A Novel Sulfonamide, Molecularly Imprinted, Upconversion Fluorescence Probe Prepared by Pickering Emulsion Polymerization and Its Adsorption and Optical Sensing Performance
Abstract
1. Introduction
2. Results and Discussion
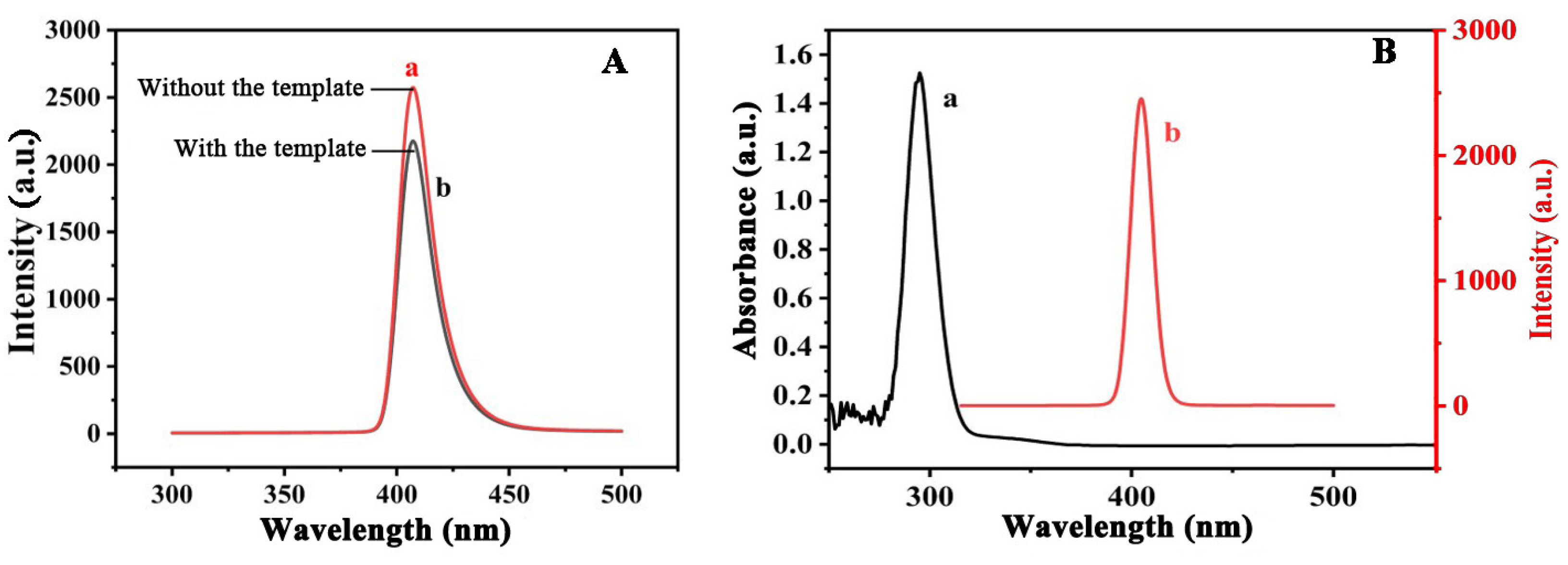
2.1. Synthesis of the UCNP@MIFPs
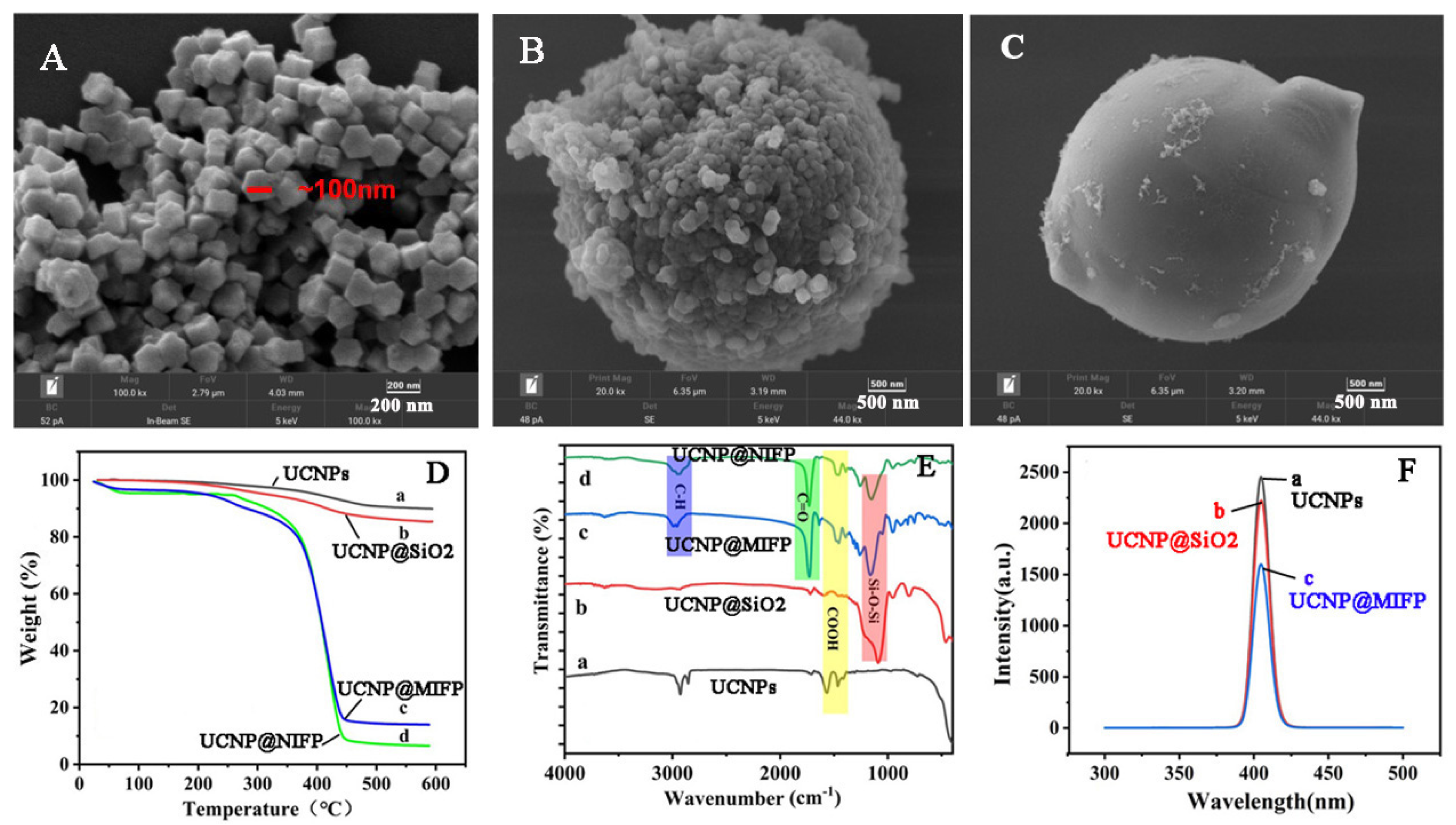
2.2. Characterization
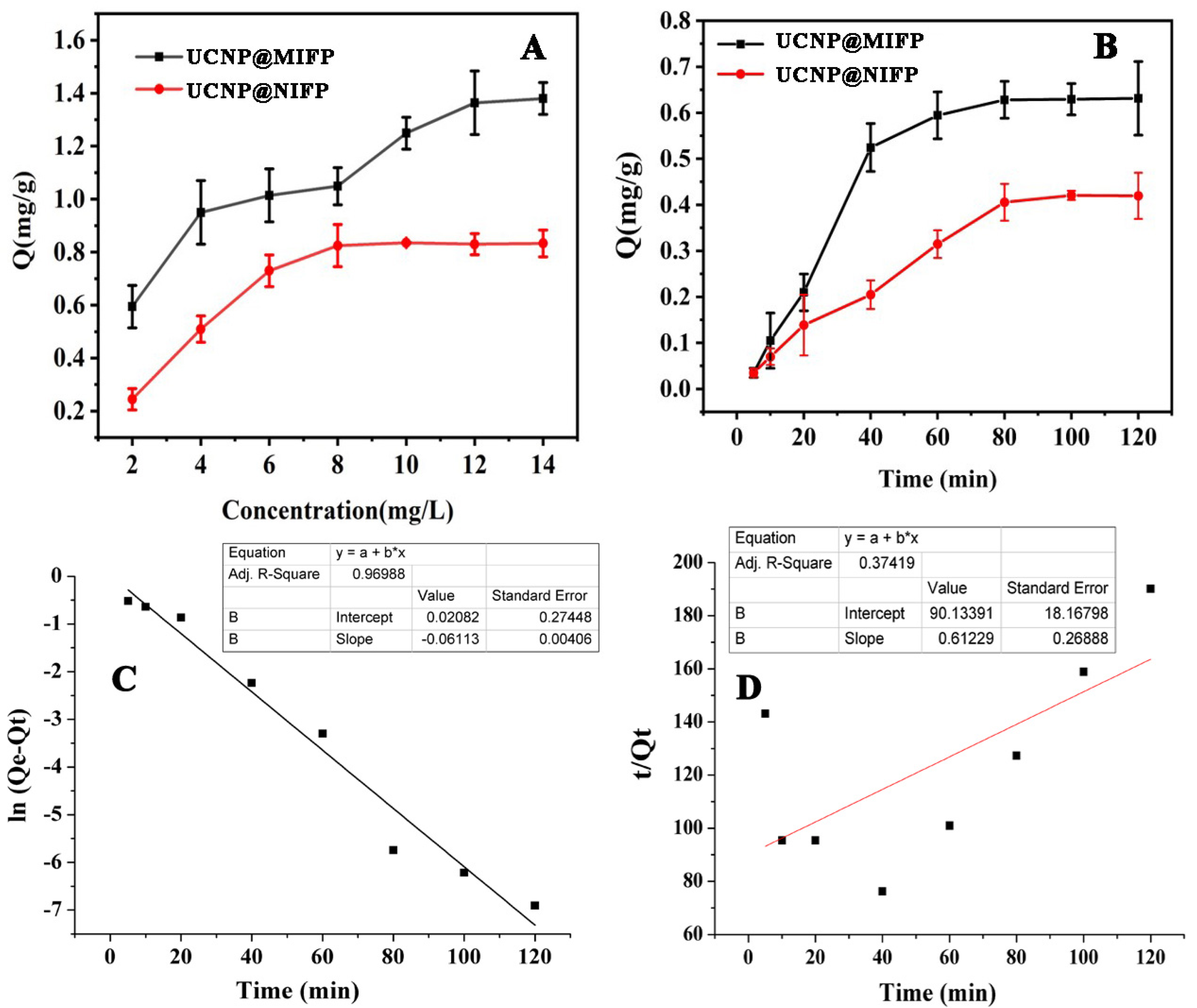
2.3. Static Adsorption
2.4. Adsorption Kinetics
2.5. Incubation Time
2.6. Possible Fluorescence Quenching Mechanism
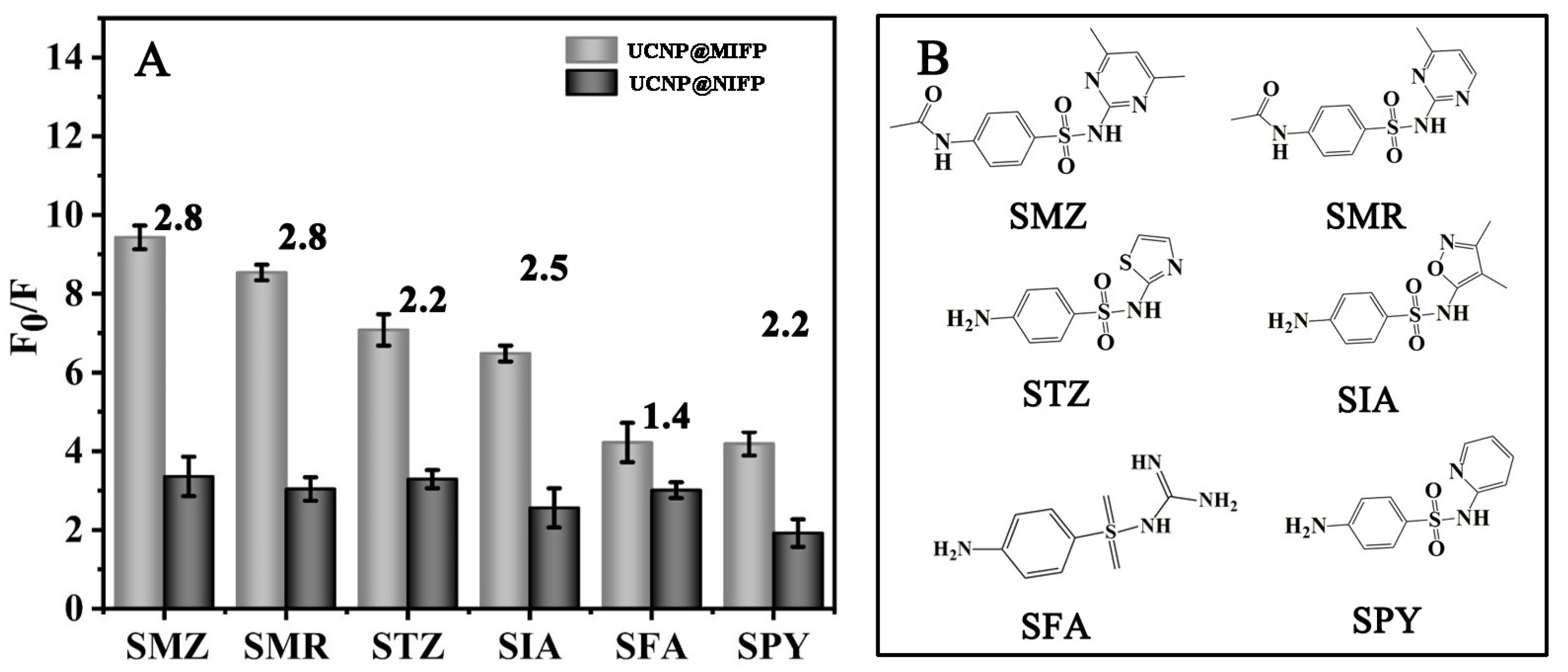
2.7. Selectivity of the UCNP@MIFP
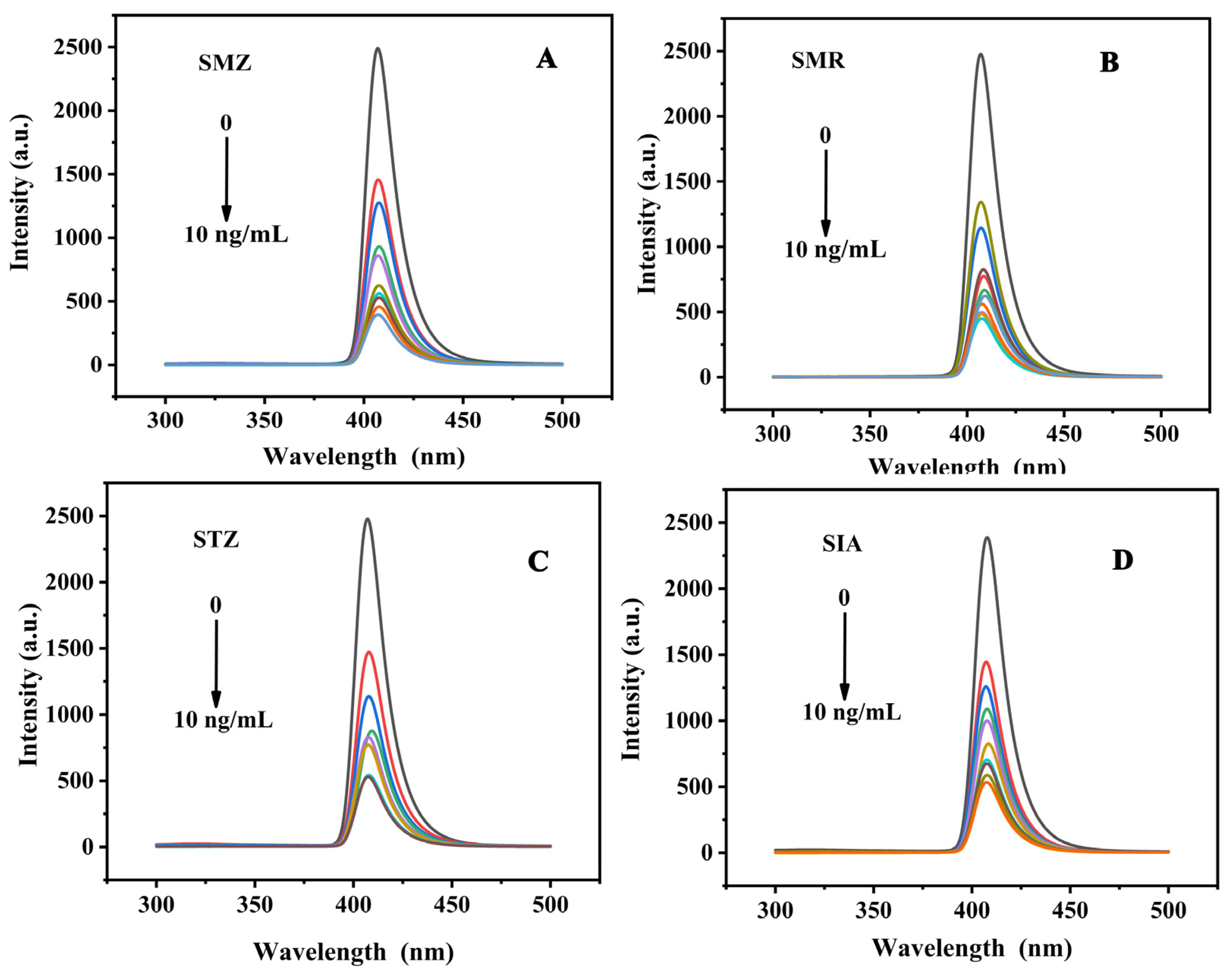
2.8. Fluorometric Analysis
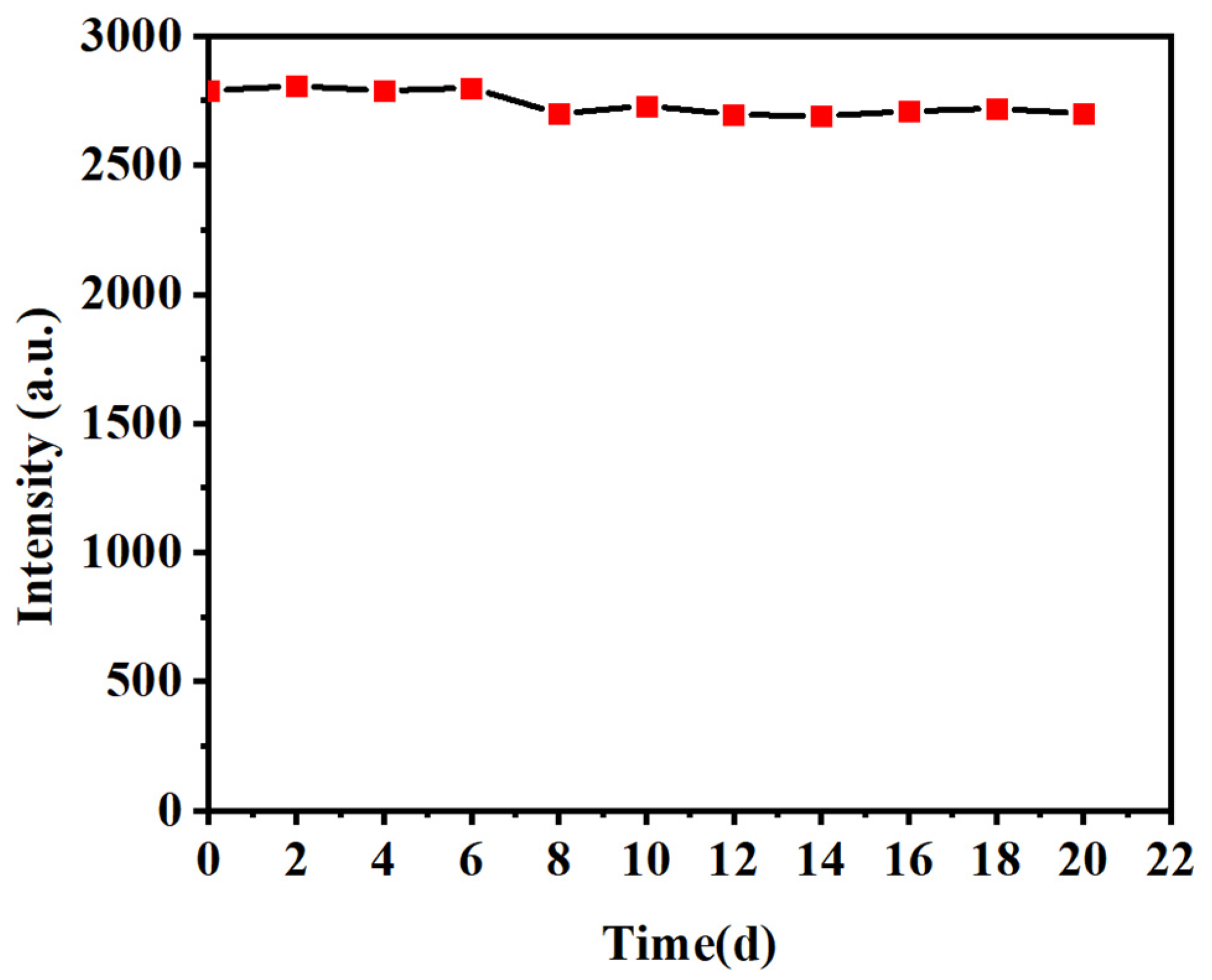
2.9. Stability of the UCNP@MIFP
2.10. Analysis of Real Samples
3. Materials and Methods
3.1. Chemicals and Material
3.2. Apparatus
3.3. Synthesis of the Upconversion Nanoparticles (UCNPs)
3.4. Fabrication of Silica-Coated Upconversion Nanoparticles (UCNP@SiO2)
3.5. Preparation of the UCNP-Based MIFPs (UCNP@MIFPs)
3.6. Adsorption Kinetic Test
3.7. Static Adsorption Test
3.8. Fluorescence Analysis
3.9. Selective Adsorption
3.10. Sample Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Bai, X.; Bai, X. Determination of sulfonamide residues in cultured sea cucumber by pre-column derivatization capillary electrophoresis with fluorescence detection. J. Indian Chem. Soc. 2022, 99, 100589. [Google Scholar] [CrossRef]
- Zhang, S.; Shao, K.; Hong, C.; Chen, S.; Lin, Z.; Huang, Z.; Lai, Z. Fluorimetric identification of sulfonamides by carbon dots embedded photonic crystal molecularly imprinted sensor array. Food Chem. 2023, 407, 135045. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.; Ramos, F. Analytical strategies for the detection and quantification of antibiotic residues in aquaculture fishes: A review. Trends Food Sci. Tech. 2016, 52, 16–30. [Google Scholar] [CrossRef]
