromination with copper (II) bromide. J. Org. Chem. 1964, 29, 3459–3461. [Google Scholar] [CrossRef]
- Sałaciak, K.; Pytka, K. Revisiting the sigma-1 receptor as a biological target to treat affective and cognitive disorders. Neurosci. Biobehav. Rev. 2022, 132, 1114–1136. [Google Scholar] [CrossRef] [PubMed]
- Naia, L.; Pinho, C.M.; Dentoni, G.; Liu, J.; Leal, N.S.; Ferreira, D.M.S.; Schreiner, B.; Filadi, R.; Fão, L.; Connolly, N.M.C.; et al. Neuronal cell-based high-throughput screen for enhancers of mitochondrial function reveals luteolin as a modulator of mitochondria-endoplasmic reticulum coupling. BMC Biol. 2021, 24, 57. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, J.; Cui, X.; Mysona, B.A.; Navneet, S.; Saul, A.; Ahuja, M.; Lambert, N.; Gazaryan, I.G.; Thomas, B.; et al. The molecular chaperone sigma 1 receptor mediates rescue of retinal cone photoreceptor cells via modulation of NRF2. Free Radic. Biol. Med. 2019, 134, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiao, H.; Barwick, S.R.; Smith, S.B. Comparison of sigma 1 receptor ligands SA4503 and PRE084 to (+)-Pentazocine in the rd10 mouse model of RP. Investig. Ophthalmol. Vis. Sci. 2020, 61, 3. [Google Scholar] [CrossRef]
- Lasbleiz, C.; Peyrel, A.; Tarot, P.; Sarniguet, J.; Crouzier, L.; Cubedo, N.; Delprat, B.; Rossel, M.; Maurice, T.; Liévens, J.C. Sigma-1 receptor agonist PRE-084 confers protection against TAR DNA-binding protein-43 toxicity through NRF2 signalling. Redox Biol. 2022, 58, 102542. [Google Scholar] [CrossRef]
- Pal, A.; Fontanilla, D.; Gopalakrishnan, A.; Chae, Y.K.; Markley, J.L.; Ruoho, A.E. The sigma-1 receptor protects against cellular oxidative stress and activates antioxidant response elements. Eur. J. Pharmacol. 2012, 682, 12–20. [Google Scholar] [CrossRef]
- Schmidt, H.R.; Zheng, S.; Gurpinar, E.; Koehl, A.; Manglik, A.; Kruse, A.C. Crystal structure of the human σ1 receptor. Nature 2016, 532, 527. [Google Scholar] [CrossRef]
- Stewart, J.J.P. Mopac-A semiempirical molecular-orbital program. J. Comput. Aided Mol. Des. 1990, 4, 1–45. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.R.; Wilter, A.; Durham, E.H.A.B.; Pascutti, P.G. Molecular dynamics simulations applied to the study of subtypes of HIV-1 protease common to Brazil, Africa, and Asia. Cell Biochem. Biophys. 2006, 44, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gustereen, W.F. Peptide folding: When simulation meets experiment. Angew. Chemie Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Pronk, S.; Pall, S.; Schulz, R.; Larrson, P.; Bjekmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kason, P.M.; Van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open-source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Gang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Rad. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar]
- Uzarski, J.S.; DiVito, M.D.; Wertheim, J.A.; Miller, W.M. Essential design considerations for the resazurin reduction assay to noninvasively quantify cell expansion within perfused extracellular matrix scaffolds. Biomaterials 2017, 129, 163–175. [Google Scholar] [CrossRef]