- Kim, D.W.; Thawng, C.N.; Lee, K.; Wellington, E.M.H.; Cha, C.J. A novel sulfonamide resistance mechanism by two-component flavin-dependent monooxygenase system in sulfonamide-degrading actinobacteria. Environ. Int. 2019, 127, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Premarathne, J.; Satharasinghe, D.; Gunasena, A.; Munasinghe, D.; Abeynayake, P. Establishment of a method to detect sulfonamide residues in chicken meat and eggs by high-performance liquid chromatography. Food Control 2017, 72, 276–282. [Google Scholar] [CrossRef]
- Ma, J.; Fan, S.; Sun, L.; He, L.; Zhang, Y.; Li, Q. Rapid analysis of fifteen sulfonamide residues in pork and fish samples by automated on-line solid phase extraction coupled to liquid chromatography–tandem mass spectrometry. Food Sci. Hum. Wellness 2020, 9, 363–369. [Google Scholar] [CrossRef]
- Jimenez, V.; Adrian, J.; Guiteras, J.; Marco, M.P.; Companyo, R. Validation of an enzyme-linked immunosorbent assay for detecting sulfonamides in feed resources. J. Agric. Food Chem. 2010, 58, 7526–7531. [Google Scholar] [CrossRef]
- Zhou, Z.M.; Zheng, H.; Liu, T.; Xie, Z.Z.; Luo, S.H.; Chen, G.Y.; Tian, Z.Q.; Liu, G.K. Improving SERS sensitivity toward trace sulfonamides: The key role of trade-off interfacial interactions among the target molecules, anions, and cations on the SERS active surface. Anal. Chem. 2021, 93, 8603–8612. [Google Scholar] [CrossRef]
- Wang, Q.X.; Xue, S.F.; Chen, Z.H.; Ma, S.H.; Zhang, S.; Shi, G.; Zhang, M. Dual lanthanide-doped complexes: The development of a time-resolved ratiometric fluorescent probe for anthrax biomarker and a paper-based visual sensor. Biosens. Bioelectron. 2017, 94, 388–393. [Google Scholar] [CrossRef]
- Wen, S.; Zhou, J.; Zheng, K.; Bednarkiewicz, A.; Liu, X.; Jin, D. Advances in highly doped upconversion nanoparticles. Nat. Commun. 2018, 9, 2415. [Google Scholar] [CrossRef]
- Rong, Y.; Hassan, M.M.; Ouyang, Q.; Zhang, Y.; Wang, L.; Chen, Q. An upconversion biosensor based on DNA hybridization and DNA-templated silver nanoclusters for the determination of acrylamide. Biosens. Bioelectron. 2022, 215, 114581. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Zhang, D.; Tan, W.; Shi, J.; Li, Z.; Liu, H.; Yu, Y.; Yang, L.; Wang, X.; et al. A fluorescence resonance energy transfer probe based on functionalized graphene oxide and upconversion nanoparticles for sensitive and rapid detection of zearalenone. LWT-Food Sci. Tech. 2021, 147, 111541. [Google Scholar] [CrossRef]
- Wang, P.; Wang, A.; Hassan, M.M.; Ouyang, Q.; Li, H.; Chen, Q. A highly sensitive upconversion nanoparticles-WS2 nanosheet sensing platform for Escherichia coli detection. Sens. Actuat. B Chem. 2020, 320, 128434. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, H.; Shi, Z.; Duan, N.; Fang, C.; Dai, S.; Wang, Z. Aptamer-based fluorescence biosensor for chloramphenicol determination using upconversion nanoparticles. Food Control 2015, 50, 597–604. [Google Scholar] [CrossRef]
- Chen, Q.; Sheng, R.; Wang, P.; Ouyang, Q.; Wang, A.; Ali, S.; Zareef, M.; Hassan, M.M. Ultra-sensitive detection of malathion residues using FRET-based upconversion fluorescence sensor in food. Spectrochim. Acta 2020, 241, 118654. [Google Scholar] [CrossRef]
- Cao, Y.; Hu, X.; Zhao, T.; Mao, Y.; Fang, G.; Wang, S. A core-shell molecularly imprinted optical sensor based on the upconversion nanoparticles decorated with Zinc-based metal-organic framework for selective and rapid detection of octopamine. Sens. Actuat. B Chem. 2021, 326, 128838. [Google Scholar] [CrossRef]
- Basak, S.; Venkatram, R.; Singhal, R.S. Recent advances in the application of molecularly imprinted polymers (MIPs) in food analysis. Food Control 2022, 139, 109074. [Google Scholar] [CrossRef]