- Lachance, V.; Bélanger, S.M.; Hay, C.; Le Corvec, V.; Banouvong, V.; Lapalme, M.; Tarmoun, K.; Beaucaire, G.; Lussier, M.P.; Kourrich, S. Overview of Sigma-1R Subcellular Specific Biological Functions and Role in Neuroprotection. Int. J. Mol. Sci. 2023, 24, 1971. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T. The Sigma-1 Receptor in Cellular Stress Signaling. Front. Neurosci. 2019, 13, 733. [Google Scholar] [CrossRef] [PubMed]
- Decuypere, J.P.; Monaco, G.; Missiaen, L.; De Smedt, H.; Parys, J.B.; Bultynck, G. IP(3) Receptors, Mitochondria, and Ca Signaling: Implications for Aging. J. Aging Res. 2011, 2011, 920178. [Google Scholar] [CrossRef] [PubMed]
- Bahar, E.; Kim, H.; Yoon, H. ER Stress-Mediated Signaling: Action Potential and Ca(2+) as Key Players. Int. J. Mol. Sci. 2016, 17, 1558. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef] [PubMed]


| Cmpd | RO5 a | MW b | HBA c | HBD d | clogP e | clogS f (mol/L) | TPSA g (Å < 140) | RO5 Violation | BBB Permeant | GI abs. |
|---|---|---|---|---|---|---|---|---|---|---|
| <500 | ≤10 | ≤5 | ≤5 | ≤5 | - | ≤1 | - | - | ||
| 5a | 322.4 | 3 | 0 | 3.19 | –4.24 | 23.55 | 0 | Yes | High | |
| 5b | 294.4 | 4 | 2 | 2.88 | –4.12 | 23.55 | 0 | Yes | High | |
| 5c | 295.4 | 2 | 0 | 4.22 | –4.54 | 20.31 | 0 | Yes | High | |
| 5d | 308.4 | 3 | 0 | 2.93 | –3.94 | 23.55 | 0 | Yes | High | |
| 5e | 352.5 | 4 | 0 | 3.22 | –4.31 | 32.78 | 0 | Yes | High | |
| 5f | 324.4 | 3 | 0 | 2.93 | –4.18 | 32.78 | 0 | Yes | High | |
| 5g | 325.4 | 3 | 0 | 4.20 | –4.61 | 29.54 | 0 | Yes | High | |
| 5h | 338.4 | 4 | 0 | 2.97 | –4.01 | 32.78 | 0 | Yes | High | |
| 5i | 338.4 | 4 | 1 | 2.79 | –4.09 | 43.78 | 0 | Yes | High | |
| 5j | 311.4 | 3 | 1 | 3.72 | –4.39 | 40.54 | 0 | Yes | High | |
| 5k | 324.4 | 4 | 1 | 2.56 | –3.80 | 43.78 | 0 | Yes | High | |
| 5l | 310.4 | 3 | 1 | 2.52 | –3.97 | 43.78 | 0 | Yes | High | |
| 5m | 297.4 | 2 | 1 | 3.94 | –4.21 | 23.47 | 0 | Yes | High | |
| 5n | 327.5 | 3 | 1 | 3.96 | –4.28 | 32.70 | 0 | Yes | High | |
| 5o | 340.5 | 3 | 1 | 2.93 | –4.16 | 35.94 | 0 | Yes | High | |
| Halo | 375.9 | 4 | 1 | 4.22 | –4.82 | 40.54 | 0 | Yes | High | |
| Ifenpr | 325.4 | 3 | 2 | 3.41 | –4.35 | 43.70 | 0 | Yes | High | |
| Donep | 379.5 | 4 | 0 | 4.00 | –4.81 | 38.77 | 0 | Yes | High |
 | ||||||
| Cmpd | R | X | 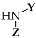 | Ki S1R (nM) a | Ki S2R (nM) a | S2R/S1R |
| 5a | H | 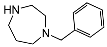 | 69 ± 14 | 108 ± 21 | 1.6 | |
| 5b | H |  | 2931 ± 1148 | 158 ± 30 | 0.05 | |
| 5c | H | 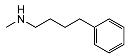 | 4721 ± 1687 | 733 ± 106 | 0.15 | |
| 5d | H | C=O | 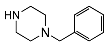 | 8.0 ± 1 | 302 ± 65 | 38 |
| 5e | OCH3 | 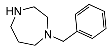 | 12 ± 2 | 97 ± 17 | 8.1 | |
| 5f | OCH3 | 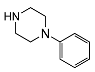 | 1858 ± 987 | 989 ± 337 | 0.53 | |