- Cui, Z.; Li, Z.; Jin, Y.; Ren, T.; Chen, J.; Wang, X.; Zhong, K.; Tang, L.; Tang, Y.; Cao, M. Novel magnetic fluorescent probe based on carbon quantum dots-doped molecularly imprinted polymer for AHLs signaling molecules sensing in fish juice and milk. Food Chem. 2020, 328, 127063. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, H.; Gao, J.; Liu, X.; Gao, X.; Lu, X.; Fang, G.; Wang, J.; Li, J. Upconversion particle@Fe3O4@molecularly imprinted polymer with controllable shell thickness as high-performance fluorescent probe for sensing quinolones. Talanta 2018, 181, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Akçapınar, R.; Özgür, E.; Goodarzi, V.; Uzun, L. Surface imprinted upconversion nanoparticles for selective albumin recognition. Colloids Surf. A 2022, 649, 129301. [Google Scholar] [CrossRef]
- Hosseinzadeh, B.; Nikfarjam, N.; Kazemi, S.H. Hollow molecularly imprinted microspheres made by w/o/w double Pickering emulsion polymerization stabilized by graphene oxide quantum dots targeted for determination of l-cysteine concentration. Colloids Surf. A 2021, 612, 125978. [Google Scholar] [CrossRef]
- Zhou, T.; Ashley, J.; Feng, X.; Sun, Y. Detection of hemoglobin using hybrid molecularly imprinted polymers/carbon quantum dots-based nanobiosensor prepared from surfactant-free Pickering emulsion. Talanta 2018, 190, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Cui, Y.; Wang, J.; Wang, J. Preparation of fluorescent molecularly imprinted polymers via Pickering emulsion interfaces and the application for visual sensing analysis of Listeria Monocytogenes. Polymers 2019, 11, 984. [Google Scholar] [CrossRef]
- Xiong, Y.; Wang, Q.; Duan, M.; Xu, J.; Chen, J.; Fang, S. Preparation of molecularly imprinted microspheres as biomimetic recognition material for in-Situ adsorption and selective chemiluminescence determination of bisphenol A. Polymers 2018, 10, 780. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Yu, Y.C.; Lan, R. Progress in application of dual/multi-template molecularly imprinted polymers. Chin. J. Anal. Chem. 2021, 49, e21205–e21215. [Google Scholar] [CrossRef]
- Surapong, N.; Pongpinyo, P.; Santaladchaiyakit, Y.; Burakham, R. A biobased magnetic dual-dummy-template molecularly imprinted polymer using a deep eutectic solvent as a coporogen for highly selective enrichment of organophosphates. Food Chem. 2023, 418, 136045. [Google Scholar] [CrossRef]
- Singh, R.; Singh, M. Highly selective and specific monitoring of pollutants using dual template imprinted MIP sensor. Electroanal. Chem. 2022, 926, 116939. [Google Scholar] [CrossRef]
- Ip, H.T.; Liu, L.; Hong, L.; Ngai, T. Synthesis of polystyrene/silica and poly(styrene-co-butyl acrylate)/silica nanocomposite particles by Pickering emulsion polymerization with non-functionalized silica nanoparticles. Colloids Surf. A 2022, 654, 130104. [Google Scholar] [CrossRef]
- Wang, L.; Ahmad, W.; Wu, J.; Wang, X.; Chen, Q.; Ouyang, Q. Selective detection of carbendazim using a upconversion fluorescence sensor modified by biomimetic molecularly imprinted polymers. Spectrochim. Acta A 2023, 284, 121457. [Google Scholar] [CrossRef]
- Sun, Y.; Yao, C.; Xie, Z.; Zhang, Y. Lotus seedpod-like molecularly imprinted polymers fabricated by MOF-808 stabilized Pickering emulsion and their specific recognition of hemoglobin. Colloids Surf. B 2021, 197, 111446. [Google Scholar] [CrossRef]
- Mehmandoust, M.; Erk, N.; Naser, M.; Soylak, M. Molecularly imprinted polymer film loaded on the metal–organic framework with improved performance using stabilized gold-doped graphite carbon nitride nanosheets for the single-step detection of Fenamiphos. Food Chem. 2023, 404, 134627. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.J.; Liu, J.Y.; Zhou, L.C.; Han, K.L. Site-selective photoinduced electron transfer from alcoholic solvents to the chromophore facilitated by hydrogen bonding: A new fluorescence quenching mechanism. J. Phys. Chem. B 2007, 111, 8940–8945. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; He, Y.; Ji, T.R.; Yan, X.P. Surface molecular imprinting on Mn-doped ZnS quantum dots for room-temperature phosphorescence optosensing of pentachlorophenol in water. Anal. Chem. 2009, 81, 1615–1621. [Google Scholar] [CrossRef]
- Wu, Q.; Fang, A.; Li, H.; Zhang, Y.; Yao, S. Enzymatic-induced upconversion photoinduced electron transfer for sensing tyrosine in human serum. Biosens. Bioelectron. 2016, 77, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Li, Q.; Wang, W.; Wen, Y.; Ji, K.; Liu, X.; He, P.; Janegitz, B.C.; Tang, K. The fabrication of a flexible and portable sensor based on home-made laser-induced porous graphene electrode for the rapid detection of sulfonamides. Microchem. J. 2022, 182, 107898. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, J.; Zhang, M.; Shi, G. Lanthanide metal-organic framework as a paper strip sensor for visual detection of sulfonamide with smartphone-based point-of-care platform. Talanta 2022, 237, 122920. [Google Scholar] [CrossRef]
- Wen, Z.; Hu, X.; Yan, R.; Wang, W.; Meng, H.; Song, Y.; Wang, S.; Wang, X.; Tang, Y. A reliable upconversion nanoparticle-based immunochromatographic assay for the highly sensitive determination of olaquindox in fish muscle and water samples. Food Chem. 2023, 406, 135081. [Google Scholar] [CrossRef]


| Fluorescence Probes | Template:MAA:EGDMA | QUCNP@MIFP ± SD a | QUCNP@NIFP ± SD | α |
|---|---|---|---|---|
| Probe 1 | 1:1:4 | 0.22 ± 0.0068 | 0.10 ± 0.0035 | 2.20 |
| Probe 2 | 1:1:6 | 0.23 ± 0.0055 | 0.09 ± 0.0021 | 2.50 |
| Probe 3 | 1:1:8 | 0.24 ± 0.0070 | 0.07 ± 0.0032 | 3.43 |