| 5g | OCH3 | 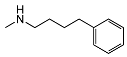 | 117 ± 18 | 386 ± 110 | 3.3 | |
| 5h | OCH3 | 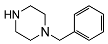 | 1.4 ± 0.2 | 84 ± 31 | 60 | |
| 5i | OH | 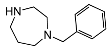 | 19 ± 3 | 22 ± 4 | 1.1 | |
| 5j | OH | 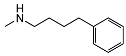 | 121 ± 22 | 45 ± 4 | 0.38 | |
| 5k | OH |  | 19 ± 2 | 129 ± 23 | 6.8 | |
| 5l | OH |  | 785 ± 226 | 147 ± 17 | 0.18 | |
| 5m | H |  | 37 ± 10 | 30 ± 8 | 0.81 | |
| 5n | OCH3 | CHOH | 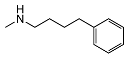 | 38 ± 9 | 479 ± 93 | 13 |
| 5o | OCH3 | 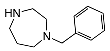 | 4.2 ± 0.6 | 128 ± 26 | 32 | |
| Halo | - | - | - | 2.6 ± 0.4 | 77 ± 18 | 30 |
| (+)-PTZ | - | - | - | 4.3 ± 0.5 | 1465 ± 224 | 312 |
| DTG | - | - | - | 124 ± 19 | 18 ± 1 | 0.14 |
| Ifenprodil | - | - | - | 125 ± 24 b | 98 ± 34 b | 0.75 |
| Donepezil | - | - | - | 14.6 c | - | - |
| Cmpd | Exp KiS1 | Docking Energy Kcal/mol | Cluster Size | Predicted Kd |
|---|---|---|---|---|
| 5d | 8.0 nM | −11.36 | 891 | 4.74 nM |
| 5o | 4.2 nM | −12.31 | 358 | 0.94 nM |
| 5h | 1.4 nM | −11.72 | 397 | 2.57 nM |
| 5i | 19 nM | −11.64 | 219 | 2.92 nM |
| 5k | 19 nM | −11.48 | 255 | 3.83 nM |
| 5e | 12 nM | −12.06 | 487 | 1.43 nM |
| Cmpd | IC50 (μM) |
|---|---|
| 5e | 74 |
| 5i | 294 |
| 5d | 118 |
| 5o | 118 |
| 5h | 117 |
| 5k | 132 |
| Haloperidol | 23 |
| NE100 | 49 |
| Siramesine | 2.0 |
| Cmpd | IC50 (μg/mL) a | |
|---|---|---|
| ABTS | H2O2 | |
| 5e | 2.495 ± 0.09 | 12.65 ± 0.21 |
| 5i | 2.603 ± 0.09 | 13.11 ± 0.24 |
| 5d | 2.854 ± 0.11 | 14.56 ± 0.18 |
| 5o | 2.481 ± 0.05 | 12.13 ± 0.25 |
| 5h | nd | 466.24 ± 1.36 |
| 5k | nd | 436.43 ± 1.17 |
| Trolox | 2.365 ± 0.19 | 15.69 ± 0.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zampieri, D.; Calabretti, A.; Romano, M.; Fortuna, S.; Collina, S.; Amata, E.; Dichiara, M.; Marrazzo, A.; Mamolo, M.G. Cytotoxicity Profiles and Neuroprotective Properties of the Novel Ifenprodil Analogues as Sigma Ligands. Molecules 2023, 28, 3431. https://doi.org/10.3390/molecules28083431
Zampieri D, Calabretti A, Romano M, Fortuna S, Collina S, Amata E, Dichiara M, Marrazzo A, Mamolo MG. Cytotoxicity Profiles and Neuroprotective Properties of the Novel Ifenprodil Analogues as Sigma Ligands. Molecules. 2023; 28(8):3431. https://doi.org/10.3390/molecules28083431
Chicago/Turabian StyleZampieri, Daniele, Antonella Calabretti, Maurizio Romano, Sara Fortuna, Simona Collina, Emanuele Amata, Maria Dichiara, Agostino Marrazzo, and Maria Grazia Mamolo. 2023. "Cytotoxicity Profiles and Neuroprotective Properties of the Novel Ifenprodil Analogues as Sigma Ligands" Molecules 28, no. 8: 3431. https://doi.org/10.3390/molecules28083431
APA StyleZampieri, D., Calabretti, A., Romano, M., Fortuna, S., Collina, S., Amata, E., Dichiara, M., Marrazzo, A., & Mamolo, M. G. (2023). Cytotoxicity Profiles and Neuroprotective Properties of the Novel Ifenprodil Analogues as Sigma Ligands. Molecules, 28(8), 3431. https://doi.org/10.3390/molecules28083431