| Probe 4 | 1:2:4 | 0.10 ± 0.0038 | 0.04 ± 0.0009 | 2.50 |
| Probe 5 | 1:2:6 | 0.78 ± 0.0120 | 0.28 ± 0.0108 | 2.79 |
| Probe 6 | 1:2:8 | 0.18 ± 0.0052 | 0.18 ± 0.0068 | 1.00 |
| Probe 7 | 1:4:4 | 0.33 ± 0.0122 | 0.09 ± 0.0040 | 3.67 |
| Probe 8 | 1:4:6 | 0.30 ± 0.0091 | 0.16 ± 0.0005 | 1.88 |
| Probe 9 | 1:4:8 | 0.22 ± 0.0072 | 0.12 ± 0.0049 | 1.83 |
| Target | Stern–Volmer Equation | R2 | Linear Range (ng/mL) | LOD (ng/mL) |
|---|---|---|---|---|
| SMZ | F0/F = 0.50 Cq + 1.13 | 0.98 | 1–10 | 1.37 |
| SMR | F0/F = 0.41 Cq + 1.54 | 0.98 | 1–10 | 1.60 |
| STZ | F0/F = 0.31 Cq + 1.61 | 0.97 | 1–10 | 2.07 |
| SIA | F0/F = 0.30 Cq + 1.27 | 0.99 | 1–10 | 2.15 |
| Target | Spiked Concentration (ng/mL or ng/g) | UNCP@MIFP-Based Fluorescent Analysis Method | HPLC | p b | |||||
|---|---|---|---|---|---|---|---|---|---|
| Mean (ng/mL or ng/g) | Recovery (%) | RSD a (%) | Mean (ng/mL or ng/g) | Recovery (%) | RSD (%) | ||||
| Water | SMR | 2 | 1.99 | 99.67 | 1.04 | 1.96 | 99.00 | 2.12 | 0.98 |
| 4 | 3.98 | 99.58 | 1.51 | 3.93 | 98.75 | 1.35 | 0.31 | ||
| 8 | 7.95 | 99.45 | 0.71 | 7.97 | 100.12 | 0.52 | 0.65 | ||
| SMZ | 2 | 2.01 | 100.33 | 1.60 | 2.01 | 101.00 | 1.04 | 0.89 | |
| 4 | 4.01 | 100.17 | 0.72 | 3.96 | 99.25 | 1.30 | 0.22 | ||
| 8 | 7.95 | 99.33 | 1.39 | 7.96 | 99.87 | 0.62 | 0.89 | ||
| STZ | 2 | 1.94 | 96.83 | 2.94 | 1.94 | 97.50 | 1.07 | 0.85 | |
| 4 | 3.92 | 98.08 | 1.64 | 3.94 | 98.50 | 0.39 | 0.63 | ||
| 8 | 7.98 | 99.79 | 0.81 | 7.88 | 99.75 | 1.06 | 0.19 | ||
| SIA | 2 | 1.94 | 97.00 | 2.36 | 1.91 | 95.50 | 1.31 | 0.43 | |
| 4 | 3.92 | 98.00 | 1.17 | 3.90 | 99.25 | 1.80 | 0.75 | ||
| 8 | 7.88 | 98.50 | 0.91 | 7.92 | 99.50 | 0.51 | 0.42 | ||
| Fish muscle | SMR | 2 | 1.92 | 96.00 | 1.56 | 1.83 | 96.00 | 5.98 | 0.25 |
| 4 | 3.92 | 97.92 | 1.21 | 3.84 | 95.50 | 0.99 | 0.08 | ||
| 8 | 7.92 | 99.00 | 0.33 | 7.89 | 98.87 | 1.60 | 0.68 | ||
| SMZ | 2 | 1.93 | 96.67 | 1.30 | 1.99 | 99.50 | 2.37 | 0.13 | |
| 4 | 3.90 | 97.50 | 1.93 | 3.86 | 96.25 | 0.93 | 0.45 | ||
| 8 | 7.87 | 98.37 | 0.71 | 7.97 | 98.87 | 0.62 | 0.09 | ||
| STZ | 2 | 1.92 | 96.17 | 2.46 | 1.95 | 96.00 | 1.36 | 0.44 | |
| 4 | 3.92 | 97.92 | 1.21 | 3.91 | 98.25 | 1.45 | 0.94 | ||
| 8 | 7.90 | 98.92 | 1.26 | 7.90 | 98.38 | 0.38 | 0.73 | ||
| SIA | 2 | 1.96 | 98.17 | 3.39 | 1.95 | 98.50 | 3.20 | 0.81 | |
| 4 | 3.91 | 97.84 | 0.64 | 3.93 | 98.75 | 0.53 | 0.46 | ||
| 8 | 7.84 | 98.00 | 0.63 | 7.90 | 98.75 | 1.59 | 0.19 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, Q.; Gao, Z.; Meng, H.; Guo, X.; Zhang, M.; Tang, Y. A Novel Sulfonamide, Molecularly Imprinted, Upconversion Fluorescence Probe Prepared by Pickering Emulsion Polymerization and Its Adsorption and Optical Sensing Performance. Molecules 2023, 28, 3391. https://doi.org/10.3390/molecules28083391
Pan Q, Gao Z, Meng H, Guo X, Zhang M, Tang Y. A Novel Sulfonamide, Molecularly Imprinted, Upconversion Fluorescence Probe Prepared by Pickering Emulsion Polymerization and Its Adsorption and Optical Sensing Performance. Molecules. 2023; 28(8):3391. https://doi.org/10.3390/molecules28083391
Chicago/Turabian StylePan, Qidi, Zhe Gao, He Meng, Xianghua Guo, Meitian Zhang, and Yiwei Tang. 2023. "A Novel Sulfonamide, Molecularly Imprinted, Upconversion Fluorescence Probe Prepared by Pickering Emulsion Polymerization and Its Adsorption and Optical Sensing Performance" Molecules 28, no. 8: 3391. https://doi.org/10.3390/molecules28083391
APA StylePan, Q., Gao, Z., Meng, H., Guo, X., Zhang, M., & Tang, Y. (2023). A Novel Sulfonamide, Molecularly Imprinted, Upconversion Fluorescence Probe Prepared by Pickering Emulsion Polymerization and Its Adsorption and Optical Sensing Performance. Molecules, 28(8), 3391. https://doi.org/10.3390/molecules28083391






